Submitted:
13 May 2024
Posted:
14 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Synergistic Evolution between Yaks and Microorganisms
3. Microbial Mechanisms of Yaks Coping with Pasture Scarcity
3.1. Microbiological Mechanisms of Yaks with High Nutrient Digestibility
3.1.1. Mechanisms of Yak Rumen Bacteria to Improve Nutrient Digestibility
3.1.2. Mechanisms by which Yak Rumen Fungi Improve Nutrient Digestibility
3.1.3. Rumen Microbial Secretion of Enzymes to Improve Nutrient Digestibility
3.2. Microbiological Strategies of Yaks to Cope with Seasonal Variation in Forage
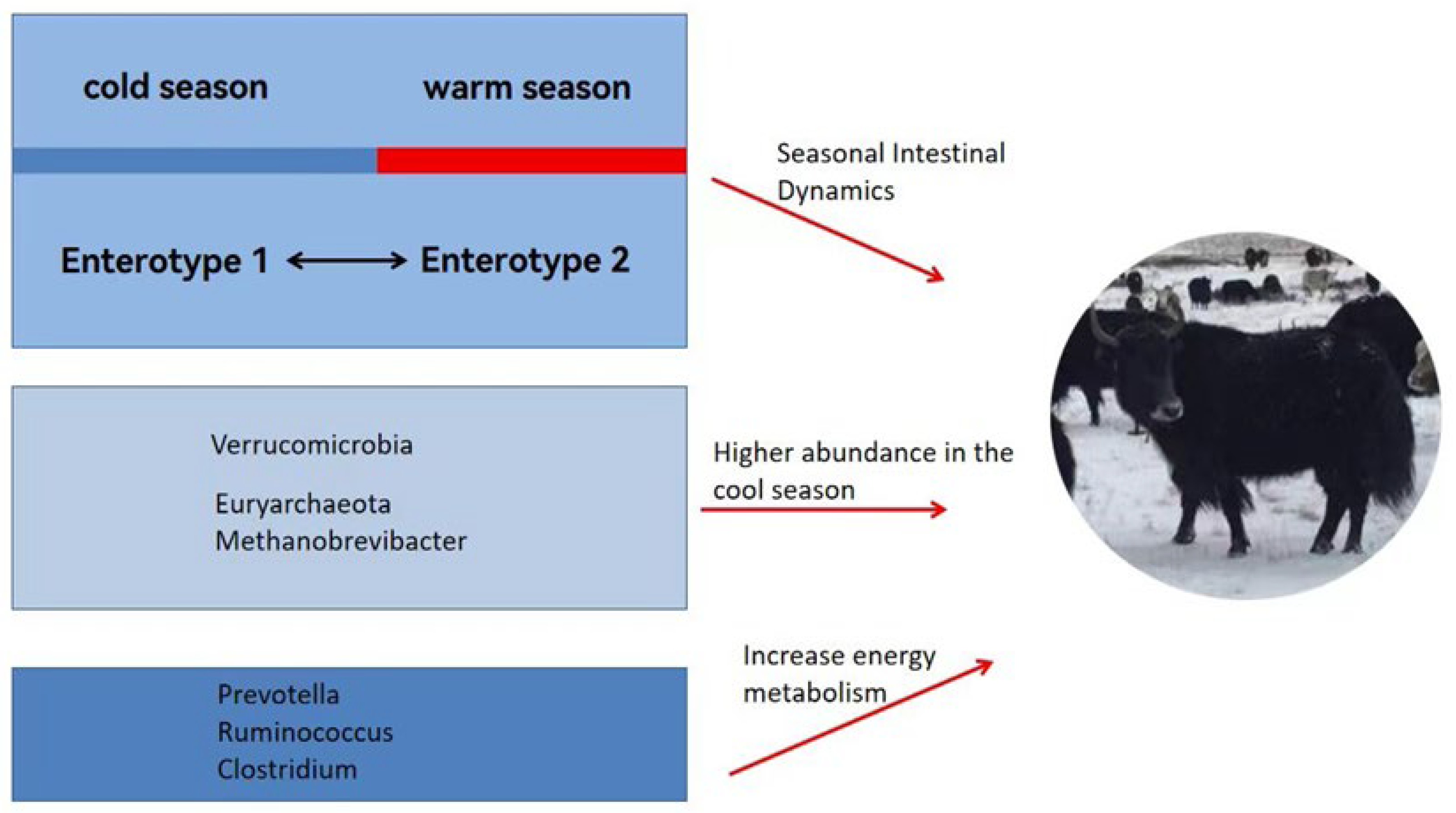
3.2.1. Seasonal Gut Patterns in Yaks
3.2.2. Dominant Flora Established by Yaks in Response to the Harsh Environment
4. Microbiological Mechanisms of Yak Health Maintenance
4.1. Interaction between Yak Health and Yak Microbes
4.1.1. Impact of Yak Gut Microbes on Yak Health
4.1.2. Impact of Yak Health on Gut Microbiota
4.2. Gut Probiotics in Yaks
5. Relationship between Microorganisms and Quality of Yak Products
5.1. Microbiological Mechanisms of High Milk Fat Content in Yak Milk
5.2. Microbiological Mechanisms of High Meat Quality in Yaks
6. The Role of Microorganisms in Yak-Environment Interactions
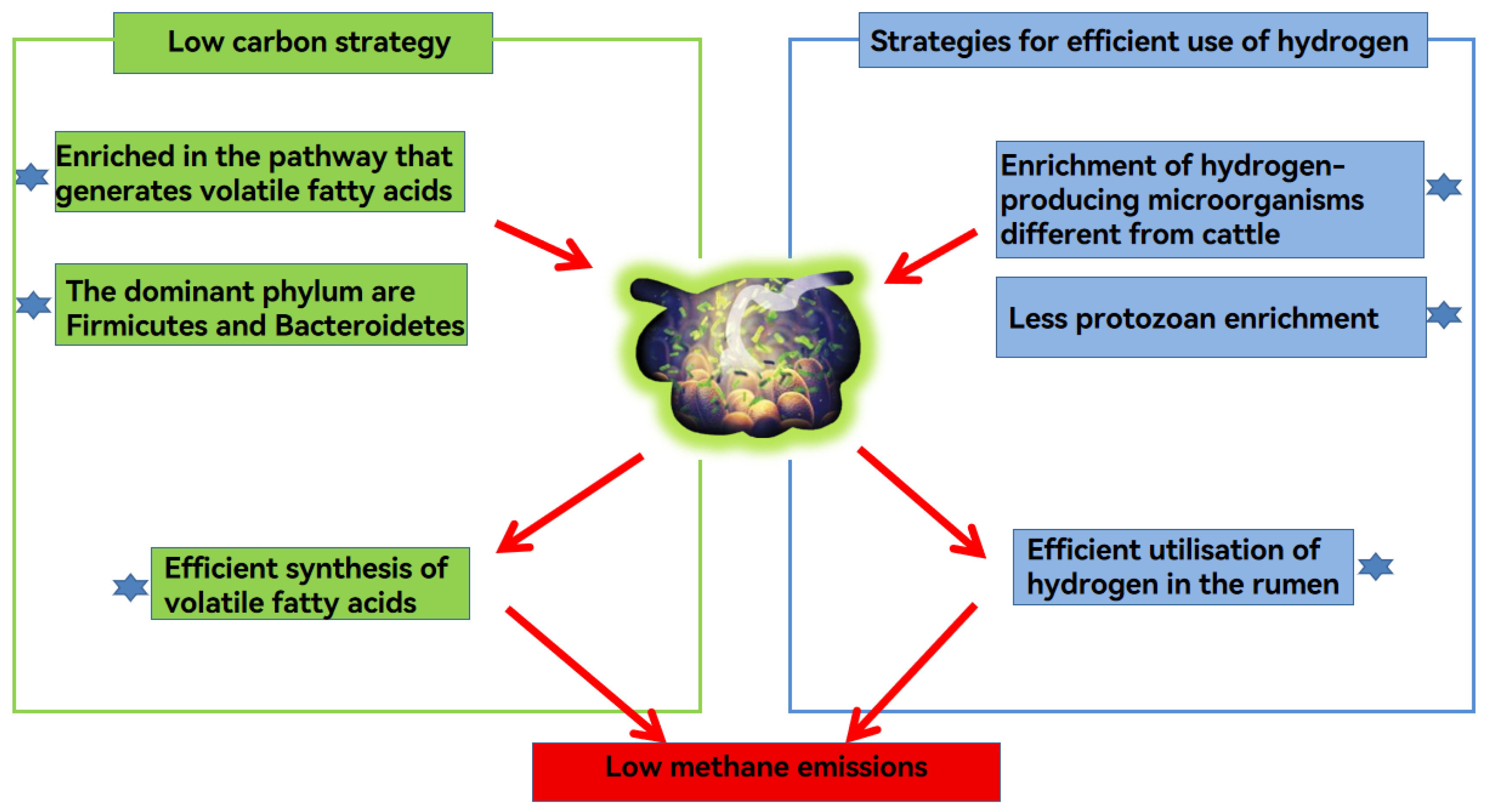
6.1. Microbiological Mechanisms of Low Methane Emissions in Yaks
6.1.1. Low Carbon Strategies
6.1.2. Microbial Strategies Competing with Methane Emission Pathways for Hydrogen
6.2. Interactions between Yak Gut Microbes and Soil Microbes
6.2.1. Effects of Yak Gut Microbes on Soil
6.2.2. Effects of Soil Micro-Organisms on Yaks
6.3. Effects of Yak Microbes on People and Plateau Pika
6.3.1. Effects of Yak Microbes on Plateau People
6.3.2. Effects of Yak Microbes on Plateau Pika
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, X.; Ju, Z.; Jiang, Q.; Zhong, J.; Liu, C.; Wang, J.; Hoff, J.L.; Schnabel, R.D.; Zhao, H.; Gao, Y.; et al. Introgression, admixture, and selection facilitate genetic adaptation to high-altitude environments in cattle. Genomics. 2021, 113, 1491-1503. [CrossRef]
