Submitted:
04 May 2024
Posted:
06 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Description of the study area
2.2. Plant material, experimental design and layout
2.3. Data collection
2.3.1. Agro-morphological traits
2.3.2. Culinary traits of selected cassava genotypes
2.4. Data analysis
3. Results
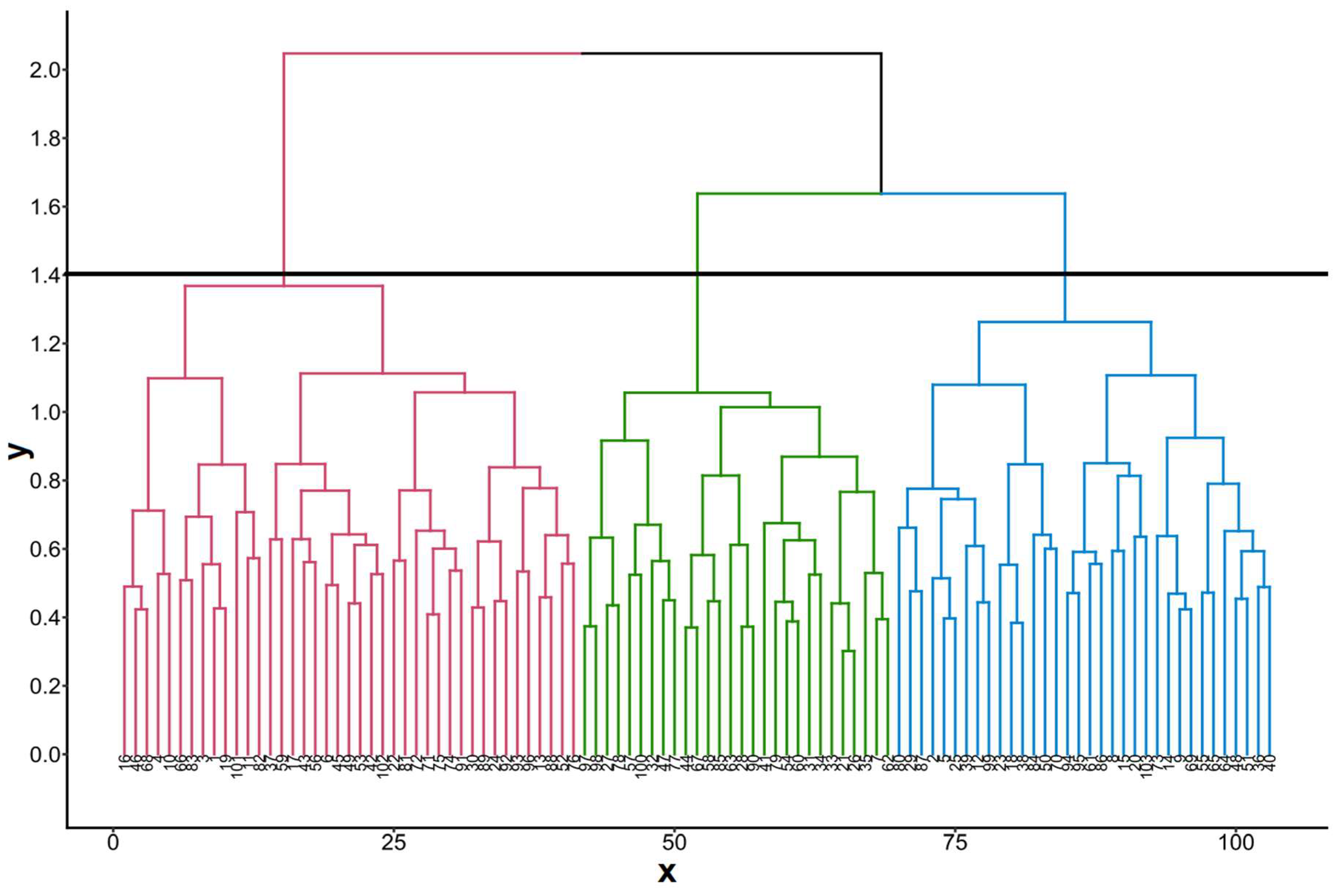
3.1. Correlation analysis, principal component analysisand genetic relationshipsamong cassava assessions based on 25 qualitative agro-morphological traits
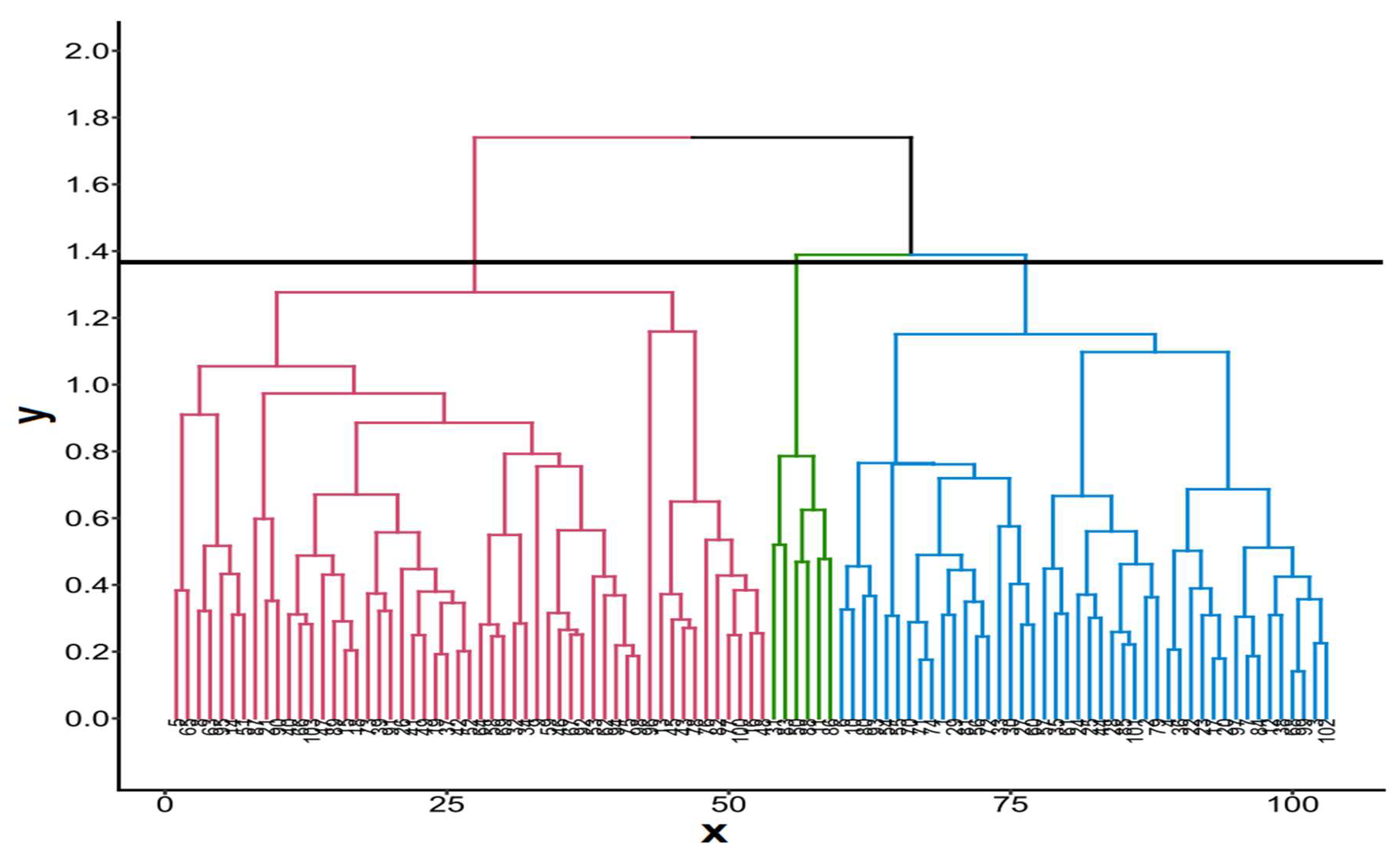
3.2. Descriptive statistics, correlation analysis, principal component analysis and genetic relationships among cassava assessions based on 14 quantitative agro-morphological traits
3.3. Culinary traits and marketable leaf assessment of 15 selected cassava genotypes
3.3. Marketable leaf assessment
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maziya-Dixon, B.; Dixon, A.G.O.; Adebowale, A.A.; Akinyele, I.O.; Dixon, J.; Egesi, C.N.; Ojo, A. Cassava: Africa's food security crop in the face of climate change. Food Security 2019, 11, 1083–1098. [Google Scholar]
- Akinola, A.; Ojo, B.; Adeyemi, C. Resilience and superior yield performance of cassava compared to other root and tuber crops. Journal of Crop Science 2020, 25, 215–224. [Google Scholar]
- Lemchi, J.; Anyanwu, C.; Eze, P. Comparative study of cassava's yield performance in nutrient-poor soils compared to other root and tuber crops. Agriculture and Environment 2020, 17, 98–107. [Google Scholar]
- Iheanacho, O.M.; Mba, C.; Chikoye, D.; Eke-Okoro, O.N.; Onyishi, G.C.; Asiegbu, J.E.; Iwuchukwu, E. Response of cassava genotypes to drought stress at different growth stages in the subhumid tropics of Nigeria. Journal of Agronomy 2021, 20, 177–187. [Google Scholar]
- Andrade, R.M.; de Jesus Sanchez, D.; Taipe-Ochoa, W.; Roca, W.M.; Beebe, S.E. The use of molecular markers in cassava breeding programs: Past, present, and future perspectives. Crop Science 2021, 61, 733–755. [Google Scholar]
- Asare, A.T.; Offei, S.K.; Danquah, E.Y.; Sarkodie-Addo, J. Genetic diversity and agro-morphological characterization of cassava (Manihot esculenta Crantz) germplasm in Ghana. African Journal of Biotechnology 2011, 10, 16642–16653. [Google Scholar]
- McKey, D.; Elias, M.; Pujol, B.; Duputié, A. Local seed systems and their importance for an improved management of genetic resources: case studies from Mali. Euphytica 2001, 120, 83–104. [Google Scholar]
- Pinton, F.; Emperaire, L. Cassava diversity and crop reproduction in traditional farming systems: a cognitive approach. Economic Botany 2001, 55, 353–368. [Google Scholar]
