Submitted:
22 April 2024
Posted:
23 April 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Case Study: LPAI or HPAI in Apparently Healthy Infected Penguins?
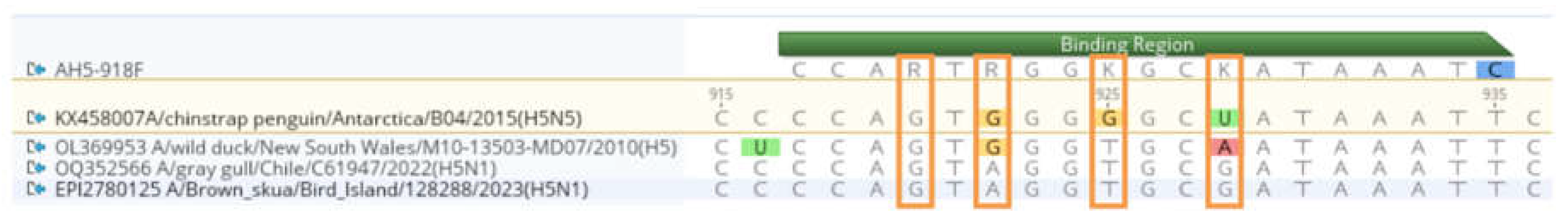

Considerations for the Detection of HPAI H5 Viruses
Conclusions
- Use validated and accredited methods to confirm the presence of the virus (whether in a field or reference laboratory),
- Provide an absolute definition of the presence of either HPAI or LPAI strains wherever detection of these pathogens are reported
- Submit all results to the World Organisation for Animal Health and the SCAR HPAI monitoring database to assist in the global monitoring and surveillance of the virus in the region and around the globe.
- Undertake full genome sequencing of any positive samples to enable genomic surveillance to assist with understanding of virus movement into and within the region and to identify any potential mutations. Submit genome sequences to a publicly-available database (e.g. GenBank) in a reasonable timeline.
| Reference | Target | Assay type | Confirmation required? | Select examples of use |
|---|---|---|---|---|
| Spackman et al (2002) [29] | 99bp region of the M Highly conserved to detect all avian influenza viruses |
rRT-PCR | Yes, H5 diagnostic and/or sequencing | 2001 citations. Detection of clade 2.3.4.4b in farmed mink in Spain [38] and wild birds in North America [39] |
| Nagy et al. (2021)[30] | 149bp region of the M Highly conserved to detect all influenza A viruses |
rRT-PCR | Yes, H5 diagnostic and/or sequencing | Detection of clade 2.3.4.4b in Gannets in UK [13] , in carnivores in Finland [40]. Is integrated into combination HA, NA, M test by Slomka et al (2023)[33] |
| Hassan et al. (2022)[41] which is updated from Hoffmann et al. (2016)[42] | All HA and NA subtypes, M. Designed to detect all avian influenza viruses |
Multiplexed rRT-PCR | Yes, sequencing to reveal H5 lineage. | Detection of clade 2.3.4.4b in Sandwich Terns in Germany [43] and in Grey Seals in Europe [44] |
| Slomka et al. (2007)[45], updated from Spackman et al (2002) [29] | 229 bp of HA segment in the HA2 region Designed to detect all H5 viruses |
rRT-PCR | Yes, sequencing to reveal H5 lineage. | Detection of clade 2.3.4.4b in Sandwich Terns in the Netherlands [46] . Integrated into combination HA, NA, M test by Slomka et al (2023)[33] |
| Slomka et al (2012)[47] | 191bp of HA segment across HA cleavage site. |
rRT-PCR | No, but best practice. | Detection of clade 2.3.4.4b in mammals in a rehabilitation centre [48] . Integrated into combination HA, NA, M test by Slomka et al (2023)[33] |
| James et al. (2022)[49] modified from an unpublished protocol based on Naguib et al. (2017)[50] | 109bp region of HA cleavage site. Designed to be specific to 2020/21 clade 2.3.4.4b H5Nx viruses Confirmed not to detect LPAI |
rRT-PCR | No, but best practice. | Detection of clade 2.3.4.4b in birds in South Georgia Island and the Falkland (Malvinas) Islands[1], Gannets in the UK[13], ducks in Botswana [51] |
| Naguib et al. (2017)[50] | 109-161bp of HA segment in the HA1 region Designed to discriminate gs/GD clades 2.2.1.2, 2.3.2.1 and 2.3.4.4 and LPAI |
Multiplexed rRT-PCR | No, but best practice. | Detection of clade 2.3.4.4b in Swedish wild birds and poultry [52], and a novel 2.3.4.4 H5N8 reassortant in Germany [53] |