- Degen A A, E.-M.S., Kam M. Milk and dung production by yaks (Poephagus grunniens): Important products for the livelihood of the herders and for carbon recycling on the Qinghai-Tibetan Plateau. Carbon Management for Promoting Local Livelihood in the Hindu Kush Himalayan (HKH) Region. 2020, 145-162. [CrossRef]
- Xue, D.; Chen, H.; Chen, F.; He, Y.; Zhao, C.; Zhu, D.; Zeng, L.; Li, W. Analysis of the rumen bacteria and methanogenic archaea of yak (Bos grunniens) steers grazing on the Qinghai-Tibetan Plateau. Livest. Sci. 2016, 188, 61-71. [CrossRef]
- Bang, C.; Dagan, T.; Deines, P.; Dubilier, N.; Duschl, W.J.; Fraune, S.; Hentschel, U.; Hirt, H.; Hülter, N.; Lachnit, T.; et al. Metaorganisms in extreme environments: do microbes play a role in organismal adaptation? Zoology. 2018, 127, 1-19. [CrossRef]
- Kolodny, O.; Callahan, B.J.; Douglas, A.E. The role of the microbiome in host evolution. Philos Trans R Soc Lond B Biol Sci 2020, 375, 20190588. [CrossRef]
- Jing, X.; Ding, L.; Zhou, J.; Huang, X.; Degen, A.; Long, R. The adaptive strategies of yaks to live in the Asian highlands. Anim Nutr 2022, 9, 249-258. [CrossRef]
- Guo, N.; Wu, Q.; Shi, F.; Niu, J.; Zhang, T.; Degen, A.A.; Fang, Q.; Ding, L.; Shang, Z.; Zhang, Z.; et al. Seasonal dynamics of diet-gut microbiota interaction in adaptation of yaks to life at high altitude. NPJ Biofilms Microbiomes 2021, 7, 38. [CrossRef]
- Knapp, J.R.; Laur, G.L.; Vadas, P.A.; Weiss, W.P.; Tricarico, J.M. Invited review: Enteric methane in dairy cattle production: quantifying the opportunities and impact of reducing emissions. J Dairy Sci. 2014, 97, 3231-3261. [CrossRef]
- YU, Q.; Han, L.; Jiang, Y.; Chen, Q.; Shen, H. Analysis of the nutritional components and flavorous substances of white yak’s milk. Yingyang Xuebao. 1956, 333-335.