- Elias, M.; Mühlen, G.S.; McKey, D.B.; Roa, A.C.; Tohme, J.; Restrepo, M.T. The influence of morphological variation on germplasm collection in cassava (Manihot esculenta Crantz) in Peru. Economic Botany 2001, 55, 256–269. [Google Scholar]
- Smykal, P.; Aubert, G.; Burstin, J.; Coyne, C.J.; Ellis, N.T.H.; Flavell, A.J.; Ford, R.; Hýbl, M.; Macas, J.; Neumann, P.; McPhee, K.; Redden, R.J.; Rubiales, D.; Weller, J.L.; Warkentin, T.D.; Thomas, W.T.B. Genetic diversity and population structure of pea (Pisum sativum L.) varieties derived from combined retrotransposon, microsatellite and morphological marker analysis. Theoretical and Applied Genetics 2008, 117, 413–424. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. Prospects for using morphological, molecular and geographic distance data to estimate gene flow and seed migration in wild Phaseolus vulgaris populations from Mexico. Crop Science 2005, 45, 336–350. [Google Scholar]
- Fukuda, W.M.G.; Guevara, C.L.; Kawuk, R.; Ferguson, M.E. Selected morphological and agronomic descriptors for the characterization of cassava; International Institute of Tropical Agriculture (IITA): Ibadan (Nigeria), 2010. [Google Scholar]
- De Oliveira, M.A.; de Moraes, P.S.B. Technological and postharvest characteristics and productivity of cassava. Ciência e Agrotecnologia 2009, 33, 837–843. [Google Scholar]
- Ngeve, J.M. Mealiness of cassava roots: A descriptive attribute for boiled root texture. Journal of Food Science 1998, 45, 215–220. [Google Scholar]
- Iwe, M.O. Sensory evaluation of cassava products: A descriptive method for assessing surface appearance, mealy (floury) texture, taste, and aroma. Journal of Food Science and Technology 2002, 35, 231–238. [Google Scholar]
- R Core Team. R: A Language and environment for statistical computing; R foundation for statistical computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/.
- Richman, M.B. A cautionary note concerning a commonly applied eigen analysis procedure. Tellus 1988, 40, 50–58. [Google Scholar] [CrossRef]
- Karim, K.Y.; Norman, P.E.; Dzidzienyo, D.; Ifie, B.; Kulakow, P.; Rabbi, I.; Omoigui, L.; Parkes, E. Gene action analysis studies for agronomic traits in cassava (Manihot esculenta Crantz) genotypes developed using line by tester mating design. African Journal of Agricultural Research 2020, 16, 1471–1479. [Google Scholar]
- Agre, A.P.; Gueye, B.; Adjatin, A.; Dansi, M.; Bathacharjee, R.; Rabbi, I.Y.; Gedil, M. Folk taxonomy and traditional management of cassava (Manihot esculenta Crantz) diversity in southern and central Benin. International Journal of Innovative Science Research 2016, 20, 500–551. [Google Scholar]
