Submitted:
26 April 2024
Posted:
28 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Metodology
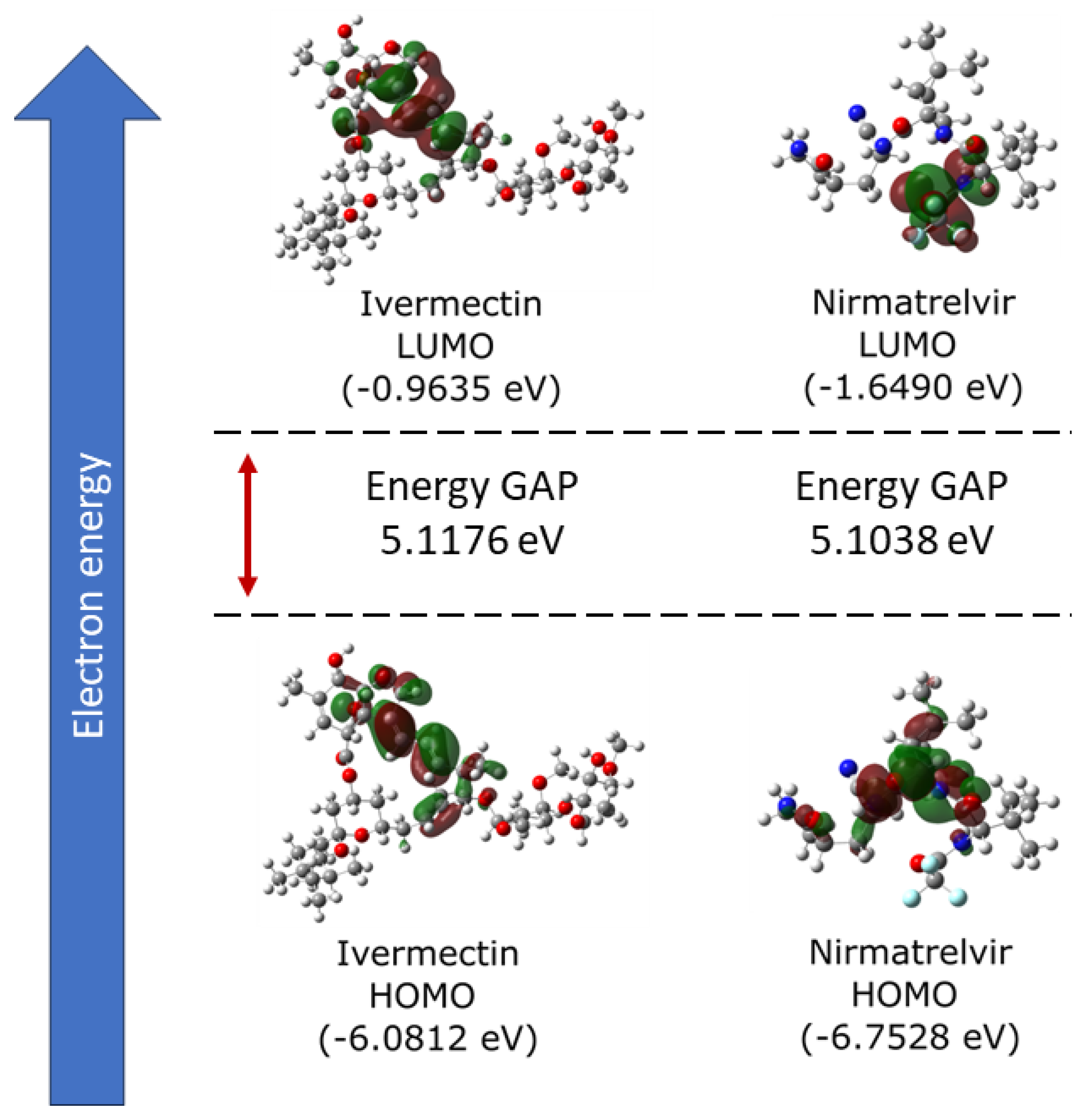
2.1. Electronic Properties
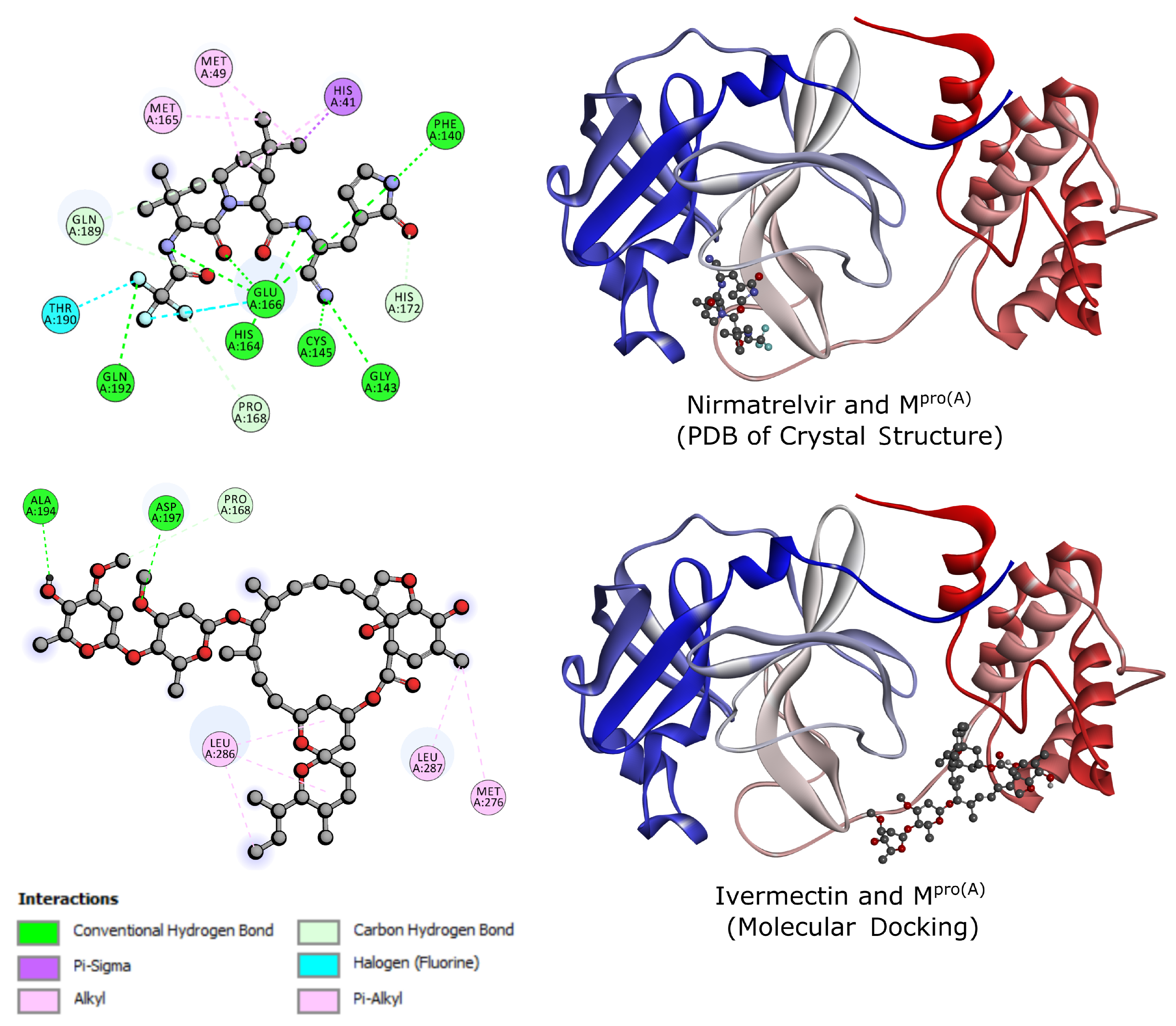
2.2. Molecular Docking
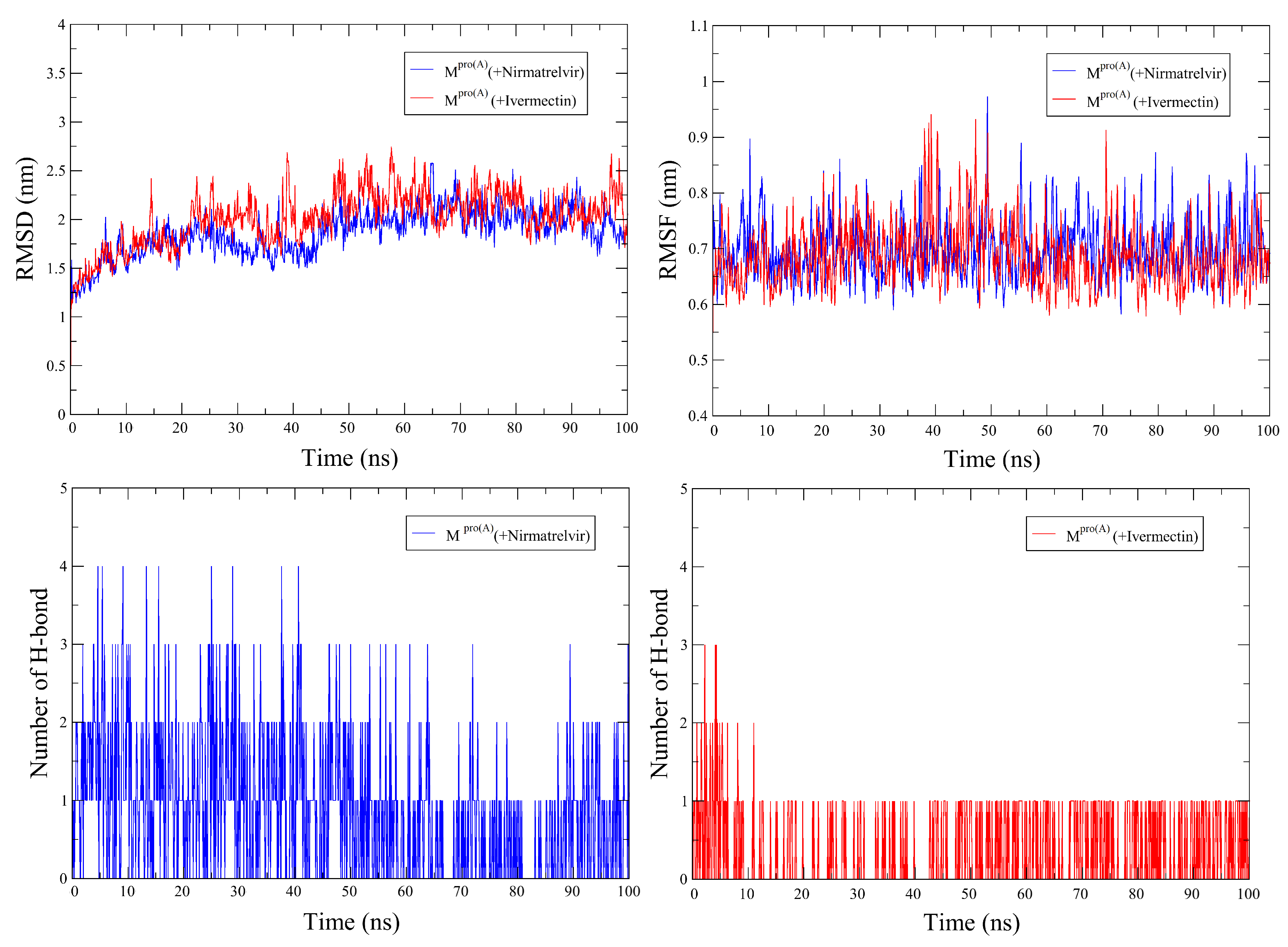
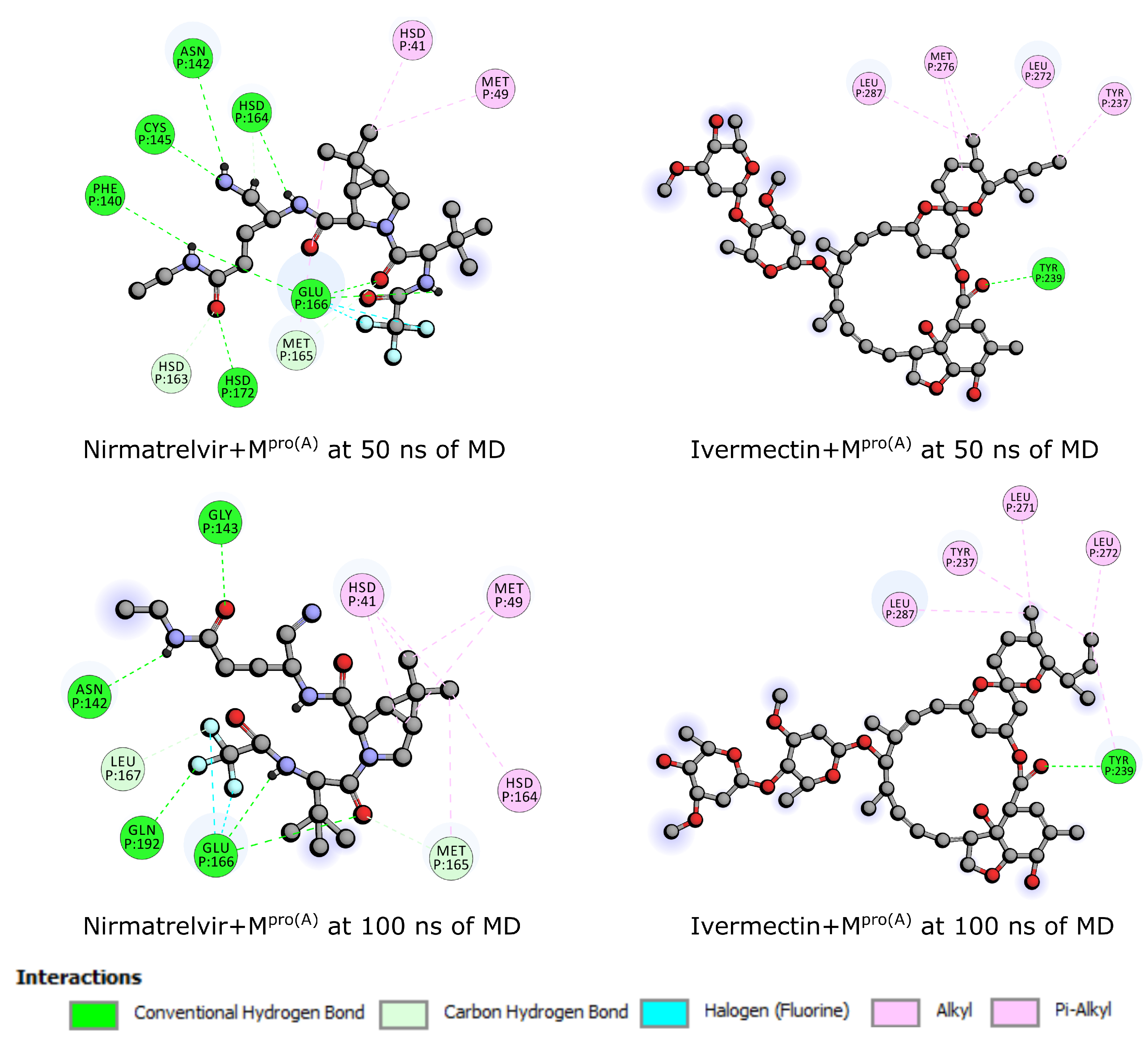
2.3. Molecular Dynamics
3. Results
4. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atzrodt, C.L.; Maknojia, I.; McCarthy, R.D.; Oldfield, T.M.; Po, J.; Ta, K.T.; Stepp, H.E.; Clements, T.P. A Guide to COVID-19: a global pandemic caused by the novel coronavirus SARS-CoV-2. Febs J. 2020, 287, 3633–3650. [Google Scholar] [CrossRef] [PubMed]
- Jayaweera, M.; Perera, H.; Gunawardana, B.; Manatunge, J. Transmission of COVID-19 virus by droplets and aerosols: A critical review on the unresolved dichotomy. Environ. Res. 2020, 188, 109819. [Google Scholar] [CrossRef] [PubMed]
- Louten, J. Virus transmission and epidemiology. Essent. Hum. Virol. 2016, 71. [Google Scholar]
- Asadi, S. Airborne Infectious Disease Transmission Via Expiratory Aerosol Particles and Aerosolized Fomites; University of California: Davis, CA, USA, 2020. [Google Scholar]
- Weiss, R.A.; Sankaran, N. Emergence of epidemic diseases: Zoonoses and other origins. Fac. Rev. 2022, 11. [Google Scholar] [CrossRef]
- Rajeev, R.; Prathiviraj, R.; Kiran, G.S.; Selvin, J. Zoonotic evolution and implications of microbiome in viral transmission and infection. Virus Res. 2020, 290, 198175. [Google Scholar] [CrossRef] [PubMed]
- WORLDOMETER. COVID-19 CORONAVIRUS PANDEMIC. 2024. Available online: https://www.worldometers.info/coronavirus (accessed on 5 March 2024).