- Malmuthuge, N.; Griebel, P.J.; Guan, L.L. The gut microbiome and its potential role in the development and function of newborn calf gastrointestinal tract. Front Vet Sci. 2015, 2, 36. [CrossRef]
- Pan, X.; Li, Z.; Li, B.; Zhao, C.; Wang, Y.; Chen, Y.; Jiang, Y. Dynamics of rumen gene expression, microbiome colonization, and their interplay in goats. BMC genomics 2021, 22, 288. [CrossRef]
- Schwaiger, K.; Storch, J.; Bauer, C.; Bauer, J. Abundance of selected bacterial groups in healthy calves and calves developing diarrhea during the first week of life: Are there differences before the manifestation of clinical symptoms? Front Microbiol. 2022, 13, 958080. [CrossRef]
- Palma-Hidalgo, J.M.; Yanez-Ruiz, D.R.; Jimenez, E.; Martin-Garcia, A.I.; Belanche, A. Presence of Adult Companion Goats Favors the Rumen Microbial and Functional Development in Artificially Reared Kids. Front Vet Sci. 2021, 8, 706592. [CrossRef]
- Han, X.; Li, B.; Wang, X.; Chen, Y.; Yang, Y. Effect of dietary concentrate to forage ratios on ruminal bacterial and anaerobic fungal populations of cashmere goats. Anaerobe. 2019, 59, 118-125. [CrossRef]
- Yang, S.; Zhang, G.; Yuan, Z.; He, S.; Wang, R.; Zheng, J.; Mao, H.; Chai, J.; Wu, D. Exploring the temporal dynamics of rumen bacterial and fungal communities in yaks (Bos grunniens) from 5 days after birth to adulthood by full-length 16S and 18S rRNA sequencing. Front Vet Sci. 2023, 10, 1166015. [CrossRef]
- Cui, X.; Liu, Y.; Wu, H.; Meng, Q.; Liu, S.; Chai, S.; Hao, L.; Zhou, Z. Dynamic changes in the yak rumen eukaryotic community and metabolome characteristics in response to feed type. Front Vet Sci. 2022, 9, 1027967. [CrossRef]
- Mao, S.Y.; Huo, W.J.; Zhu, W.Y. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ Microbiol. 2016, 18, 525-541. [CrossRef]
- Guo, W.; Zhou, M.; Ma, T.; Bi, S.; Wang, W.; Zhang, Y.; Huang, X.; Guan, L.L.; Long, R. Survey of rumen microbiota of domestic grazing yak during different growth stages revealed novel maturation patterns of four key microbial groups and their dynamic interactions. Anim. Microbiome. 2020, 2, 1-20. [CrossRef]
- Wang, X.; Zhang, Z.; Li, B.; Hao, W.; Yin, W.; Ai, S.; Han, J.; Wang, R.; Duan, Z. Depicting Fecal Microbiota Characteristic in Yak, Cattle, Yak-Cattle Hybrid and Tibetan Sheep in Different Eco-Regions of Qinghai-Tibetan Plateau. Microbiol Spectr. 2022, 10, e0002122. [CrossRef]
- Cao, Y.; Yang, H.J.A.f.s.; technology. Effect of roughage fibre content on fibrolytic activities and volatile fatty acid profiles of Neocallimastix sp. YAK11 isolated from rumen fluids of yak (Bos grunniens). Anim. Feed Sci. Technol. 2011, 170, 284-290. [CrossRef]
- Dai, X.; Zhu, Y.; Luo, Y.; Song, L.; Liu, D.; Liu, L.; Chen, F.; Wang, M.; Li, J.; Zeng, X.J.P.o. Metagenomic insights into the fibrolytic microbiome in yak rumen. PLoS One. 2012, 7, e40430. [CrossRef]
- Chen, Y.-B.; Lan, D.-L.; Tang, C.; Yang, X.-N.; Li, J. Effect of DNA extraction methods on the apparent structure of yak rumen microbial communities as revealed by 16S rDNA sequencing. Pol J Microbiol 2015, 64, 29-36. [CrossRef]
- Sha, Y.; Hu, J.; Shi, B.; Dingkao, R.; Wang, J.; Li, S.; Zhang, W.; Luo, Y.; Liu, X. Characteristics and Functions of the Rumen Microbial Community of Cattle-Yak at Different Ages. BioMed research international 2020, 2020, 3482692. [CrossRef]
- Dai, X.; Tian, Y.; Li, J.; Su, X.; Wang, X.; Zhao, S.; Liu, L.; Luo, Y.; Liu, D.; Zheng, H.J.A.; et al. Metatranscriptomic analyses of plant cell wall polysaccharide degradation by microorganisms in the cow rumen. Appl. Environ. Microbiol. 2015, 81, 1375-1386. [CrossRef]
- Gong, G.; Zhou, S.; Luo, R.; Gesang, Z.; Suolang, S. Metagenomic insights into the diversity of carbohydrate-degrading enzymes in the yak fecal microbial community. BMC Microbiol. 2020, 20, 302. [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105-108. [CrossRef]