| Fereidouni et al. (2009)[54] | 126-250bp of NA segment | End point RT-PCR | Yes | Detection of clade 2.3.4.4b in carnivores in Finland [40], emergence 2.3.4.4b in wild birds in South Korea[55] |
| James et al. (2018)[34] updated from Hoffman (2016) [42] | ~150bp of NA segment | rRT-PCR | Yes | Detection of clade 2.3.4.4b in ducks in Botswana [51], in birds in South Georgia Island and the Falkland (Malvinas) Islands[1]. Integrated into combination HA, NA, M test by Slomka et al (2023)[33] |
References
- Bennison, A.; Byrne, A.M.; Reid, S.M.; Lynton-Jenkins, J.G.; Mollett, B.; Sliva, D.D.; Peers-Dent, J.; Finlayson, K.E.; Hall, R.; Blockley, F.; et al. Detection and spread of high pathogenicity avian influenza virus H5N1 in the Antarctic Region. bioRxiv 2023. [Google Scholar]
- Dewar, M.; Wille, M.; Gamble, A.; Vanstreels, R.; Boulinier, T.; Smith, A.; Varsani, A.; Ratcliffe, N.; Black, J.; Lynnes, A. The risk of avian influenza in the Southern Ocean: A practical guide for operators interacting with wildlife. Antarctic Science 2023. [Google Scholar] [CrossRef]
- Bennet, B. Confirmation of highly pathogenic avian influenza (HPAI) H5N1 associated with an unexpected mortality event in South Polar Skuas (Stercorarius maccormicki) during 2023-2024 surveillance activities in Antarctica. bioRxiv, 2024. [Google Scholar]
- León, F.; Bohec, C.L.; Pizarro, E.J.; Baille, L.; Cristofari, R.; Houstin, A.; Zitterbart, D.P.; Barriga, G.; Poulin, E.; Vianna, J.A. Highly Pathogenic Avian Influenza A (H5N1) Suspected in penguins and shags on the Antarctic Peninsula and West Antarctic Coast. bioRxiv 2024. [Google Scholar]
- Lisovski, S. No evidence for highly pathogenic avian influenza H5N1 (clade 2.3.4.4b) in the Antarctic region due the austral summer 2022/23. bioRxiv, 2023. [Google Scholar]
- The Guardian. ‘Cautious optimism’ as penguins test positive for bird flu but show no symptoms. (2024). Available online: https://www.theguardian.com/environment/2024/mar/2026/bird-flu-asymptomatic-penguins-adelie-penguins-antarctic.
- INACH. Chile detecta casos positivos de gripe aviar en pingüinos, cormoranes y skuas en la Antártica. (2024). Available online: https://www.inach.cl/chile-detecta-casos-positivos-de-gripe-aviar-en-pinguinos-cormoranes-y-skuas-en-la-antartica/.
- Reuters. Antarctic scientists warn of bird flu spread as penguin cases confirmed. (2024). Available online: https://www.reuters.com/business/healthcare-pharmaceuticals/antarctic-scientists-warn-bird-flu-spread-penguin-cases-confirmed-2024-2003-2014/.
- Lv, X.; et al. Highly pathogenic avian influenza A(H5N8) clade 2.3.4.4b viruses in satellite-tracked wild ducks, Ningxia, China, 2020. Emerg. Infect. Dis. 2022, 28, 1039–1042. [Google Scholar] [CrossRef]
- Teitelbaum, C.S.; et al. North American wintering mallards infected with highly pathogenic avian influenza show few signs of altered local or migratory movements. Sci Rep-Uk 2023, 13, 14473. [Google Scholar] [CrossRef]
- Spackman, E.; Pantin-Jackwood, M.J.; Lee, S.A.; Prosser, D. The pathogenesis of a 2022 North American highly pathogenic clade 2.3.4.4b H5N1 avian influenza virus in mallards (Anas platyrhynchos). Avian Pathology 2023, 52, 219–228. [Google Scholar] [CrossRef]
- Banyard, A.C. Continued expansion of high pathogenicity avian influenza H5 in wildlife in South America and incursions in to the Antarctic Region. OFFLU statement, 2023. Available online: https://www.offlu.org/wp-content/uploads/2023/2012/OFFLU-wildlife-statement-no.-II.pdf.