- Afonso, S.D.J.; Ledo, C.D.S.; Moreira, R.F.C.; Silva, S.D.O.; Leal, V.D.J.; Conceição, A.D.S. Selection of descriptors in a morphological characteristic considered in cassava accessions by means of multivariate techniques. IOSR Journal of Agriculture and Veterinary Science 2014, 7, 13–20. [Google Scholar] [CrossRef]
- Agre, A.P.; Dansi, A.; Rabbi, I.Y.; Battachargee, R.; Dansi, M.; Melaku, G.; Augusto, B.; Sanni, A.; Akouegninou, A.; Akpagana, K. Agro morphological characterization of elite cassava (Manihot esculenta Crantz) cultivars collected in Benin. International Journal Current Research Bioscience Plant Biology 2015, 2, 1–14. [Google Scholar]
- Karim, K.Y. Genetic characterization of exotic and landraces of cassava in Ghan; Kwame Nkrumah University of Science and Technology, College of Agriculture and Natural Resources, Faculty of Agriculture, Department of Crop and Soil Science, 2012. [Google Scholar]
- De Pedri, E.C.M.; Rossi, A.A.B.; Cardoso, E.S.; Tiago, A.V.; Hoogerheide, E.S.S.; Yamashita, O.M. Morphological characteristics and culinary quality of cassava ethno-varieties at different harvesting times. Brazilian Journal of Food Technology 2018, 21, e2018073. [Google Scholar]
- Santos, H.G.; Jacomine, P.K.T.; dos Anjos, L.H.C.; de Oliveira, V.A.; Lumbreras, J.F.; Coelho, M.R.; Cunha, T.J.F. Sistema Brasileiro de Classificação de Solos, rev. e ampl., 5th ed.; Embrapa: Brasília, DF, 2018. [Google Scholar]
- Mugalavai, E.; Patel, R.; Singh, S. Evaluation of cassava accessions for cooking qualities: Texture, taste, and mealiness. Journal of Crop Science and Food Technology 2018, 22, 215–224. [Google Scholar]
| Trait | Code | Scoring | Sampling time (MAP) |
|---|---|---|---|
| Color of apical leaves | CAL | (3) Light green; (5) Dark green; (7) Purplish green; (9) Purple |
3 |
| Leaf color | LC | 3 = light green; 5 = dark green; 7 = purple green; 9 = purple | 6 |
| Presence of fruit | PFRT | (0) Absent; (1) Presence | 6 |
| Presence of seeds | PSE | (0) Absent; (1) Presence | 9 |
| Lobe margins | LM | (3) Smooth; (7) Winding | 6 |
| Color of leaf vein | CLV | (3) Green; (5) Green-reddish; (7) Red | 6 |
| Petiole color | PEC | (1) Yellowish-green; (2) Green; (3) Reddish-green; (5) Greenish-red; (7) Red; (9) Purple | 6 |
| Shape of central leaflet | SCL | (1) Ovoid; (2) Elliptic-lanceolate; (3) Obovate-lanceolate; (4) Oblong-lanceolate; (5) Lanceolate; (6) Straight or linear; (7) Pandurate; (8) Linera-piramidal; (9) Linear-pandurate; (10) Linear-hostatilobalate | 6 |
| Orientation of petiole | OPE | (1) Inclined-upwards; (3) Horizontal; (5) inclined-downwards; (7) Irregular | 6 |
| Leaf retention | LRE | (1) Very poor retention; (2) Less than average retention; (3) Average; (4) Better than average retention | 6 |
| Stipule margin | STM | (1) Entire; (2) Split | 9 |