- Yuan, N.; Wang, Y.; Pan, Q.; Zhao, L.; Qi, X.; Sun, S.; Suolang, Q.; Ciren, L.; Danzeng, L.; Liu, Y.; et al. From the perspective of rumen microbiome and host metabolome, revealing the effects of feeding strategies on Jersey Cows on the Tibetan Plateau. PeerJ. 2023, 11, e16010. [CrossRef]
- Wen, Y.; Li, S.; Wang, Z.; Feng, H.; Yao, X.; Liu, M.; Chang, J.; Ding, X.; Zhao, H.; Ma, W. Intestinal Microbial Diversity of Free-Range and Captive Yak in Qinghai Province. Microorganisms. 2022, 10, 754. [CrossRef]
- Long, R.J.; Ding, L.M.; Shang, Z.H.; Guo, X.H. The yak grazing system on the Qinghai-Tibetan plateau and its status. Rangeland J 2008, 30, 241-246. [CrossRef]
- Wei, Y.Q.; Yang, H.J.; Luan, Y.; Long, R.J.; Wu, Y.J.; Wang, Z.Y. Isolation, identification and fibrolytic characteristics of rumen fungi grown with indigenous methanogen from yaks (Bos grunniens) grazing on the Qinghai-Tibetan Plateau. J Appl Microbiol. 2016, 120, 571-587. [CrossRef]
- Auffret, M.; Yergeau, E.; Pilote, A.; Proulx, E.; Proulx, D.; Greer, C.W.; Vandenberg, G.; Villemur, R. Impact of water quality on the bacterial populations and off-flavours in recirculating aquaculture systems. FEMS Microbiol Ecol. 2013, 84, 235-247. [CrossRef]
- Yang, X.; Fan, X.; Jiang, H.; Zhang, Q.; Basangwangdui; Zhang, Q.; Dang, S.; Long, R.; Huang, X. Simulated seasonal diets alter yak rumen microbiota structure and metabolic function. Front Microbiol. 2022, 13, 1006285. [CrossRef]
- Gong, G.; Zhou, S.; Luo, R.; Gesang, Z.; Suolang, S. Metagenomic insights into the diversity of carbohydrate-degrading enzymes in the yak fecal microbial community. BMC microbiology 2020, 20, 302. [CrossRef]
- Lammens, W.; Le Roy, K.; Schroeven, L.; Van Laere, A.; Rabijns, A.; Van den Ende, W. Structural insights into glycoside hydrolase family 32 and 68 enzymes: functional implications. J Exp Bot 2009, 60, 727-740. [CrossRef]
- Guillen, D.; Sanchez, S.; Rodriguez-Sanoja, R. Carbohydrate-binding domains: multiplicity of biological roles. Appl Microbiol Biotechnol 2010, 85, 1241-1249. [CrossRef]
- Zhao, C.; Wang, L.; Ke, S.; Chen, X.; Kenez, A.; Xu, W.; Wang, D.; Zhang, F.; Li, Y.; Cui, Z.; et al. Yak rumen microbiome elevates fiber degradation ability and alters rumen fermentation pattern to increase feed efficiency. Anim Nutr. 2022, 11, 201-214. [CrossRef]
- Zhang, X.L.; Xu, T.W.; Wang, X.G.; Geng, Y.Y.; Liu, H.J.; Hu, L.Y.; Zhao, N.; Kang, S.P.; Zhang, W.M.; Xu, S.X. The Effect of Transitioning between Feeding Methods on the Gut Microbiota Dynamics of Yaks on the Qinghai-Tibet Plateau. Animals (Basel). 2020, 10, 1641. [CrossRef]
- Zhou, Z., Tran, P. Q., Kieft, K., & Anantharaman, K. Genome diversification in globally distributed novel marine Proteobacteria is linked to environmental adaptation. ISME J. 2020, 14, 2060-2077. [CrossRef]
- Ma, L.; Xu, S.; Liu, H.; Xu, T.; Hu, L.; Zhao, N.; Han, X.; Zhang, X. Yak rumen microbial diversity at different forage growth stages of an alpine meadow on the Qinghai-Tibet Plateau. PeerJ. 2019, 7, e7645. [CrossRef]
- Han, X.; Liu, H.; Hu, L.; Zhao, N.; Xu, S.; Lin, Z.; Chen, Y. Bacterial Community Characteristics in the Gastrointestinal Tract of Yak (Bos grunniens) Fully Grazed on Pasture of the Qinghai-Tibetan Plateau of China. Animals : an open access journal from MDPI 2021, 11, 2243. [CrossRef]
- Chevalier, C.; Stojanovic, O.; Colin, D.J.; Suarez-Zamorano, N.; Tarallo, V.; Veyrat-Durebex, C.; Rigo, D.; Fabbiano, S.; Stevanovic, A.; Hagemann, S.; et al. Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell 2015, 163, 1360-1374. [CrossRef]
- Zou, H.; Hu, R.; Wang, Z.; Shah, A.M.; Zeng, S.; Peng, Q.; Xue, B.; Wang, L.; Zhang, X.; Wang, X.; et al. Effects of Nutritional Deprivation and Re-Alimentation on the Feed Efficiency, Blood Biochemistry, and Rumen Microflora in Yaks (Bos grunniens). Animals (Basel). 2019, 9, 807. [CrossRef]
- Mi, J. Dynamics in rumen and distribution along gastrointestinal tracts of bacteria and methanogen in yak. Lanzhou: Lanzhou University. 2016.
