Submitted:
26 April 2024
Posted:
26 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
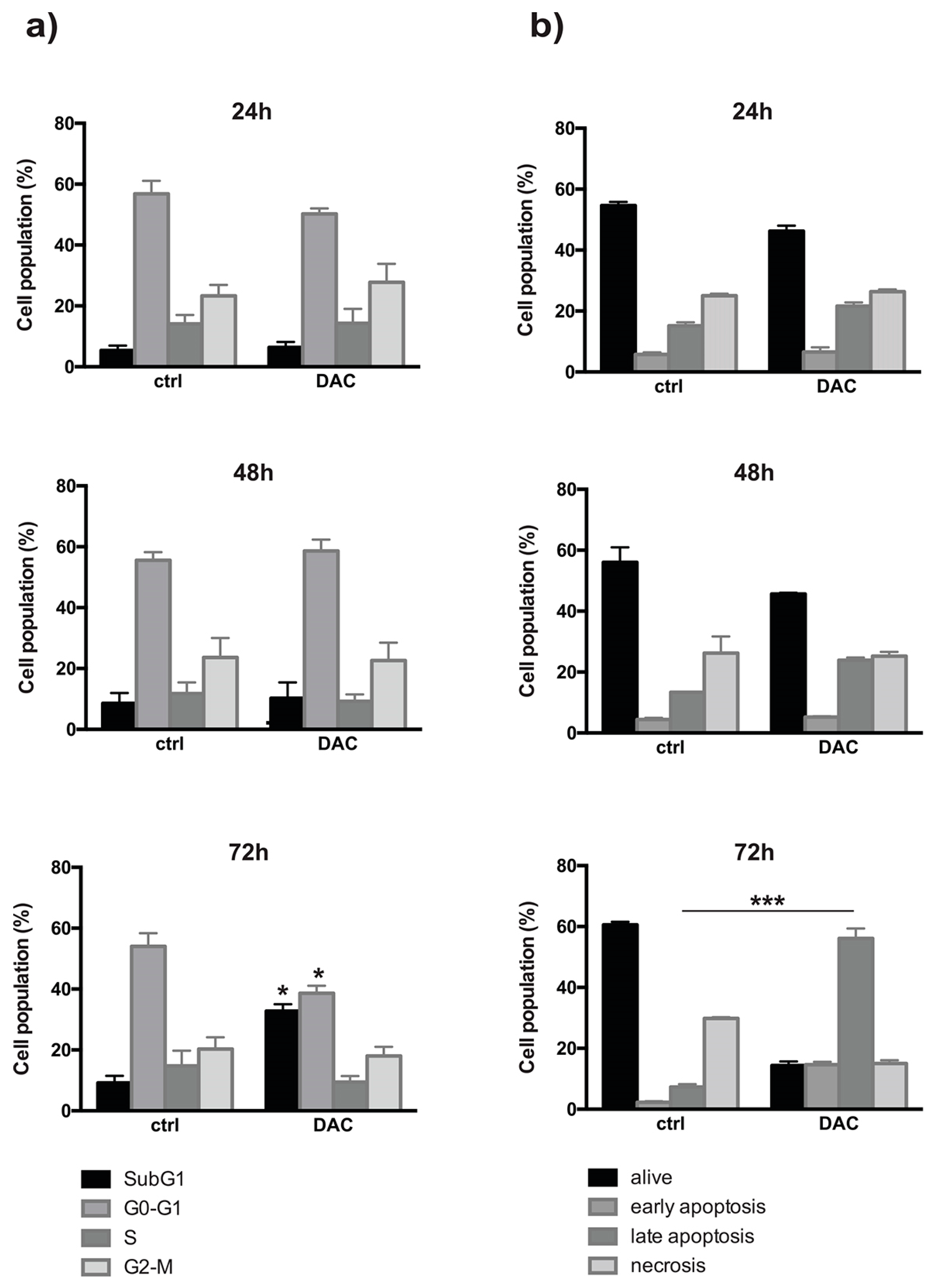
2.1. In Vitro Anti-Proliferative Effect of DAC on Weri-Rb-1 Cells
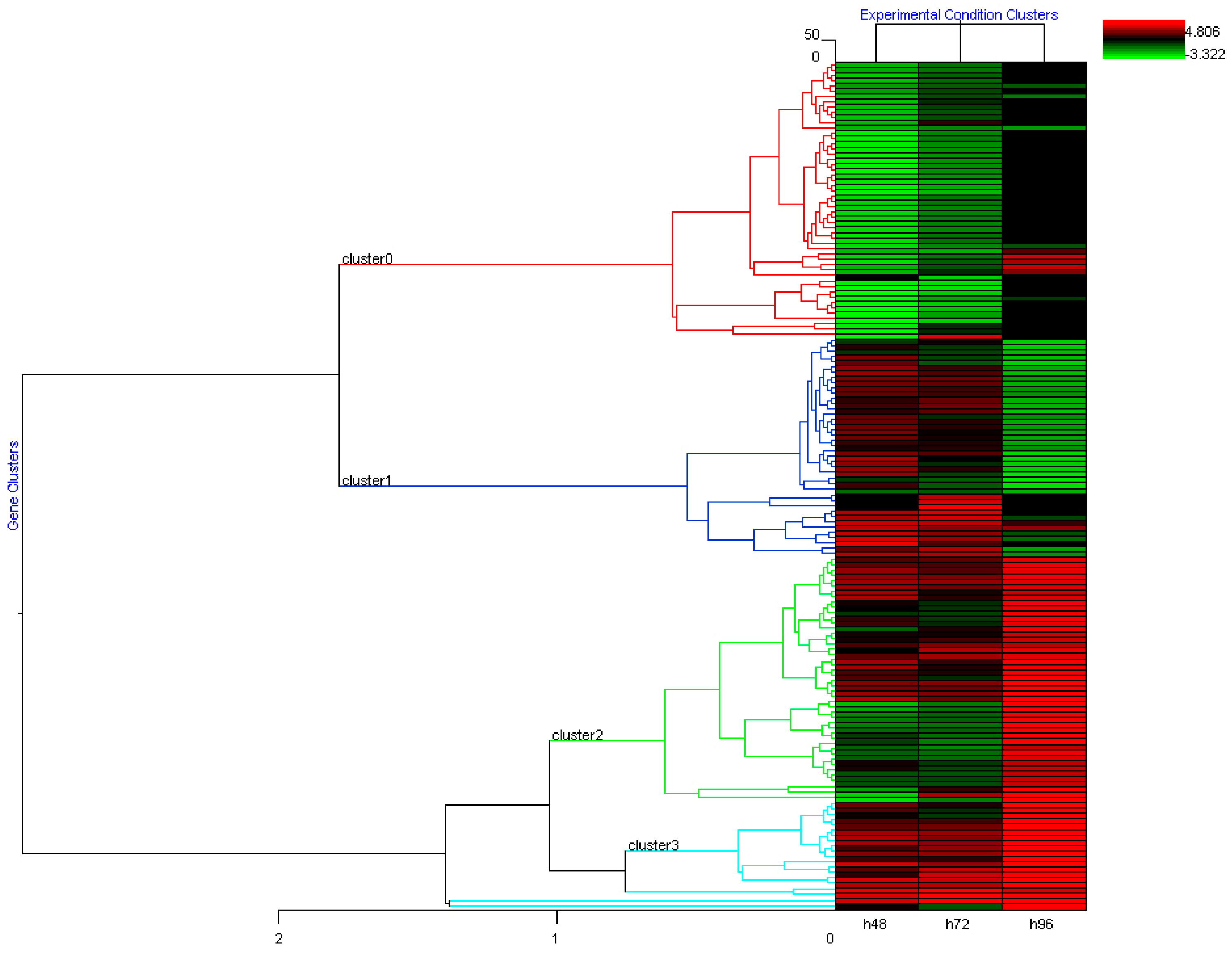