| Color of stem epidermis | CSTE | (1) Creme; (2) Light brown; (3) Dark brown; (4) Orange | 9 |
| Color of end branches | CEBR | (3) Green; (5) Green-purple; (7) purple | 9 |
| Color of stem cortex | CSC | (1) Orange; (2) Light green; (3) Green | 9 |
| Color of stem exterior | CSE | (3) Orange; (4) Greeny-yellowish (5) Golden; (6) Light brown; (7) Silver; (8) Gray; (9) Dark brown. | 9 |
| Prominence of foliar scar | PFS | (3) Semi-prominent; (5) Prominent | 9 |
| Branching habit | BRH | (1) Erect; (2) Dichotomous; (3) Trichotomous; (4) Tetrachomotous | 12 |
| Root constrictions | RCO | (1) Few to none; (2) Some; (3) Many | 12 |
| Color of root cortex | CRC | (1) White or cream; (2) Yellow; (3) Pink; (4) Purple | 12 |
| Color of root pulp | CRP | (1) White; (2) Cream; (3) Yellow; (4) Orange; (5) Pink | 12 |
| External color of storage root | ECSR | (1) White or cream; (2) Yellow; (3) Light brown; (4) Dark brown | 12 |
| Extent of root peduncle | ERP | (0) Sessile; (3) Pedunculate; (5) Mixed | 12 |
| Root taste | RT | (1) Sweet; (2) Intermediate; (3) Bitter | 12 |
| Root shape | RS | (1) Conical; (2) Conical-cylindrical; (3) Cylindrical; (4) Irregular | 12 |
| Plant shape | PLNS | (1) Compact; (2) Open; (3) Umbrella; (4) Cylindrical | 12 |
| Trait | Code | Techniques of measurement | Sampling time (MAP) |
|---|---|---|---|
| Width of leaf robe | WLL | Two leaves from the middle of the plant measured from the widest part of the middle lobe | 6 |
| Length of leaf lobe | LLL | Measured from the intersection of all lobes to the end of the middle lobe | 6 |
| Petiole length | PLEN | Measured on two leaves per plant | 6 |
| Number of leaf robes | NLL | Counted on five leaves per plant with consideration of the predominant number of lobes | 6 |
| Distance between foliar scar | DSL | Measuring the distance between two foliar scars | 6 |
| Length of stipule | LST | Measured using meter rule. | 9 |
| Height at first branching | HFB | Measured vertically from ground to first primary branch | 12 |
| Level of branching | LBR | Number of branching points or levels | 12 |
| Plant height | PHT | Measured vertically from the ground to the top of the canopy | 12 |
| Number of commercial roots | NCR | Recorded on root with length greater than 20 cm from three plants | 12 |
| Number of storage root | NSR | Number of roots with length greater than 20 cm from three plant | 12 |
| Harvest index | HI | Measured as ratio of fresh root yield to the total fresh biomass | 12 |
| Root yield per plant | RYPP | All the root showing length greater than 20 cm are weighted | 12 |
| Root dry matter content | RDMC | Weighed the dry roots | 12 |
| Trait | RT | CLV | LC | CAL | SCL | LM | PEC | RS | PFS | CRP | CSC | CSE | ECR | CRC |
| RT | 1 | |||||||||||||