- Shah, T.; Ding, L.; Ud Din, A.; Hassan, F.U.; Ahmad, A.A.; Wei, H.; Wang, X.; Yan, Q.; Ishaq, M.; Ali, N.; et al. Differential Effects of Natural Grazing and Feedlot Feeding on Yak Fecal Microbiota. Front Vet Sci. 2022, 9, 791245. [CrossRef]
- Guo, X.; Xia, X.; Tang, R.; Wang, K. Real-time PCR quantification of the predominant bacterial divisions in the distal gut of Meishan and Landrace pigs. Anaerobe 2008, 14, 224-228. [CrossRef]
- Zhang, Z.; Xu, D.; Wang, L.; Hao, J.; Wang, J.; Zhou, X.; Wang, W.; Qiu, Q.; Huang, X.; Zhou, J.; et al. Convergent Evolution of Rumen Microbiomes in High-Altitude Mammals. Curr Biol. 2016, 26, 1873-1879. [CrossRef]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E.J.N.R.E. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577-591. [CrossRef]
- Baothman, O.A.; Zamzami, M.A.; Taher, I.; Abubaker, J.; Abu-Farha, M.J.L.i.H.; Disease. The role of Gut Microbiota in the development of obesity and Diabetes. Lipids Health Dis. 2016, 15, 1-8. [CrossRef]
- Yang, T.; Richards, E.M.; Pepine, C.J.; Raizada, M.K. The gut microbiota and the brain-gut-kidney axis in hypertension and chronic kidney disease. Nat Rev Nephrol 2018, 14, 442-456. [CrossRef]
- Noureldein, M.H.; Bitar, S.; Youssef, N.; Azar, S.; Eid, A.A. Butyrate modulates diabetes-linked gut dysbiosis: epigenetic and mechanistic modifications. J. Mol. Endocrinol. 2020, 64, 29-42. [CrossRef]
- Pratt, V.C.; Tappenden, K.A.; McBurney, M.I.; Field, C.J. Short-chain fatty acid-supplemented total parenteral nutrition improves nonspecific immunity after intestinal resection in rats. JPEN J Parenter Enteral Nutr. 1996, 20, 264-271. [CrossRef]
- Glazko, V.I.; Zybaylov, B.L.; Kosovsky, Y.G.; Glazko, G.V.; Glazko, T.T. Domestication and microbiome. Holocene. 2021, 31, 1635-1645. [CrossRef]
- Bodogai, M.; O’Connell, J.; Kim, K.; Kim, Y.; Moritoh, K.; Chen, C.; Gusev, F.; Vaughan, K.; Shulzhenko, N.; Mattison, J.A.J.S.t.m. Commensal bacteria contribute to insulin resistance in aging by activating innate B1a cells. Sci. Transl. Med. 2018, 10, eaat4271. [CrossRef]
- Ansaldo, E.; Slayden, L.C.; Ching, K.L.; Koch, M.A.; Wolf, N.K.; Plichta, D.R.; Brown, E.M.; Graham, D.B.; Xavier, R.J.; Moon, J.J.J.S. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science. 2019, 364, 1179-1184. [CrossRef]
- Kuczma, M.P.; Szurek, E.A.; Cebula, A.; Chassaing, B.; Jung, Y.J.; Kang, S.M.; Fox, J.G.; Stecher, B.; Ignatowicz, L. Commensal epitopes drive differentiation of colonic T(regs). Sci Adv. 2020, 6, eaaz3186. [CrossRef]
- McVey Neufeld, K.A.; Perez-Burgos, A.; Mao, Y.K.; Bienenstock, J.; Kunze, W.A. The gut microbiome restores intrinsic and extrinsic nerve function in germ-free mice accompanied by changes in calbindin. Neurogastroenterol Motil 2015, 27, 627-636. [CrossRef]
- Bercik, P.; Park, A.J.; Sinclair, D.; Khoshdel, A.; Lu, J.; Huang, X.; Deng, Y.; Blennerhassett, P.A.; Fahnestock, M.; Moine, D.; et al. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol Motil 2011, 23, 1132-1139. [CrossRef]
- Liu, J.; Wang, X.; Zhang, W.; Kulyar, M.F.; Ullah, K.; Han, Z.; Qin, J.; Bi, C.; Wang, Y.; Li, K. Comparative analysis of gut microbiota in healthy and diarrheic yaks. Microb Cell Fact. 2022, 21, 111. [CrossRef]
- Li, F.; Shah, A.M.; Wang, Z.; Peng, Q.; Hu, R.; Zou, H.; Tan, C.; Zhang, X.; Liao, Y.; Wang, Y.; et al. Effects of Land Transport Stress on Variations in Ruminal Microbe Diversity and Immune Functions in Different Breeds of Cattle. Animals (Basel). 2019, 9. [CrossRef]
- Li, Y.; Li, X.; Liu, Y.; Nie, C.; Chen, C.; Niu, J.; Zhang, W. Comparison of Bacterial and Fungal Community Structure and Potential Function Analysis of Yak Feces before and after Weaning. Biomed Res Int. 2022, 2022, 6297231. [CrossRef]
- Zhenda, S.; Qinghui, K.; Jiakui, L.; Suozhu, L.; Zhankun, T.; Peng, S.; Honghui, W. Characterization of Bacterial Microbial Diversity in Wild Yak and Domestic Yak in Qiangtang Region of Tibet. Pak J Zool 2022, 54. [CrossRef]