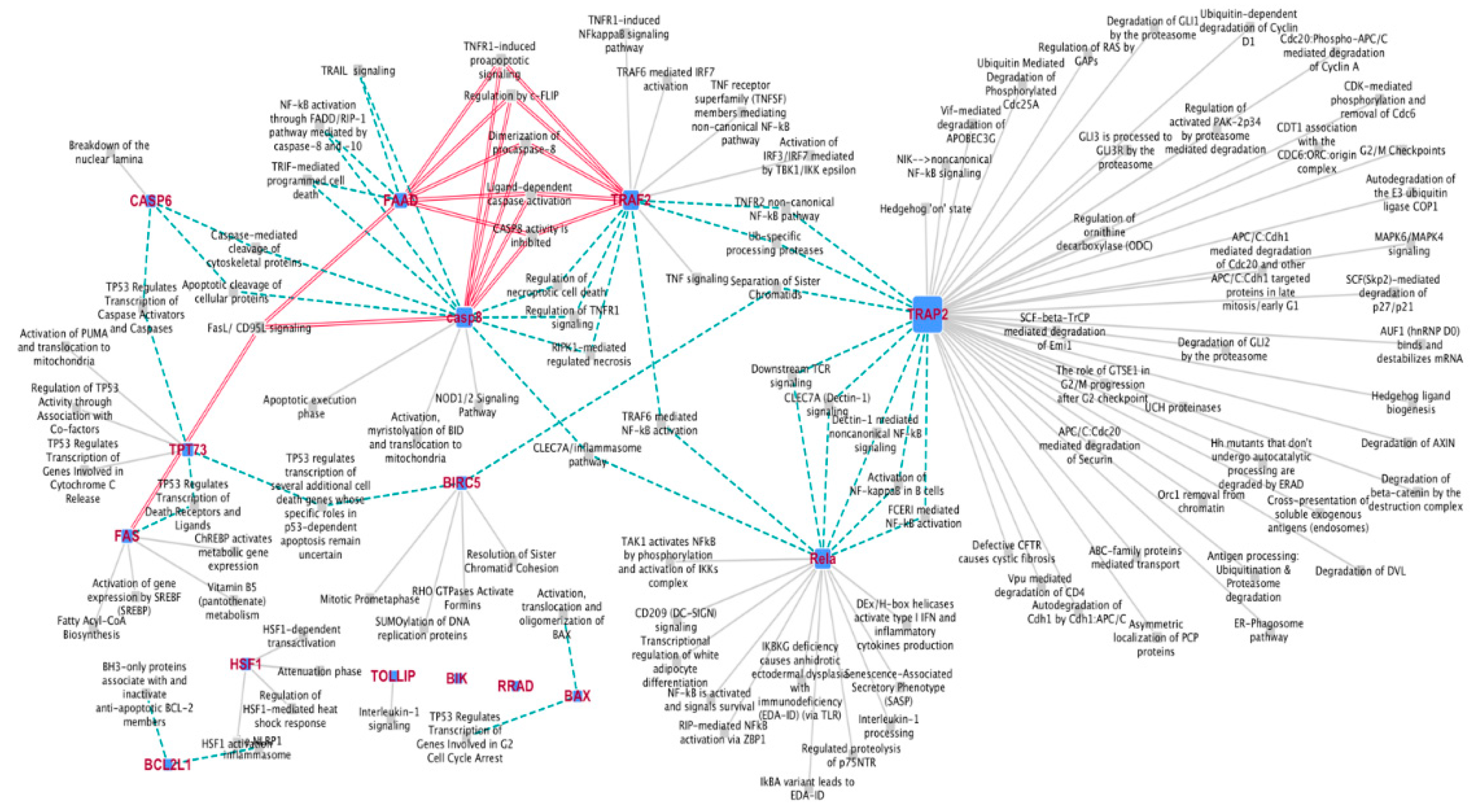
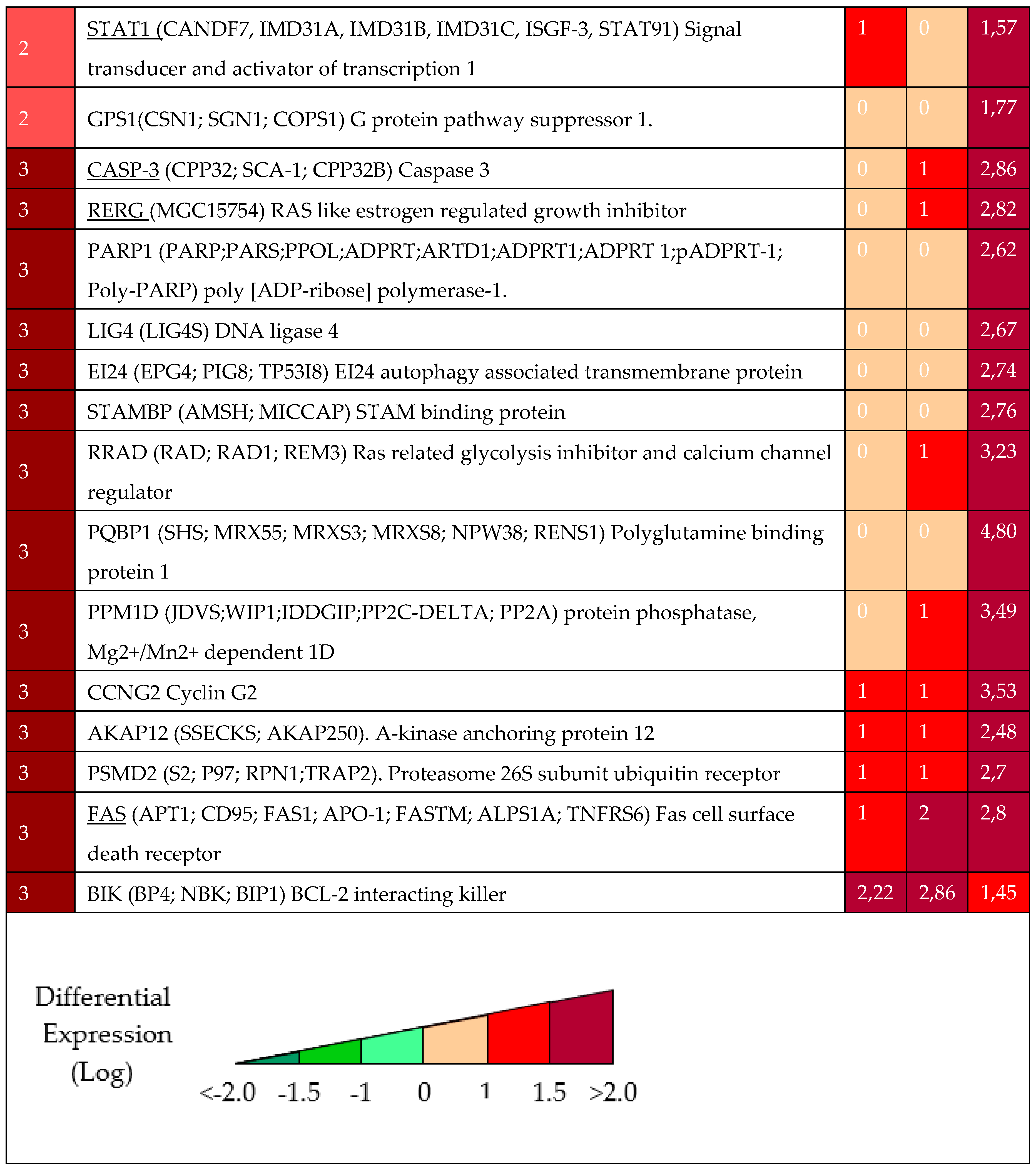
2.1.1. Gene Expression Profile after DAC Treatment in Weri-Rb-1 Cells
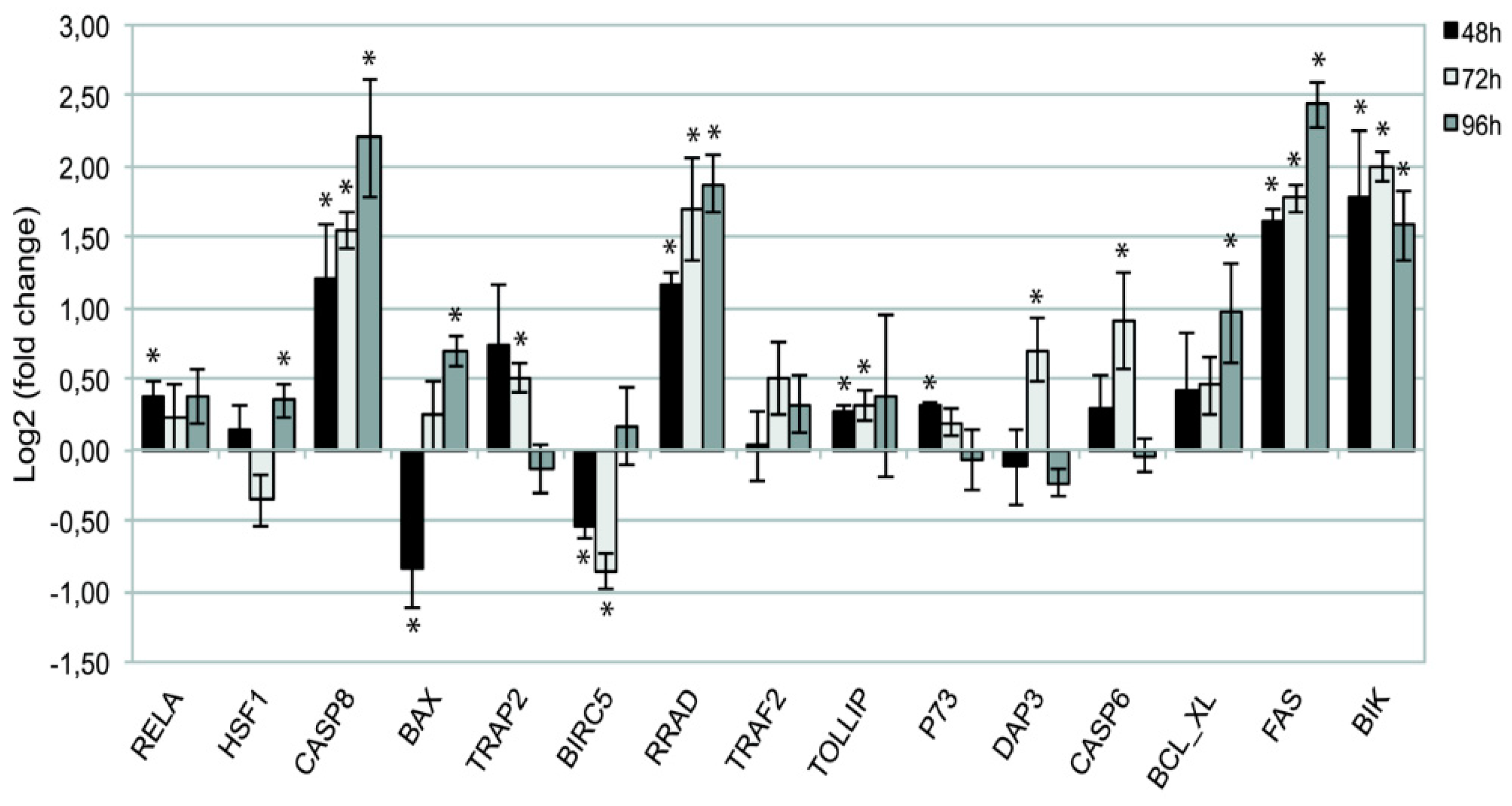