- Pellis, L.; Scarabel, F.; Stage, H.B.; Overton, C.E.; Chappell, L.H.; Fearon, E.; Bennett, E.; Lythgoe, K.A.; House, T.A.; Hall, I. Challenges in control of COVID-19: short doubling time and long delay to effect of interventions. Philos. Trans. R. Soc. 2021, 376, 20200264. [Google Scholar] [CrossRef] [PubMed]
- Cannalire, R.; Cerchia, C.; Beccari, A.R.; Di Leva, F.S.; Summa, V. Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities. J. Med. Chem. 2020, 65, 2716–2746. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Perera, L.; Tillekeratne, L.V. Potential SARS-CoV-2 main protease inhibitors. Drug Discov. Today 2021, 26, 804–816. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Agost-Beltrán, L.; de la Hoz-Rodríguez, S.; Bou-Iserte, L.; Rodríguez, S.; Fernández-de-la Pradilla, A.; González, F.V. Advances in the development of SARS-CoV-2 Mpro inhibitors. Molecules 2022, 27, 2523. [Google Scholar] [CrossRef]
- Alugubelli, Y.R.; Geng, Z.Z.; Yang, K.S.; Shaabani, N.; Khatua, K.; Ma, X.R.; Vatansever, E.C.; Cho, C.C.; Ma, Y.; Xiao, J. A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals. Eur. J. Med. Chem. 2022, 240, 114596. [Google Scholar] [CrossRef] [PubMed]
- Andi, B.; Kumaran, D.; Kreitler, D.F.; Soares, A.S.; Keereetaweep, J.; Jakoncic, J.; Lazo, E.O.; Shi, W.; Fuchs, M.R.; Sweet, R.M. Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease. Sci. Rep. 2022, 12, 12197. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.Z.; Yasuo, N.; Sekijima, M. Screening for inhibitors of main protease in sars-cov-2: In silico and in vitro approach avoiding peptidyl secondary amides. J. Chem. Inf. Model. 2022, 62, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Zeng, X.; Duan, Y.; Yang, Z.; Ma, Y.; Yang, H.; Yang, X.; Liu, X. Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants. Commun. Biol. 2023, 6, 694. [Google Scholar] [CrossRef] [PubMed]
- Di Chio, C.; Previti, S.; Amendola, G.; Ravichandran, R.; Wagner, A.; Cosconati, S.; Hellmich, U.A.; Schirmeister, T.; Zappalà, M.; Ettari, R. Development of novel dipeptide nitriles as inhibitors of rhodesain of Trypanosoma brucei rhodesiense. Eur. J. Med. Chem. 2022, 236, 114328. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Malla, T.R.; Owen, C.D.; Tumber, A.; Brewitz, L.; McDonough, M.A.; Salah, E.; Terasaka, N.; Katoh, T.; Lukacik, P. In vitro selection of macrocyclic peptide inhibitors containing cyclic γ2, 4-amino acids targeting the SARS-CoV-2 main protease. Nat. Chem. 2023, 1–8. [Google Scholar]
- Tyndall, J.D. S-217622, a 3CL protease inhibitor and clinical candidate for SARS-CoV-2. J. Med. Chem. 2022, 65, 6496–6498. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Fang, C.; Zhang, Q.; Zhang, R.; Zhao, X.; Duan, Y.; Wang, H.; Zhu, Y.; Feng, L.; Zhao, J. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell 2022, 13, 689–693. [Google Scholar] [CrossRef]
- Ahmad, B.; Batool, M.; Ain, Q.u.; Kim, M.S.; Choi, S. Exploring the binding mechanism of PF-07321332 SARS-CoV-2 protease inhibitor through molecular dynamics and binding free energy simulations. Int. J. Mol. Sci. 2021, 22, 9124. [Google Scholar] [CrossRef]