- Garneau, J.E.; Tremblay, D.M.; Moineau, S. Characterization of 1706, a virulent phage from Lactococcus lactis with similarities to prophages from other Firmicutes. Virology. 2008, 373, 298-309. [CrossRef]
- Li, A.; Wang, M.; Zhang, Y.; Lin, Z.; Xu, M.; Wang, L.; Kulyar, M.F.; Li, J. Complete genome analysis of Bacillus subtilis derived from yaks and its probiotic characteristics. Front Vet Sci. 2022, 9, 1099150. [CrossRef]
- Hamana, K.; Itoh, T.; Sakamoto, M.; Hayashi, H. Covalently linked polyamines in the cell wall peptidoglycan of the anaerobes belonging to the order Selenomonadales. J Gen Appl Microbiol. 2012, 58, 339-347. [CrossRef]
- Zhang, Y.; Hu, N.; Cai, Q.; Zhang, F.; Zou, J.; Liu, Y.; Wei, D.; Zhu, Q.; Chen, K.; Zeng, L.; et al. Treatment with the traditional Chinese medicine BuYang HuanWu Tang induces alterations that normalize the microbiome in ASD patients. Biosci Microbiota Food Health. 2020, 39, 109-116. [CrossRef]
- Lei, J.; Ran, X.; Guo, M.; Liu, J.; Yang, F.; Chen, D. Screening, Identification, and Probiotic Properties of Bacillus Pumilus From Yak. Probiotics Antimicrob Proteins. 2023, 1-10. [CrossRef]
- Dong, H.; Liu, B.; Li, A.; Iqbal, M.; Wu, Q.J.F.i.C.; Microbiology, I. Microbiome Analysis Reveals the Attenuation Effect of Lactobacillus From Yaks on Diarrhea via Modulation of Gut Microbiota. 2021, 10, 610781. [CrossRef]
- Miao, V.; Davies, J. Actinobacteria: the good, the bad, and the ugly. Anton Van Leeuw. 2010, 98, 143-150. [CrossRef]
- Wang, Y.; Li, A.; Jiang, X.; Zhang, H.; Mehmood, K.; Zhang, L.; Jiang, J.; Waqas, M.; Iqbal, M.; Li, J. Probiotic Potential of Leuconostoc pseudomesenteroides and Lactobacillus Strains Isolated From Yaks. Frontiers in microbiology 2018, 9, 2987. [CrossRef]
- Zhang, Q.; Pan, Y.; Wang, M.; Sun, L.; Xi, Y.; Li, M.; Zeng, Q. In vitro evaluation of probiotic properties of lactic acid bacteria isolated from the vagina of yak (Bos grunniens). Peerj 2022, 10, e13177. [CrossRef]
- Dong, H.L.; Liu, B.X.; Li, A.Y.; Iqbal, M.; Mehmood, K.; Jamil, T.; Chang, Y.F.; Zhang, H.; Wu, Q.X. Microbiome Analysis Reveals the Attenuation Effect of From Yaks on Diarrhea Modulation of Gut Microbiota. Front. Cell. Infect. Microbiol. 2021, 10, 610781. [CrossRef]
- Mizrahi, I. The role of the rumen microbiota in determining the feed efficiency of dairy cows; Springer: Berlin Heidelberg, 2011; pp. 203-210.
- Council, N.R. Nutrient requirements of dairy cattle; National Academy Press: Washington, D.C., 2001.
- Gaffney, J.; Embree, J.; Gilmore, S.; Embree, M. Ruminococcus bovis sp. nov., a novel species of amylolytic Ruminococcus isolated from the rumen of a dairy cow. Int J Syst Evol Microbiol. 2021, 71. [CrossRef]
- Colin, R.; Sourjik, V. Emergent properties of bacterial chemotaxis pathway. Curr Opin Microbiol 2017, 39, 24-33. [CrossRef]
- Liu, L.; Wu, P.F.; Chen, F.F.; Zhou, J.L.; Guo, A.W.; Shi, K.R.; Zhang, Q. Multi-omics analyses reveal that the gut microbiome and its metabolites promote milk fat synthesis in Zhongdian yak cows. Peerj. 2022, 10, e14444. [CrossRef]
- Guan, L.L.; Nkrumah, J.D.; Basarab, J.A.; Moore, S.S. Linkage of microbial ecology to phenotype: correlation of rumen microbial ecology to cattle’s feed efficiency. FEMS Microbiol Lett. 2008, 288, 85-91. [CrossRef]
- Zhang, Y.P.; Zhao, H.W.; Li, Q.Q.; Tsechoe, D.; Yuan, H.L.; Su, G.J.; Yang, J.S. Environmental factors influence yak milk composition by modulating short-chain fatty acid metabolism in intestinal microorganisms. Lwt-Food Science and Technology. 2022, 163, 113608. [CrossRef]
- Backhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004, 101, 15718-15723. [CrossRef]
- Xulan., G. Determination of the number and relative abundance of Sclerotinia and Mycobacterium species in the intestinal tract of pigs and their correlation with fat deposition. Sichuan Agricultural University, Chengdu, 2009.