| CLV | -0.08 | 1 | ||||||||||||
| LC | 0.03 | 0.22* | 1 | |||||||||||
| CAL | 0.11 | 0.03 | 0.39*** | 1 | ||||||||||
| SCL | 0.1 | -0.14 | -0.30** | -0.1 | 1 | |||||||||
| LM | 0.14 | -0.08 | -0.1 | 0.06 | -0.16 | 1 | ||||||||
| PEC | 0.08 | 0.62*** | 0.43*** | 0.04 | -0.42*** | 0.02 | 1 | |||||||
| RS | -0.13 | -0.09 | -0.02 | 0.02 | 0 | 0.06 | 0.01 | 1 | ||||||
| PFS | 0.13 | -0.14 | -0.11 | 0.09 | 0.03 | 0.29** | -0.18 | 0 | 1 | |||||
| CRP | 0.10 | 0.05 | -0.03 | -0.01 | 0.02 | 0.08 | -0.06 | 0.03 | -0.09 | 1 | ||||
| CSC | 0.01 | -0.07 | -0.03 | -0.03 | -0.05 | 0.12 | -0.05 | 0.10 | 0 | -0.09 | 1 | |||
| CSE | 0.11 | -0.12 | -0.1 | -0.06 | 0.13 | 0 | -0.15 | -0.15 | 0.07 | -0.03 | -0.02 | 1 | ||
| ECSR | 0.12 | -0.04 | -0.05 | -0.18 | 0.08 | -0.14 | 0.02 | 0.07 | -0.01 | 0.11 | -0.05 | -0.05 | 1 | |
| CRC | -0.17 | 0.01 | 0.05 | -0.14 | -0.06 | -0.06 | -0.01 | -0.24* | -0.24* | -0.05 | 0.12 | 0.08 | 0.07 | 1 |
| LR | 0.07 | 0.03 | 0.22* | 0.23* | -0.12 | 0.13 | 0.24* | -0.01 | -0.05 | 0.06 | -0.27** | -0.19 | -0.14 | -0.1 |
| PO | 0.10 | 0.11 | 0.1 | 0.15 | 0.04 | 0 | 0.04 | -0.03 | 0.10 | 0.13 | -0.05 | -0.14 | -0.06 | -0.2 |
| CSE | 0.04 | -0.13 | -0.01 | -0.02 | 0.23* | -0.11 | -0.25* | -0.01 | 0.12 | -0.21* | -0.08 | 0.18 | -0.03 | 0.15 |
| CEBR | 0.04 | 0.06 | -0.07 | -0.03 | -0.16 | 0.24* | 0.16 | 0.07 | -0.17 | 0.02 | 0.09 | -0.12 | -0.03 | -0.04 |
| STM | -0.01 | 0.03 | -0.08 | -0.10 | 0.06 | -0.03 | 0.04 | -0.05 | -0.12 | -0.05 | -0.12 | 0.06 | 0.04 | 0.09 |
| BRH | -0.03 | -0.08 | -0.20* | 0 | 0.07 | 0.10 | -0.14 | 0.10 | 0.04 | -0.11 | 0.03 | -0.02 | 0.01 | -0.1 |
| PLNS | -0.03 | 0 | 0.16 | 0.05 | 0.01 | -0.12 | 0.09 | -0.13 | -0.10 | -0.12 | -0.09 | 0.21* | -0.06 | 0.12 |
| ERP | -0.02 | 0 | -0.18 | -0.07 | 0.09 | -0.10 | -0.07 | 0 | 0.04 | 0.08 | -0.12 | -0.10 | 0.10 | -0.1 |
| RCO | -0.04 | 0.08 | 0.11 | -0.15 | -0.29** | 0.02 | 0.13 | 0.16 | -0.01 | 0.13 | 0.01 | -0.04 | 0.05 | 0.07 |
| PFRT | 0.02 | -0.11 | -0.04 | 0.16 | 0.01 | 0.06 | -0.03 | 0.25* | -0.03 | -0.03 | 0.03 | -0.18 | 0.03 | -0.1 |
| PSE | 0 | -0.05 | -0.07 | 0.14 | 0.05 | 0.03 | 0 | 0.14 | 0.01 | -0.08 | 0.01 | -0.24* | 0.04 | -0.1 |
| Trait | LR | OPE | CSE | CEBR | STM | BRH | PS | EXT | RCO | PFRT | PSE |
| LR | 1 | ||||||||||
| PO | 0.19 | 1 | |||||||||
| CSE | -0.08 | -0.22* | 1 | ||||||||
| CEBR | -0.05 | -0.12 | -0.19 | 1 | |||||||
| STM | 0.24* | -0.07 | -0.04 | 0.11 | 1 | ||||||
| BRH | 0.06 | 0.18 | -0.05 | 0.07 | -0.06 | 1 | |||||
| PLNS | -0.03 | -0.23* | 0.05 | -0.04 | 0.16 | -0.53*** | 1 | ||||
| ERP | 0.06 | 0.06 | -0.08 | -0.18 | -0.09 | 0.06 | -0.18 | 1 | |||
| RCO | -0.05 | -0.05 | -0.08 | -0.11 | -0.19* | 0 | -0.11 | 0.09 | 1 | ||