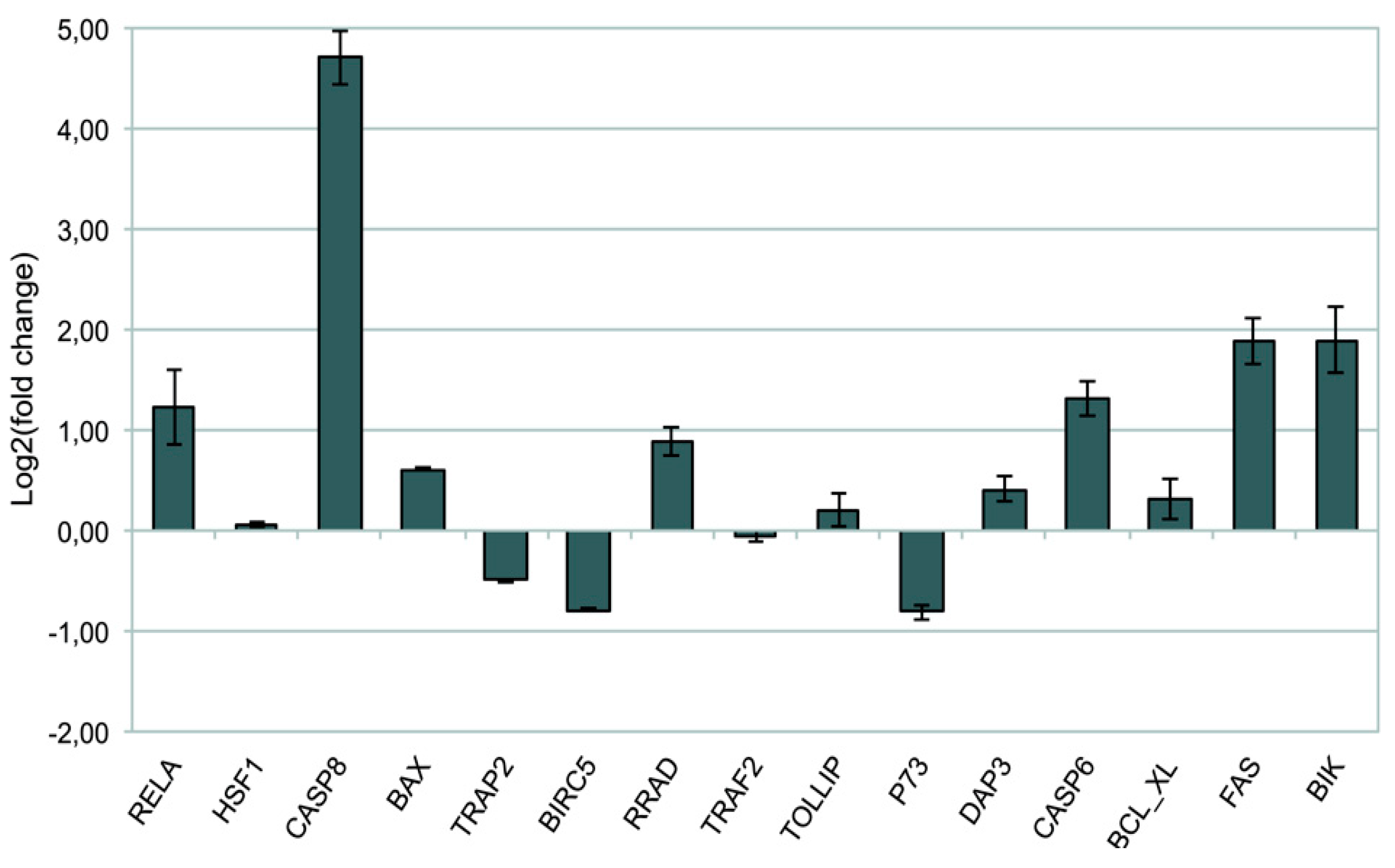
2.1.2. Validation of 15 Hub Genes Expression by qPCR Analysis
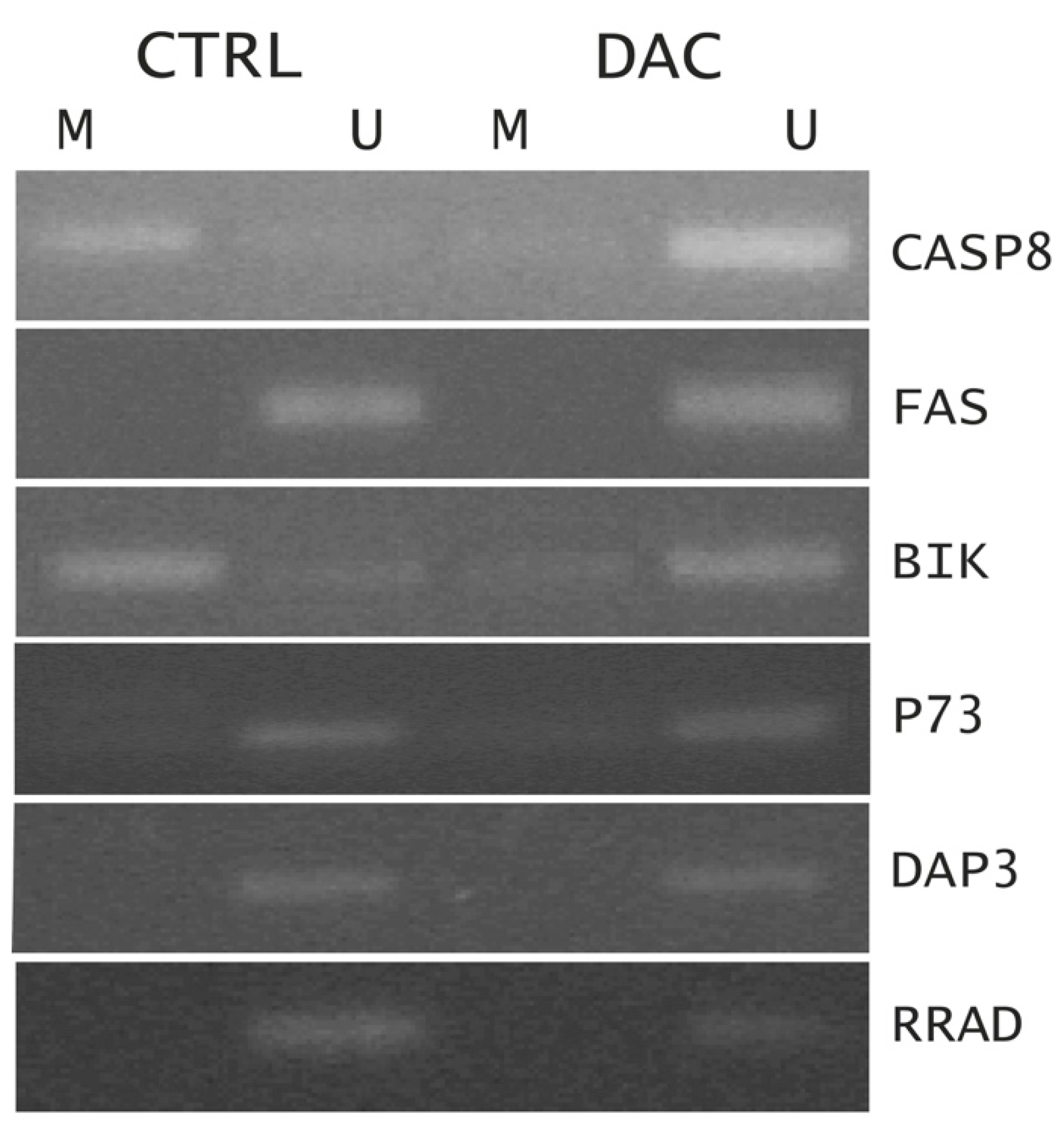
2.1.3. Methylation Status of Selected Genes after DAC on Weri-Rb-1