- Zaidi, A.K.; Dehgani-Mobaraki, P. The mechanisms of action of ivermectin against SARS-CoV-2—an extensive review. J. Antibiot. 2022, 75, 60–71. [Google Scholar] [CrossRef]
- Hill, A.; Garratt, A.; Levi, J.; Falconer, J.; Ellis, L.; McCann, K.; Pilkington, V.; Qavi, A.; Wang, J.; Wentzel, H. Retracted: meta-analysis of randomized trials of ivermectin to treat SARS-CoV-2 infection. In Open Forum Infectious Diseases; Oxford University Press: Oxford, MI, USA, 2021; Volume 8, p. ofab358. [Google Scholar]
- Sharun, K.; Dhama, K.; Patel, S.K.; Pathak, M.; Tiwari, R.; Singh, B.R.; Sah, R.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J.; Leblebicioglu, H. Ivermectin, a new candidate therapeutic against SARS-CoV-2/COVID-19, 2020.
- Cobos-Campos, R.; Apiñaniz, A.; Parraza, N.; Cordero, J.; García, S.; Orruño, E. Potential use of ivermectin for the treatment and prophylaxis of SARS-CoV-2 infection. Curr. Res. Transl. Med. 2021, 69, 103309. [Google Scholar] [CrossRef] [PubMed]
- Buonfrate, D.; Chesini, F.; Martini, D.; Roncaglioni, M.C.; Fernandez, M.L.O.; Alvisi, M.F.; De Simone, I.; Rulli, E.; Nobili, A.; Casalini, G. High-dose ivermectin for early treatment of COVID-19 (COVER study): a randomised, double-blind, multicentre, phase II, dose-finding, proof-of-concept clinical trial. Int. J. Antimicrob. Agents 2022, 59, 106516. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.e.; Trucks, G.; Schlegel, H.B.; Scuseria, G.; Robb, M.; Cheeseman, J.; Scalmani, G.; Barone, V.; Petersson, G.; Nakatsuji, H. Gaussian 16, 2016.
- Stewart, J.J. Application of the PM6 method to modeling proteins. J. Mol. Model. 2009, 15, 765–805. [Google Scholar] [CrossRef] [PubMed]
- Parr, R.G. Density functional theory. Annual Review of Physical Chemistry 1983, 34, 631–656. [Google Scholar] [CrossRef]
- Sham, L.J.; Schlüter, M. Density-functional theory of the energy gap. Phys. Rev. Lett. 1983, 51, 1888. [Google Scholar] [CrossRef]
- Becke, A.D. A new mixing of Hartree–Fock and local density-functional theories. J. Chem. Phys. 1993, 98, 1372–1377. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Huey, R.; Morris, G.M.; Forli, S. Using AutoDock 4 and AutoDock vina with AutoDockTools: a tutorial. Scripps Res. Inst. Mol. Graph. Lab. 2012, 10550, 1000. [Google Scholar]
- Phillips, J.C.; Zheng, G.; Kumar, S.; Kalé, L.V. NAMD: Biomolecular simulation on thousands of processors. SC’02: Proceedings of the 2002 ACM/IEEE Conference on Supercomputing. IEEE, 2002, pp. 36–36. [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell Jr, A.D.; Pastor, R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. 2010, 114, 7830–7843. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Fukunishi, Y.; Yamashita, Y.; Mashimo, T.; Nakamura, H. Prediction of protein- compound binding energies from known activity data: docking-score-based method and its applications. Mol. Inform. 2018, 37, 1700120. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