- Lane, J.V.; Jeglinski, J.W.; Avery-Gomm, S.; Ballstaedt, E.; Banyard, A.C.; Barychka, T.; Brown, I.H.; Brugger, B.; Burt, T.V.; Careen, N.; et al. High pathogenicity avian influenza (H5N1) in Northern Gannets: Global spread, clinical signs, and demographic consequences. Ibis 2023. [Google Scholar] [CrossRef]
- Knief, U. Highly pathogenic avian influenza causes mass mortality in Sandwich tern (Thalasseus sandvicensis) breeding colonies across northwestern Europe. bioRxiv, 2023. Available online: https://www.biorxiv.org/content/10.1101/2023.1105.1112.540367v540361.
- Molini, U.; et al. Avian influenza H5N8 outbreak in African Penguins (Spheniscus demersus), Namibia, 2019. Journal of Wildlfe Diseases 2020, 56, 214–218. [Google Scholar] [CrossRef]
- Roberts, L.C. Descriptive Epidemiology of and Response to the High Pathogenicity Avian Influenza (H5N8) Epidemic in South African Coastal Seabirds, 2018. Transbound Emerg Dis, 2708458; https://doi.org/2708410.2701155/2702023/2708458. [Google Scholar]
- Abolnik, C. The Molecular Epidemiology of Clade 2.3.4.4B H5N1 High Pathogenicity Avian Influenza in Southern Africa, 2021–2022. Viruses, 2023; 15, 1383, https://doi.org/1310.3390/v15061383. [Google Scholar]
- Muñoz, G. Stranding and Mass Mortality in Humboldt Penguins (Spheniscus humboldti), Associated to HPAIV H5N1 Outbreak in Chile. Preventative Veterinary Medicine, 2024. [Google Scholar] [CrossRef]
- Roberts, L.C. Vaccination of African penguins (Spheniscus demersus) against high-pathogenicity avian influenza. VetRecord, 2023. [Google Scholar] [CrossRef]
- Molini, U.; et al. Highly pathogenic avian influenza H5N1 virus outbreak among Cape cormorants (Phalacrocorax capensis) in Namibia, 2022. Emerging Microbes & Infections 2023, 12, 2167610. [Google Scholar]
- Barriga, G.P.; et al. Avian influenza virus H5 strain with North American and Eurasian Genes in an Antarctic Penguin. Emerg. Infect. Dis. 2016, 22, 2221–2223. [Google Scholar] [CrossRef]
- Hurt, A.C.; Su, Y.C.F.; Aban, M.; Peck, H.; Lau, H.; Baas, C.; Deng, Y.-M.; Spirason, N.; Ellström, P.; Hernandez, J.; et al. Evidence for the Introduction, Reassortment, and Persistence of Diverse Influenza A Viruses in Antarctica. J. Virol. 2016, 90, 9674–9682. [Google Scholar] [CrossRef]
- Kuiken, T. Is low pathogenic avian influenza virus virulent for wild waterbirds? Proceedings. Biological sciences 2013, 280, 20130990, doi: 20130910.20131098/rspb.20132013.20130990. [Google Scholar]
- Bengtsson, D.; Avril, A.; Gunnarsson, G.; Elmberg, J.; Söderquist, P.; Norevik, G.; Tolf, C.; Safi, K.; Fiedler, W.; Wikelski, M.; et al. Movements, Home-Range Size and Habitat Selection of Mallards during Autumn Migration. PLOS ONE 2014, 9, e100764. [Google Scholar] [CrossRef]
- van Toor, M.L.; Avril, A.; Wu, G.; Holan, S.H.; Waldenström, J. As the Duck Flies—Estimating the Dispersal of Low-Pathogenic Avian Influenza Viruses by Migrating Mallards. Front. Ecol. Evol. 2018, 6. [Google Scholar] [CrossRef]
- van Dijk, J. et al. Weak negative associations between avian influenza virus infection and movement behaviour in a key host species, the mallard Anas platyrhynchos. Oikos 10, 1293–1303.
- van Gils, J.A.; Munster, V.J.; Radersma, R.; Liefhebber, D.; Fouchier, R.A.; Klaassen, M. Hampered Foraging and Migratory Performance in Swans Infected with Low-Pathogenic Avian Influenza A Virus. PLOS ONE 2007, 2, e184. [Google Scholar] [CrossRef]
- World Organisation for Animal Health. Chapter 3.3.4. AVIAN INFLUENZA (INCLUDING INFECTION WITH HIGH PATHOGENICITY AVIAN INFLUENZA VIRUSES) WOAH Terrestrial Manual, 2025, p 28 pages.