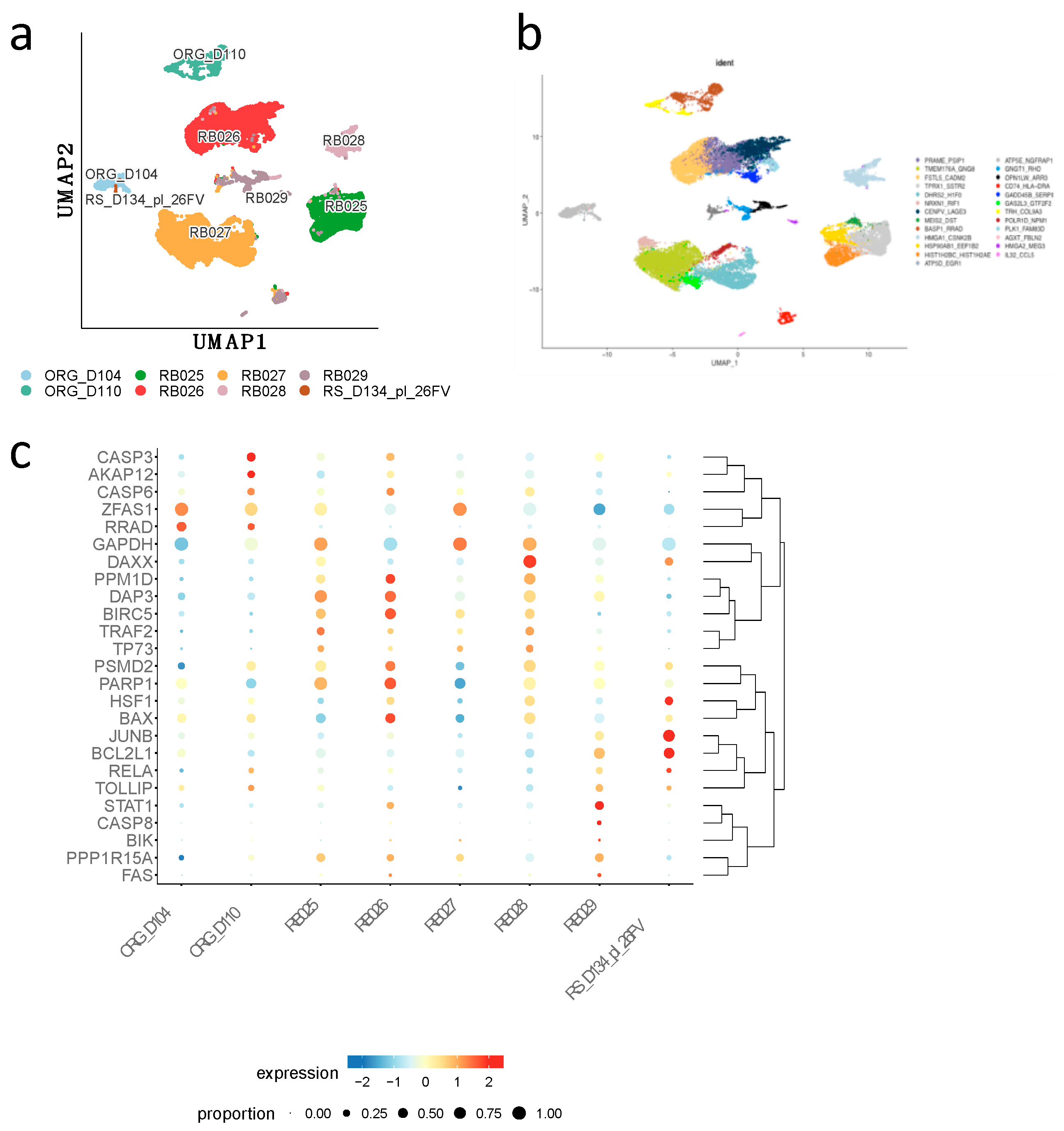
2.2. Comparison of Differential Expressed Genes in Primary Tumors vs Normal Retinal Cells with Weri-Rb-1 DEGs
2.3. DAC Anti-Cancer Effect in Preclinical Model
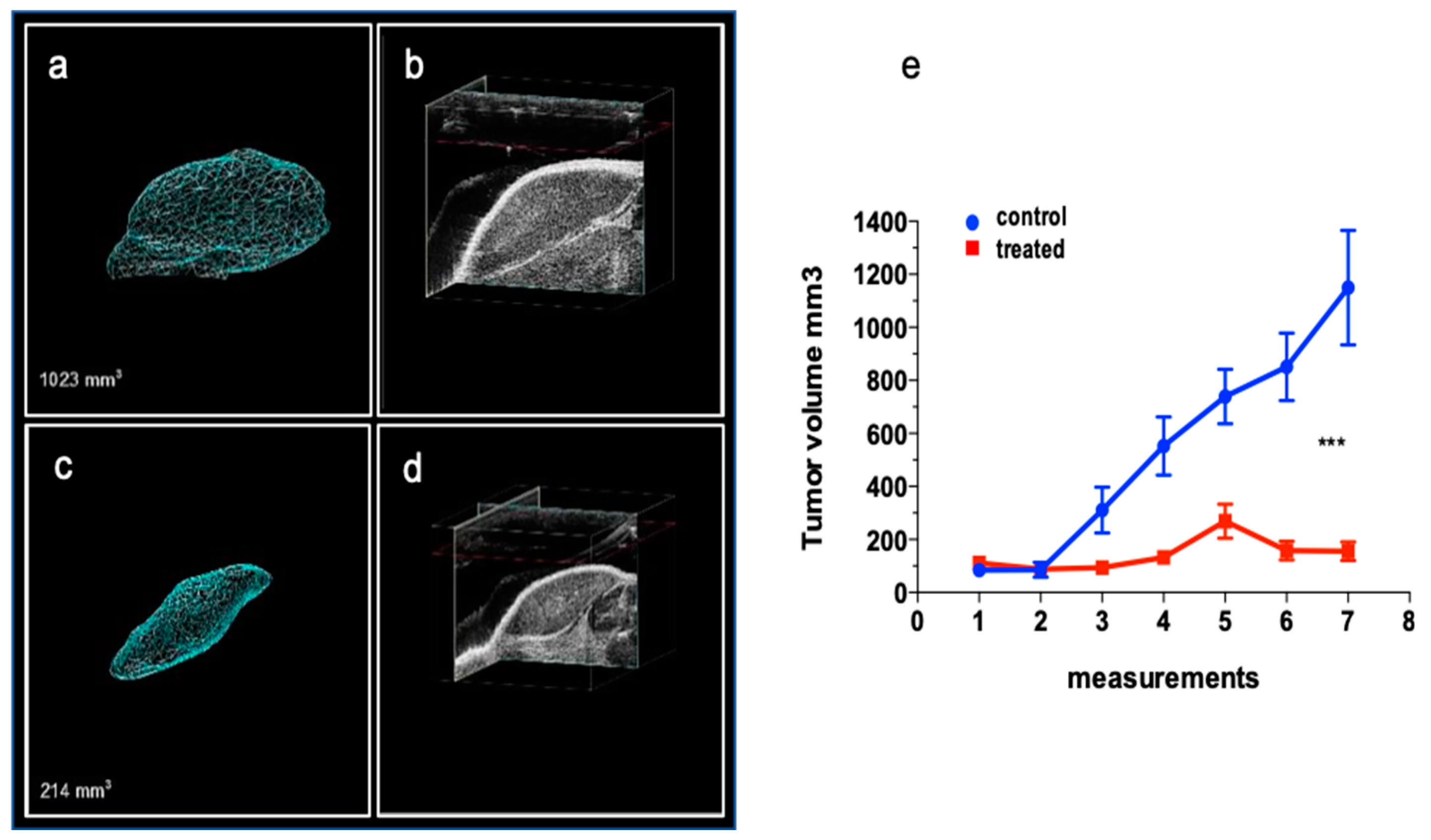