- Liu, H.; Li, Z.G.; Pei, C.F.; Degen, A.; Hao, L.Z.; Cao, X.L.; Liu, H.S.; Zhou, J.W.; Long, R.J. A comparison between yaks and Qaidam cattle in rumen fermentation, methane emission, and bacterial community composition with poor quality substrate. Animal Feed Science and Technology 2022, 291, 115395. [CrossRef]
- Wang, Y.; Fu, Y.; He, Y.; Kulyar, M.F.; Iqbal, M.; Li, K.; Liu, J. Longitudinal Characterization of the Gut Bacterial and Fungal Communities in Yaks. J Fungi (Basel) 2021, 7, 559. [CrossRef]
- Krause, T.R.; Lourenco, J.M.; Welch, C.B.; Rothrock, M.J.; Callaway, T.R.; Pringle, T.D. The relationship between the rumen microbiome and carcass merit in Angus steers. J Anim Sci 2020, 98, skaa287. [CrossRef]
- Holman, D.B.; Gzyl, K.E. A meta-analysis of the bovine gastrointestinal tract microbiota. FEMS Microbiol Ecol. 2019, 95. [CrossRef]
- Sun, J.; Xu, J.; Ge, R.; Wang, M.; Yu, L.; Wang, H. Effects of different dietary ratio of metabolizable glucose and metabolizable protein on growth performance, rumen fermentation, blood biochemical indices and ruminal microbiota of 8 to 10-month-old dairy heifers. Asian-Australas J Anim Sci. 2018, 31, 1205-1212. [CrossRef]
- Zhang, Q.; Guo, T.; Wang, X.; Zhang, X.; Geng, Y.; Liu, H.; Xu, T.; Hu, L.; Zhao, N.; Xu, S. Rumen Microbiome Reveals the Differential Response of CO(2) and CH(4) Emissions of Yaks to Feeding Regimes on the Qinghai-Tibet Plateau. Animals (Basel). 2022, 12. [CrossRef]
- Greening, C.; Geier, R.; Wang, C.; Woods, L.C.; Morales, S.E.; McDonald, M.J.; Rushton-Green, R.; Morgan, X.C.; Koike, S.; Leahy, S.C.; et al. Diverse hydrogen production and consumption pathways influence methane production in ruminants. Isme j. 2019, 13, 2617-2632. [CrossRef]
- Kumar, S. Physiology of rumen bacteria associated with low methane emitting sheep: a thesis presented in partial fulfilment of the requirements for the degree of Doctor of Philosophy in Microbiology and Genetics at Massey University, Palmerston North, New Zealand. Massey University, New Zealand, 2017.
- Yarlett, N.; Coleman, G.S.; Williams, A.G.; Lloyd, D. Hydrogenosomes in known species of rumen entodiniomorphid protozoa. FEMS Microbiology Letters. 1984, 21, 15-19. [CrossRef]
- Andersen, T.O.; Altshuler, I.; Vera-Ponce de Leon, A.; Walter, J.M.; McGovern, E.; Keogh, K.; Martin, C.; Bernard, L.; Morgavi, D.P.; Park, T.; et al. Metabolic influence of core ciliates within the rumen microbiome. The ISME journal 2023, 17, 1128-1140. [CrossRef]
- Mi, J.; Zhou, J.; Huang, X.; Long, R. Lower Methane Emissions from Yak Compared with Cattle in Rusitec Fermenters. PLoS One. 2017, 12, e0170044. [CrossRef]
- Dan, X.; Chen, H.; Chen, F.; He, Y.; Zhao, C.; Zhu, D.; Zeng, L.; Li, W. Analysis of the rumen bacteria and methanogenic archaea of yak (Bos grunniens) steers grazing on the Qinghai-Tibetan Plateau. Livest Sci. 2016, 188, 61-71. [CrossRef]
- Huang, X.D.; Tan, H.Y.; Long, R.; Liang, J.B.; Wright, A.D. Comparison of methanogen diversity of yak (Bos grunniens) and cattle (Bos taurus) from the Qinghai-Tibetan plateau, China. BMC microbiology 2012, 12, 237. [CrossRef]
- Fan, Q.; Xie, K.; Cui, X.; Zhang, G.; Zheng, H.; Chang, S.; Hou, F. Microecosystem of yak rumen on the Qinghai-Tibetan Plateau is stable and is unaffected by soil or grass microbiota. Environ. Microbiol. 2022, 24, 5760-5773. [CrossRef]
- Shang, Z.; Wang, Y.; An, M.; Chen, X.; Kulyar, M.F.; Tan, Z.; Liu, S.; Li, K. The successional trajectory of bacterial and fungal communities in soil are fabricated by yaks’ excrement contamination in plateau, China. Front Microbiol. 2022, 13, 1016852. [CrossRef]
- Lai, L.; Kumar, S. A global meta-analysis of livestock grazing impacts on soil properties. PLoS One. 2020, 15, e0236638. [CrossRef]
- Huang, X.; Liu, L.; Wen, T.; Zhu, R.; Zhang, J.; Cai, Z. Illumina MiSeq investigations on the changes of microbial community in the Fusarium oxysporum f.sp. cubense infected soil during and after reductive soil disinfestation. Microbiol Res. 2015, 181, 33-42. [CrossRef]
- Yao, F.; Yang, S.; Wang, Z.; Wang, X.; Ye, J.; Wang, X.; DeBruyn, J.M.; Feng, X.; Jiang, Y.; Li, H. Microbial Taxa Distribution Is Associated with Ecological Trophic Cascades along an Elevation Gradient. Front. Microbiol. 2017, 8, 2071. [CrossRef]
- Hu, J.; Wu, J.; Ma, M.; Nielsen, U.N.; Wang, J.; Du, G. Nematode communities response to long-term grazing disturbance on Tibetan plateau. Eur. J. Soil Biol. 2015, 69, 24-32. [CrossRef]
- Wang, Y.H.T., L.M.; Ai, R.Y.; Chen; S.Y; Zejan; D.K. Effects of short-term yak grazing on soil fungal communities in alpine grasslands of the Tibetan Plateau. caoyexuebao. 2022, 31, 41-52. [CrossRef]
- Gill, S.R.; Pop, M.; Deboy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic analysis of the human distal gut microbiome. Science. 2006, 312, 1355-1359. [CrossRef]
- Ricaboni, D.; Mailhe, M.; Khelaifia, S.; Raoult, D.; Million, M. Romboutsia timonensis, a new species isolated from human gut. New Microbes New Infect. 2016, 12, 6-7. [CrossRef]
- De Donder, L.; Uyttebroek, O.; Van Cleven, S.; Berrevoet, F.J.A.C.B. A subcutaneous infection mimicking necrotizing fasciitis due to Butyricimonas virosa. Acta Chir. Belg. 2020, 120, 425-428. [CrossRef]
- Yang, H.; Xiao, Y.; Gui, G.; Li, J.; Wang, J.; Li, D. Microbial community and short-chain fatty acid profile in gastrointestinal tract of goose. Poult Sci. 2018, 97, 1420-1428. [CrossRef]
- Wang, Y.; Li, A.; Zhang, L.; Waqas, M.; Mehmood, K.; Iqbal, M.; Muyou, C.; Li, Z.; Lian, Y.; Sizhu, S.; et al. Probiotic potential of Lactobacillus on the intestinal microflora against Escherichia coli induced mice model through high-throughput sequencing. Microb Pathog. 2019, 137, 103760. [CrossRef]
- Li, H.; Ma, Y.; Li, Q.; Wang, J.; Cheng, J.; Xue, J.; Shi, J. The Chemical Composition and Nitrogen Distribution of Chinese Yak (Maiwa) Milk. Int. J. Mol. Sci. 2011, 12, 4885-4895. [CrossRef]
- Fan, Q.; Wanapat, M.; Hou, F. Chemical Composition of Milk and Rumen Microbiome Diversity of Yak, Impacting by Herbage Grown at Different Phenological Periods on the Qinghai-Tibet Plateau. Animals (Basel). 2020, 10, 1030. [CrossRef]
- Goldstein, M.C.; Beall, C.M.J.H., the Journal of the Association for Nepal; Studies, H. Anthropological fieldwork in Tibet studying nomadic pastoralists on the Changtang. HIMALAYA, the Journal of the Association for Nepal and Himalayan Studies 1987, 7, 4.