| PFRT | 0.14 | 0.17 | 0 | 0.01 | -0.14 | 0.32** | 0.47*** | 0.08 | 0.07 | 1 | |
| PSE | 0.09 | 0.14 | 0.07 | -0.01 | -0.17 | 0.31** | 0.44*** | 0.13 | 0.04 | 0.86*** | 1 |
| Trait | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | PC9 | PC10 |
| RT | 0.053 | -0.003 | 0.384 | 0.184 | -0.230 | -0.115 | 0.574 | 0.402 | -0.007 | 0.052 |
| CLV | -0.150 | 0.581 | -0.149 | -0.185 | -0.111 | 0.002 | 0.246 | -0.293 | -0.083 | 0.356 |
| LC | -0.181 | 0.628 | 0.230 | -0.141 | 0.339 | -0.228 | 0.077 | 0.105 | -0.085 | -0.129 |
| CAL | 0.144 | 0.327 | 0.573 | 0.048 | 0.301 | -0.081 | -0.032 | 0.025 | -0.212 | -0.243 |
| SCL | 0.057 | -0.594 | 0.206 | -0.266 | -0.134 | 0.199 | 0.137 | -0.025 | -0.314 | -0.003 |
| LM | 0.188 | 0.081 | 0.135 | 0.704 | -0.114 | -0.021 | -0.061 | 0.108 | 0.340 | 0.010 |
| PEC | -0.143 | 0.805 | -0.124 | -0.032 | -0.004 | 0.02 | 0.197 | 0.049 | 0.012 | 0.347 |
| RT | 0.327 | 0.036 | -0.222 | 0.113 | 0.079 | -0.082 | -0.482 | 0.349 | -0.385 | 0.117 |
| PFS | 0.168 | -0.237 | 0.401 | 0.295 | -0.084 | -0.418 | -0.080 | -0.042 | 0.158 | 0.369 |
| CRP | 0.056 | 0.095 | -0.046 | -0.031 | -0.568 | -0.159 | 0.032 | 0.216 | -0.039 | -0.536 |
| COX | 0.055 | -0.089 | -0.313 | 0.440 | 0.248 | -0.144 | 0.216 | -0.211 | -0.236 | -0.216 |
| CSE | -0.331 | -0.374 | 0.167 | 0.096 | 0.055 | -0.112 | 0.210 | 0.019 | 0.222 | 0.047 |
| ECSR | 0.037 | -0.115 | -0.292 | -0.248 | -0.262 | -0.054 | 0.323 | 0.488 | -0.110 | 0.123 |
| CRC | -0.315 | -0.057 | -0.338 | -0.152 | 0.316 | 0.135 | 0.230 | -0.040 | 0.441 | -0.372 |
| LR | 0.142 | 0.429 | 0.424 | -0.152 | -0.090 | 0.336 | -0.250 | 0.168 | 0.350 | -0.104 |
| PO | 0.354 | 0.228 | 0.336 | -0.124 | -0.296 | -0.050 | 0.146 | -0.371 | -0.093 | -0.177 |
| CSE | -0.089 | -0.419 | 0.182 | -0.207 | 0.487 | -0.053 | 0.083 | 0.206 | 0.130 | 0.212 |
| CEBR | 0.008 | 0.204 | -0.264 | 0.550 | -0.070 | 0.438 | 0.090 | 0.118 | -0.169 | 0.033 |
| STM | -0.273 | 0.010 | 0.073 | -0.032 | -0.165 | 0.651 | -0.122 | 0.177 | 0.157 | 0.097 |
| BRH | 0.594 | -0.130 | -0.070 | 0.099 | -0.016 | 0.218 | 0.098 | -0.257 | 0.193 | 0.112 |
| PLNS | -0.749 | 0.018 | 0.171 | -0.025 | 0.120 | 0.002 | -0.169 | 0.145 | -0.161 | -0.012 |
| ERP | 0.248 | -0.099 | -0.066 | -0.394 | -0.374 | -0.158 | -0.181 | -0.105 | 0.124 | 0.118 |
| RCO | 0.099 | 0.212 | -0.409 | -0.071 | -0.025 | -0.540 | -0.148 | 0.190 | 0.341 | -0.034 |
| PFRT | 0.804 | 0.073 | -0.048 | -0.128 | 0.305 | 0.128 | 0.094 | 0.187 | 0.034 | -0.079 |
| PSE | 0.788 | 0.047 | -0.039 | -0.182 | 0.307 | 0.119 | 0.128 | 0.126 | 0.003 | 0.040 |
| Eigenvalue | 2.97 | 2.61 | 1.77 | 1.65 | 1.58 | 1.50 | 1.18 | 1.17 | 1.16 | 1.10 |
| Proportion of variance (%) | 11.88 | 10.44 | 7.1 | 6.6 | 6.31 | 6.00 | 4.71 | 4.67 | 4.63 | 4.4 |