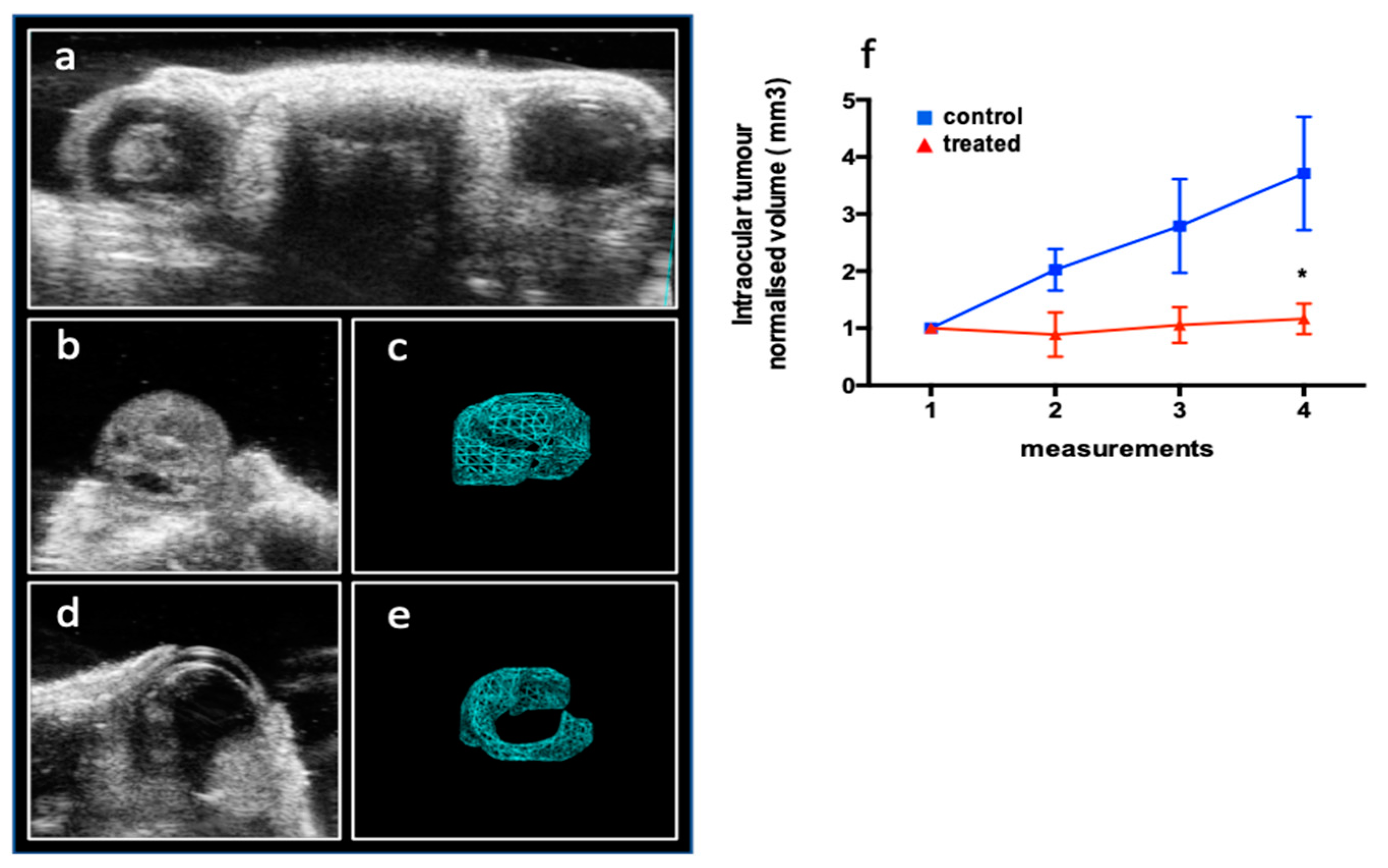
2.3.1. The Effect of DAC on the Rb Xenograft Model
2.3.2. The Effect of DAC on the Intraocular Rb Model
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- J. Wise et al., “Eye-related quality of life and activities of daily living in pediatric retinoblastoma patients: A single-center, non-controlled, cross-sectional analysis.,” Pediatr Blood Cancer, vol. 70, no. 8, pp. e30479, 2023.