- Spackman, E.; Senne, D.A.; Myers, T.J.; Bulaga, L.L.; Garber, L.P.; Perdue, M.L.; Lohman, K.; Daum, L.T.; Suarez, D.L. Development of a Real-Time Reverse Transcriptase PCR Assay for Type A Influenza Virus and the Avian H5 and H7 Hemagglutinin Subtypes. J. Clin. Microbiol. 2002, 40, 3256–3260. [Google Scholar] [CrossRef]
- Nagy, A. et al. A universal RT-qPCR assay for "One Health" detection of influenza A viruses. Plos One 16 (2021).
- Laconi, A.; et al. Detection of avian influenza virus: a comparative study of the in silico and in vitro performances of current RT-qPCR assays. Sci Rep-Uk, 2020; 10, 8441. [Google Scholar]
- Xie, R.; Edwards, K.M.; Wille, M.; Wei, X.; Wong, S.-S.; Zanin, M.; El-Shesheny, R.; Ducatez, M.; Poon, L.L.M.; Kayali, G.; et al. The episodic resurgence of highly pathogenic avian influenza H5 virus. Nature 2023, 622, 810–817. [Google Scholar] [CrossRef]
- Slomka, M.J.; Reid, S.M.; Byrne, A.M.P.; Coward, V.J.; Seekings, J.; Cooper, J.L.; Peers-Dent, J.; Agyeman-Dua, E.; de Silva, D.; Hansen, R.D.E.; et al. Efficient and Informative Laboratory Testing for Rapid Confirmation of H5N1 (Clade 2.3.4.4) High-Pathogenicity Avian Influenza Outbreaks in the United Kingdom. Viruses 2023, 15, 1344. [Google Scholar] [CrossRef]
- James, J.; Slomka, M.J.; Reid, S.M.; Thomas, S.S.; Mahmood, S.; Byrne, A.M.P.; Cooper, J.; Russell, C.; Mollett, B.C.; Agyeman-Dua, E.; et al. Development and Application of Real-Time PCR Assays for Specific Detection of Contemporary Avian Influenza Virus Subtypes N5, N6, N7, N8, and N9. Avian Dis. 2018, 63, 209–218. [Google Scholar] [CrossRef]
- Ulloa, M.; et al. Mass mortality event in South American sea lions correlated to highly pathogenic avian influenza (HPAI) H5N1 outbreak in Chile. 2023; 43, 8–18. [Google Scholar]
- Ariyama, N.; et al. Emergence and rapid dissemination of highly pathogenic avian influenza virus H5N1 clade 2.3.4.4b in wild birds, Chile. bioRxiv (2023).
- World Organisation for Animal Health. Wildlife under threat as avian influenza reaches Antarctica. 2024. Available online: https://www.woah.org/en/wildlife-under-threat-as-avian-influenza-reaches-antarctica/.
- Agüero, M.; Monne, I.; Sánchez, A.; Zecchin, B.; Fusaro, A.; Ruano, M.J.; Arrojo, M.d.V.; Fernández-Antonio, R.; Souto, A.M.; Tordable, P.; et al. Highly pathogenic avian influenza A(H5N1) virus infection in farmed minks, Spain, October 2022. Eurosurveillance 2023, 28, 2300001–10. [Google Scholar] [CrossRef]
- Alkie, T.N.; Lopes, S.; Hisanaga, T.; Xu, W.; Suderman, M.; Koziuk, J.; Fisher, M.; Redford, T.; Lung, O.; Joseph, T.; et al. A threat from both sides: Multiple introductions of genetically distinct H5 HPAI viruses into Canada via both East Asia-Australasia/Pacific and Atlantic flyways. Virus Evol. 2022, 8, veac077. [Google Scholar] [CrossRef] [PubMed]
- Tammiranta, N.; Isomursu, M.; Fusaro, A.; Nylund, M.; Nokireki, T.; Giussani, E.; Zecchin, B.; Terregino, C.; Gadd, T. Highly pathogenic avian influenza A (H5N1) virus infections in wild carnivores connected to mass mortalities of pheasants in Finland. Infect. Genet. Evol. 2023, 111, 105423. [Google Scholar] [CrossRef] [PubMed]