| Cumulative variance (%) | 11.88 | 22.33 | 29.43 | 36.03 | 42.34 | 48.34 | 53.05 | 57.72 | 62.34 | 66.74 |
| Traits | Minimum | Maximum | Mean | Standard deviation | Coefficient of variance (%) |
| Distance between leaf scar (cm) | 1.0 | 5.0 | 3.0 | 0.5 | 18.7 |
| Height at first branching (cm) | 0.0 | 165.9 | 78.8 | 45.4 | 57.6 |
| Harvest index | 0.2 | 0.6 | 0.4 | 0.1 | 23.9 |
| Level of branching | 0.0 | 20.0 | 2.8 | 2.3 | 81.2 |
| Length of leaf lobe (cm) | 5.7 | 21.8 | 14.2 | 2.8 | 20.1 |
| Length of stipule (cm) | 1.0 | 5.0 | 3.3 | 1.1 | 34.8 |
| Number of commercial roots | 1.0 | 24.0 | 8.8 | 5.1 | 57.9 |
| Number of leaf lobe | 3.0 | 9.0 | 6.3 | 1.1 | 17.8 |
| Number of storage root | 2.0 | 53.0 | 15.9 | 9.7 | 61.1 |
| Plant height (cm) | 41.0 | 346.0 | 178.1 | 43.1 | 24.2 |
| Petiole length (cm) | 4.7 | 37.3 | 20.9 | 6.1 | 29.2 |
| Roots dry matter content (%) | 15.0 | 36.0 | 27.8 | 4.1 | 14.8 |
| Root yield per plant (kg) | 1.0 | 19.0 | 6.4 | 4.2 | 65.0 |
| Width of leaf lobe (cm) | 1.5 | 6.0 | 3.3 | 0.8 | 24.9 |
| HI | RYPP | PLEN | LLL | WLL | RDMC | NSR | PHT | HFB | NLL | DLS | LST | LBR | NCR | |
| HI | 1 | |||||||||||||
| RYPP | 0.40*** | 1 | ||||||||||||
| PLEN | -0.06 | -0.12 | 1 | |||||||||||
| LLL | 0.02 | 0.02 | 0.22* | 1 | ||||||||||
| WLL | -0.05 | 0.08 | 0.05 | 0.66*** | 1 | |||||||||
| RDMC | 0.01 | 0.07 | 0.11 | 0.05 | 0.05 | 1 | ||||||||
| NSR | 0.24* | 0.76*** | -0.03 | 0.09 | 0.12 | -0.08 | 1 | |||||||
| PHT | 0.03 | 0.09 | 0.09 | 0.17 | 0.20* | 0.08 | 0.08 | 1 | ||||||
| HFB | 0.08 | 0.04 | 0.04 | 0.11 | 0.04 | 0.08 | -0.02 | 0.11 | 1 | |||||
| NLL | 0.06 | 0 | 0.26** | 0.41*** | 0.14 | 0.02 | 0.08 | 0.24* | -0.12 | 1 | ||||
| DLS | -0.20* | -0.01 | 0.19 | 0.03 | -0.04 | 0.07 | 0.02 | 0.06 | 0.16 | -0.05 | 1 | |||
| LST | 0.16 | 0.08 | 0 | 0.13 | 0.06 | 0.02 | 0.11 | 0.09 | 0 | 0.01 | -0.07 | 1 | ||
| LBR | 0.08 | 0.08 | 0.02 | -0.21* | -0.21* | -0.05 | 0.06 | 0.04 | 0.25* | 0.09 | 0.02 | -0.05 | 1 | |
| NCR | 0.17 | 0.68*** | -0.09 | 0.02 | 0.08 | -0.05 | 0.84*** | 0.04 | 0.01 | 0.05 | 0.01 | 0.06 | 0.04 | 1 |
| Variable | Prin1 | Prin2 | Prin3 | Prin4 | Prin5 | Prin6 |
| Harvest index | 0.43 | -0.13 | 0.07 | 0.24 | 0.54 | 0.27 |
| Root yield per plant | 0.86 | -0.20 | 0.06 | -0.01 | -0.05 | 0.08 |
| Petiole length | -0.08 | 0.43 | 0.44 | -0.11 | 0.05 | -0.13 |
| Length of leaflet | 0.23 | 0.83 | 0.01 | -0.06 | 0.01 | 0.07 |
| Width of leaflet | 0.23 | 0.70 | -0.48 | 0.13 | -0.30 | -0.01 |
| Root dry matter content | -0.01 | 0.17 | 0.21 | 0.12 | -0.09 | 0.54 |
| Number of storage root | 0.90 | -0.13 | 0.07 | -0.18 | -0.11 | -0.09 |
| Plant height | 0.19 | 0.40 | 0.15 | 0.30 | 0.04 | -0.09 |
| Height at first branching | 0.05 | 0.05 | 0.35 | 0.66 | -0.32 | 0.16 |