- H. Dimaras et al., “Retinoblastoma.,” Nat Rev Dis Primers, vol. 1, pp. 15021, 2015.
- Singh, U.; Malik, M.A.; Goswami, S.; Shukla, S.; Kaur, J. Epigenetic regulation of human retinoblastoma. Tumor Biol. 2016, 37, 14427–14441, . [CrossRef]
- G. M. Tosi et al., “Genetic and epigenetic alterations of RB2/p130 tumor suppressor gene in human sporadic retinoblastoma: implications for pathogenesis and therapeutic approach.,” Oncogene, vol. 24, no. 38, pp. 5827-5836, 2005.
- Poeta, L.; Drongitis, D.; Verrillo, L.; Miano, M.G. DNA Hypermethylation and Unstable Repeat Diseases: A Paradigm of Transcriptional Silencing to Decipher the Basis of Pathogenic Mechanisms. Genes 2020, 11, 684, . [CrossRef]
- Costa, P.M.d.S.; Sales, S.L.A.; Pinheiro, D.P.; Pontes, L.Q.; Maranhão, S.S.; Pessoa, C.D..; Furtado, G.P.; Furtado, C.L.M. Epigenetic reprogramming in cancer: From diagnosis to treatment. Front. Cell Dev. Biol. 2023, 11, 1116805, . [CrossRef]
- U. Chianese et al., “Epigenomic machinery regulating pediatric AML: Clonal expansion mechanisms, therapies, and future perspectives.,” Semin Cancer Biol, vol. 92, pp. 84-101, 2023.
- Gherardini, L.; Sharma, A.; Capobianco, E.; Cinti, C. Targeting cancer with epi-drugs: a precision medicine perspective.. Curr. Pharm. Biotechnol. 2016, 17, 1–1, . [CrossRef]
- Katarzyna, R.; Lucyna, B. Epigenetic therapies in patients with solid tumors: Focus on monotherapy with deoxyribonucleic acid methyltransferase inhibitors and histone deacetylase inhibitors. J. Cancer Res. Ther. 2019, 15, 961–970, . [CrossRef]
- Song, Y.; Yang, G.; Ma, D.; Wang, W.; Leung, C. The role and prospect of lysine-specific demethylases in cancer chemoresistance. Med. Res. Rev. 2023, 43, 1438–1469, . [CrossRef]
- Li, F.; Xia, Q.; Ren, L.; Nie, Y.; Ren, H.; Guo, X.; Yu, J.; Xing, Y.; Chen, Z. GSDME Increases Chemotherapeutic Drug Sensitivity by Inducing Pyroptosis in Retinoblastoma Cells. Oxidative Med. Cell. Longev. 2022, 2022, 1–29, . [CrossRef]
- Salahuddin, A.; Ghanem, H.; Omran, G.A.; Helmy, M.W. Epigenetic restoration and activation of ERβ: an inspiring approach for treatment of triple-negative breast cancer. Med Oncol. 2022, 39, 1–11, . [CrossRef]
- Z. Zheng et al., “5-Aza-2′-deoxycytidine reactivates gene expression via degradation of pRb pocket proteins.,” FASEB J, vol. 26, no. 1, pp. 449-459, 2012.
- Greger, V.; Passarge, E.; Messmer, E.; Horsthemke, B.; Höpping, W. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum. Genet. 1989, 83, 155–158, . [CrossRef]
- Karmakar, A.; Khan, M.A.; Kumari, N.; Devarajan, N.; Ganesan, S.K. Identification of Epigenetically Modified Hub Genes and Altered Pathways Associated With Retinoblastoma. Front. Cell Dev. Biol. 2022, 10, 743224, . [CrossRef]
- Li, H.-T.; Xu, L.; Weisenberger, D.J.; Li, M.; Zhou, W.; Peng, C.-C.; Stachelek, K.; Cobrinik, D.; Liang, G.; Berry, J.L. Characterizing DNA methylation signatures of retinoblastoma using aqueous humor liquid biopsy. Nat. Commun. 2022, 13, 1–14, . [CrossRef]
- Livide, G.; Epistolato, M.C.; Amenduni, M.; Disciglio, V.; Marozza, A.; Mencarelli, M.A.; Toti, P.; Lazzi, S.; Hadjistilianou, T.; De Francesco, S.; et al. Epigenetic and Copy Number Variation Analysis in Retinoblastoma by MS-MLPA. Pathol. Oncol. Res. 2012, 18, 703–712, . [CrossRef]
- Malusa, F.; Taranta, M.; Zaki, N.; Cinti, C.; Capobianco, E. Time-course gene profiling and networks in demethylated retinoblastoma cell line. Oncotarget 2015, 6, 23688–23707, . [CrossRef]
- Song, J.; VanBuskirk, J.A.; Merbs, S.L. Regulation of Opsin Gene Expression by DNA Methylation and Histone Acetylation. Int. J. Mol. Sci. 2022, 23, 1408, . [CrossRef]
- Nguyen, C.T.; Weisenberger, D.J.; Velicescu, M.; A Gonzales, F.; Lin, J.C.Y.; Liang, G.; A Jones, P. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2'-deoxycytidine.. 2002, 62, 6456–61.