- Ding, W.; Wang, L.; Zhang, J.; Ke, W.; Zhou, J.; Zhu, J.; Guo, X.; Long, R. Characterization of antioxidant properties of lactic acid bacteria isolated from spontaneously fermented yak milk in the Tibetan Plateau. J. Funct. Foods 2017, 35, 481-488. [CrossRef]
- Zheng, J.; Du, M.; Jiang, W.; Zhang, J.; Shen, W.; Ma, X.; Liang, Z.; Shen, J.; Wu, X.; Ding, X. In Vitro Probiotic Characteristics and Whole Genome Sequence Analysis of Lactobacillus Strains Isolated from Cattle-Yak Milk. Biology (Basel). 2021, 11. [CrossRef]
- Zhang, H.; Xu, J.; Wang, J.; Menghebilige; Sun, T.; Li, H.; Guo, M. A survey on chemical and microbiological composition of kurut, naturally fermented yak milk from Qinghai in China. Food Control. 2008, 19, 578-586. [CrossRef]
- Ding, W.; Shi, C.; Chen, M.; Zhou, J.; Long, R.; Guo, X.J.J.o.F.F. Screening for lactic acid bacteria in traditional fermented Tibetan yak milk and evaluating their probiotic and cholesterol-lowering potentials in rats fed a high-cholesterol diet. J. Funct. Foods. 2017, 32, 324-332. [CrossRef]
- Chao, Y.; Xuezhi, D.; Ruijun, L. Nutritional value and microbial composition of yak fresh milk and yogurt in the Tibetan Plateau region . J.Anim.Nutr. 2018, 30, 1262-1270.
- Ayeni, F.A.; Sánchez, B.; Adeniyi, B.A.; De los Reyes-Gavilan, C.G.; Margolles, A.; Ruas-Madiedo, P. Evaluation of the functional potential of Weissella and Lactobacillus isolates obtained from Nigerian traditional fermented foods and cow’s intestine. Int. J. Food Microbiol. 2011, 147, 97-104. [CrossRef]
- Qu, J.; Russell, J.C.; Ji, W.; Yang, M.; Chen, Q.; Li, W.; Zhang, Y. Five-year population dynamics of plateau pikas (Ochotona curzoniae) on the east of Tibetan Plateau. European Journal of Wildlife Research 2017, 63, 1-10. [CrossRef]
- Speakman, J.R.; Chi, Q.; Oldakowski, L.; Fu, H.; Fletcher, Q.E.; Hambly, C.; Togo, J.; Liu, X.; Piertney, S.B.; Wang, X.; et al. Surviving winter on the Qinghai-Tibetan Plateau: Pikas suppress energy demands and exploit yak feces to survive winter. Proc Natl Acad Sci U S A. 2021, 118, e2100707118. [CrossRef]
- Liu, Y.; Fan, J.; Shi, Z.; Yang, X.; Harris, W. Relationships between plateau pika ( Ochotona curzoniae ) densities and biomass and biodiversity indices of alpine meadow steppe on the Qinghai–Tibet Plateau China. Ecol. Eng. 2017, 102, 509-518. [CrossRef]
- Fu, H.; Zhang, L.; Fan, C.; Li, W.; Liu, C.; Zhang, H.; Cheng, Q.; Zhang, Y. Sympatric Yaks and Plateau Pikas Promote Microbial Diversity and Similarity by the Mutual Utilization of Gut Microbiota. Microorganisms. 2021, 9, 1890. [CrossRef]
| Family | Main functions |
|---|---|
| Ruminococcaceae | Degrade fiber and proteins |
| Succinivibrionaceae | Degrade starch and fiber |
| Lachnospiraceae | Promote growth of fiber-degrading bacteria |
| Rikenellaceae | Degrade fiber |
| Bacteroidaceae | Degrade starch and fiber and improve fiber digestibility |
| Prevotellaceae | One of the major glycolytic flora of the rumen, known for its protein binding capacity and digestion of a wide range of carbohydrate substrates |
| Christensenellaceae | Quickly respond to changes in feed components and participate in protein catabolism |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