| Number of leaflet | 0.16 | 0.53 | 0.23 | -0.09 | 0.44 | -0.42 |
| Distance between leaf scar | -0.06 | 0.08 | 0.55 | -0.15 | -0.56 | 0.05 |
| Length of stipule | 0.20 | 0.12 | -0.11 | 0.15 | 0.32 | 0.44 |
| Level of branching | 0.06 | -0.26 | 0.30 | 0.61 | 0.06 | -0.49 |
| Number of commercial root | 0.85 | -0.19 | 0.05 | -0.18 | -0.18 | -0.09 |
| Eigenvalue | 2.75 | 2.09 | 1.73 | 1.24 | 1.22 | 1.11 |
| Proportion of variance (%) | 18.34 | 13.94 | 11.53 | 8.30 | 8.18 | 7.40 |
| Cumulative variance (%) | 18.34 | 32.29 | 43.83 | 52.13 | 60.31 | 67.72 |
|
Genotypes |
Cooking time | Cooking percent | Surface appearance | Mealiness | Taste | Aroma | Texture |
| Mean ± SE | Mean ± SE | Mean ± SE | Mean ± SE | Mean± SE | Mean± SE | Mean± SE | |
| Cook soon | 15.2±1.2 | 96.4±2.4 | 4.5±0.50 | 4.75±0.80 | 4.8±0.50 | 4.8±0.50 | 4.5±0.58 |
| Butter cassava | 15.4±1.2 | 95.0±1.8 | 4.5±0.58 | 4.50±0.50 | 4.9±0.43 | 4.8±0.50 | 4.5±0.50 |
| Ndiamonya-malo | 21.0±1.4 | 83.3±1.6 | 4.3±0.50 | 4.25±0.48 | 4.5±0.50 | 4.3±0.50 | 4.3±0.50 |
| Cocoa cassada | 20.0±1.7 | 88.2±2.1 | 4.5±0.50 | 4.25±0.80 | 4.5±0.58 | 4.5±0.58 | 4.3±0.58 |
| Tangagboi | 28.7±1.8 | 76.2±2.3 | 4.3±0.58 | 4.0±0.40 | 4.5±0.50 | 4.3±0.58 | 4.3±0.58 |
| Mende Tangai | 28.6±1.5 | 78.4±2.0 | 4.0±0.65 | 4.0±0.65 | 4.0±0.65 | 4.0±0.65 | 4.0±0.65 |
| Nikaneh | 27.0±1.5 | 80.2±2.5 | 4.3±0.58 | 4.3±0.58 | 4.3±0.58 | 4.3±0.50 | 4.0±0.50 |
| Yaa Kanu | 27.0±1.7 | 80.2±3.0 | 4.0±0.50 | 4.0±0.60 | 4.3±0.50 | 4.3±0.82 | 3.5±0.58 |
| Kendemeh | 28.6±1.6 | 73.4±1.9 | 3.5±0.58 | 4.0±0.63 | 4.3±0.50 | 4.0±0.58 | 3.5±0.50 |
| Tapiyoka | 25.7±1.4 | 82.0±3.2 | 4.3±0.55 | 4.3±0.50 | 4.5±0.80 | 4.3±0.50 | 4.0±0.60 |
| Soja color | 29.8±1.5 | 70.3±2.8 | 3.0±0.50 | 3.0±0.58 | 2.0±0.50 | 3.0±0.65 | 2.0±0.82 |
| Pink lady | 30.0±1.5 | 63.2±2.6 | 3.5±0.82 | 3.0±0.80 | 3.5±0.65 | 3.0±0.50 | 3.0±0.58 |
| SLICASS 7 | 29.8±1.8 | 58.3±1.7 | 4.0±0.50 | 3.5±0.40 | 3.0±0.45 | 4.0±0.60 | 3.0±0.50 |
| SLICASS 6 | 30.0±1.7 | 30.2±3.1 | 3.5±0.58 | 3.0±0.55 | 2.0±0.50 | 3.0±0.50 | 3.0±0.50 |
| SLICASS 4 | 30.0±1.3 | 27.9±1.8 | 2.0±0.40 | 2.0±0.82 | 2.0±0.50 | 2.0±0.82 | 2.0±0.82 |
| Varieties/Genotypes | Rating |
| Tangaigboi | Highly preferred |
| Coco cassada | Highly preferred |
| Cooksoon | Highly preferred |
| Butter cassava | Highly preferred |
| Ndiamonyamalo | Highly preferred |
| Tapiyoka | Moderately preferred |
| Yaa kanu | Moderately preferred |
| Nikaneh | Moderately preferred |
| Mende tangai | Moderately preferred |
| Kendemeh | Moderately preferred |
| SLICASS 4 | Not good |
| SLICASS 6 | Not good |
| SLICASS 7 | Not good |
| Pink lady | Not good |
| Soja colour | Not good |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