- Kooi, I.E.; Mol, B.M.; Moll, A.C.; van der Valk, P.; de Jong, M.C.; de Graaf, P.; van Mil, S.E.; Schouten-van Meeteren, A.Y.; Meijers-Heijboer, H.; Kaspers, G.L.; et al. Loss of photoreceptorness and gain of genomic alterations in retinoblastoma reveal tumor progression. eBioMedicine 2015, 2, 660–670, . [CrossRef]
- Milošević, D.; Medeiros, A.S.; Piperac, M.S.; Cvijanović, D.; Soininen, J.; Milosavljević, A.; Predić, B. The application of Uniform Manifold Approximation and Projection (UMAP) for unconstrained ordination and classification of biological indicators in aquatic ecology. Sci. Total. Environ. 2021, 815, 152365, . [CrossRef]
- W. Zhang et al., “Comparative Study of Subcutaneous and Orthotopic Mouse Models of Prostate Cancer: Vascular Perfusion, Vasculature Density, Hypoxic Burden and BB2r-Targeting Efficacy.,” Sci Rep, vol. 9, no. 1, pp. 11117, 2019.
- Hazazi, A.; A AlShehah, A.; Khan, F.R.; Hakami, M.A.; Almarshadi, F.; Abalkhail, A.; Nassar, S.A.; Almasoudi, H.H.; Al Ali, A.; Abu-Alghayth, M.H.; et al. From Diagnosis to Therapy: The Transformative Role of lncRNAs in Eye Cancer Management. Pathol. - Res. Pr. 2024, 254, 155081, . [CrossRef]
- Al-Ghazzawi, K.; Wessolly, M.; Dalbah, S.; Ketteler, P.; Kiefer, T.; Bechrakis, N.; Leyla, J.; Ting, S.; Biewald, E.; Mairinger, F.D. PDGF, NGF, and EGF as main contributors to tumorigenesis in high-risk retinoblastoma. Front. Oncol. 2023, 13, 1144951, . [CrossRef]
- Rathore, S.; Verma, A.; Ratna, R.; Marwa, N.; Ghiya, Y.; Honavar, S.G.; Tiwari, A.; Das, S.; Varshney, A. Retinoblastoma: A review of the molecular basis of tumor development and its clinical correlation in shaping future targeted treatment strategies. Indian J. Ophthalmol. 2023, 71, 2662–2676, . [CrossRef]
- Zhang, X.; Jiang, Y.; Cai, Y.; Fu, Q.; Chen, Y. Epigenetics research in eye diseases: a bibliometric analysis from 2000 to 2023. Clin. Exp. Optom. 2023, 1–8, . [CrossRef]
- M. Kulis and M. Esteller, “DNA methylation and cancer.,” Adv Genet, vol. 70, pp. 27-56, 2010.
- Feinberg, A.P.; Koldobskiy, M.A.; Göndör, A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 2016, 17, 284–299, . [CrossRef]
- Jiang, Y.; Zheng, G.; Sun, X. PRMT5 promotes retinoblastoma development. Hum. Cell 2022, 36, 1–13, . [CrossRef]
- M. L. de Groote et al., “Epigenetic Editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes.,” Nucleic Acids Res, vol. 40, no. 21, pp. 10596-10613, 2012.
- M. Berdasco et al., “DNA Methylomes Reveal Biological Networks Involved in Human Eye Development, Functions and Associated Disorders.,” Sci Rep, vol. 7, no. 1, pp. 11762, 2017.
- Choi, Y.K.; Kim, J.H.; Kim, W.J.; Lee, H.Y.; Park, J.A.; Lee, S.-W.; Yoon, D.-K.; Kim, H.H.; Chung, H.; Yu, Y.S.; et al. AKAP12 Regulates Human Blood–Retinal Barrier Formation by Downregulation of Hypoxia-Inducible Factor-1α. J. Neurosci. 2007, 27, 4472–4481, . [CrossRef]
- E. Jabbour et al., “Evolution of decitabine development: accomplishments, ongoing investigations, and future strategies.,” Cancer, vol. 112, no. 11, pp. 2341-2351, 2008.
- Yang, D.; Torres, C.M.; Bardhan, K.; Zimmerman, M.; McGaha, T.L.; Liu, K. Decitabine and Vorinostat Cooperate To Sensitize Colon Carcinoma Cells to Fas Ligand-Induced Apoptosis In Vitro and Tumor Suppression In Vivo. J. Immunol. 2012, 188, 4441–4449, . [CrossRef]
- Field, M.G.; Kuznetsoff, J.N.; Zhang, M.G.; Dollar, J.J.; Durante, M.A.; Sayegh, Y.; Decatur, C.L.; Kurtenbach, S.; Pelaez, D.; Harbour, J.W. RB1 loss triggers dependence on ESRRG in retinoblastoma. Sci. Adv. 2022, 8, eabm8466, . [CrossRef]
- Kolesnikov, A.V.; Luu, J.; Jin, H.; Palczewski, K.; Kefalov, V.J. Deletion of Protein Phosphatase 2A Accelerates Retinal Degeneration in GRK1- and Arr1-Deficient Mice. Investig. Opthalmology Vis. Sci. 2022, 63, 18–18, . [CrossRef]
- A. Sridhar et al., “Single-Cell Transcriptomic Comparison of Human Fetal Retina, hPSC-Derived Retinal Organoids, and Long-Term Retinal Cultures.,” Cell Rep, vol. 30, no. 5, pp. 1644-1659.e4, 2020.
- Almet, A.A.; Cang, Z.; Jin, S.; Nie, Q. The landscape of cell–cell communication through single-cell transcriptomics. Curr. Opin. Syst. Biol. 2021, 26, 12–23, . [CrossRef]
- Armingol, E.; Officer, A.; Harismendy, O.; Lewis, N.E. Deciphering cell–cell interactions and communication from gene expression. Nat. Rev. Genet. 2020, 22, 71–88, . [CrossRef]
- Arnol, D.; Schapiro, D.; Bodenmiller, B.; Saez-Rodriguez, J.; Stegle, O. Modeling Cell-Cell Interactions from Spatial Molecular Data with Spatial Variance Component Analysis., . [CrossRef]
- G. Dennis et al., “DAVID: Database for Annotation, Visualization, and Integrated Discovery.,” Genome Biol, vol. 4, no. 5, pp. P3, 2003.
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 2005, 21, 3448–3449, . [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., II; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196, doi:10.1126/science.aad0501.
- Davis, S.; Meltzer, P.S. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847, doi:10.1093/bioinformatics/btm254.
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, Article17–Article17, . [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559, . [CrossRef]
- N. Percie du Sert et al., “Reporting animal research: Explanation and elaboration for the ARRIVE guidelines 2.0.,” PLoS Biol, vol. 18, no. 7, pp. e3000411, 2020.

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