- Hassan, K.E.; Ahrens, A.K.; Ali, A.; El-Kady, M.F.; Hafez, H.M.; Mettenleiter, T.C.; Beer, M.; Harder, T. Improved Subtyping of Avian Influenza Viruses Using an RT-qPCR-Based Low Density Array: ‘Riems Influenza a Typing Array’, Version 2 (RITA-2). Viruses 2022, 14, 415. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, B.; Hoffmann, D.; Henritzi, D.; Beer, M.; Harder, T.C. Riems influenza a typing array (RITA): An RT-qPCR-based low density array for subtyping avian and mammalian influenza a viruses. Sci Rep-Uk, 2016; 6, 27211. [Google Scholar]
- Pohlmann, A.; Stejskal, O.; King, J.; Bouwhuis, S.; Packmor, F.; Ballstaedt, E.; Hälterlein, B.; Hennig, V.; Stacker, L.; Graaf, A.; et al. Mass mortality among colony-breeding seabirds in the German Wadden Sea in 2022 due to distinct genotypes of HPAIV H5N1 clade 2.3.4.4b. J. Gen. Virol. 2023, 104, 001834. [Google Scholar] [CrossRef] [PubMed]
- Mirolo, M.; Pohlmann, A.; Ahrens, A.K.; Kühl, B.; Rubio-Garcìa, A.; Kramer, K.; Meinfelder, U.; Rosenberger, T.; Morito, H.L.; Beer, M.; et al. Highly pathogenic avian influenza A virus (HPAIV) H5N1 infection in two European grey seals ( Halichoerus grypus ) with encephalitis. Emerg. Microbes Infect. 2023, 12, e2257810. [Google Scholar] [CrossRef] [PubMed]
- Slomka, M.J.; Pavlidis, T.; Banks, J.; Shell, W.; McNally, A.; Essen, S.; Brown, I.H. Validated H5 Eurasian Real-Time Reverse Transcriptase–Polymerase Chain Reaction and Its Application in H5N1 Outbreaks in 2005–2006. Avian Dis. 2007, 51, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Rijks, J.M.; Leopold, M.F.; Kühn, S.; Veld, R.I.; Schenk, F.; Brenninkmeijer, A.; Lilipaly, S.J.; Ballmann, M.Z.; Kelder, L.; de Jong, J.W.; et al. Mass Mortality Caused by Highly Pathogenic Influenza A(H5N1) Virus in Sandwich Terns, the Netherlands, 2022. Emerg. Infect. Dis. 2022, 28, 2538–2542. [Google Scholar] [CrossRef]
- Slomka, M.J.; To, T.L.; Tong, H.H.; Coward, V.J.; Hanna, A.; Shell, W.; Pavlidis, T.; Densham, A.L.E.; Kargiolakis, G.; Arnold, M.E.; et al. Challenges for accurate and prompt molecular diagnosis of clades of highly pathogenic avian influenza H5N1 viruses emerging in Vietnam. Avian Pathol. 2012, 41, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Floyd, T.; Banyard, A.C.; Lean, F.Z.; Byrne, A.M.; Fullick, E.; Whittard, E.; Mollett, B.C.; Bexton, S.; Swinson, V.; Macrelli, M.; et al. Encephalitis and Death in Wild Mammals at a Rehabilitation Center after Infection with Highly Pathogenic Avian Influenza A(H5N8) Virus, United Kingdom. Emerg. Infect. Dis. 2021, 27, 2856–2863. [Google Scholar] [CrossRef]
- James, J.; Seekings, A.H.; Skinner, P.; Purchase, K.; Mahmood, S.; Brown, I.H.; Hansen, R.D.; Banyard, A.C.; Reid, S.M. Rapid and sensitive detection of high pathogenicity Eurasian clade 2.3.4.4b avian influenza viruses in wild birds and poultry. J. Virol. Methods 2022, 301, 114454. [Google Scholar] [CrossRef]
- Naguib, M.M.; Graaf, A.; Fortin, A.; Luttermann, C.; Wernery, U.; Amarin, N.; A Hussein, H.; Sultan, H.; Al Adhadh, B.; Hassan, M.K.; et al. Novel real-time PCR-based patho- and phylotyping of potentially zoonotic avian influenza A subtype H5 viruses at risk of incursion into Europe in 2017. Eurosurveillance 2017, 22, 15–26. [Google Scholar] [CrossRef]
- Letsholo, S.L.; James, J.; Meyer, S.M.; Byrne, A.M.P.; Reid, S.M.; Settypalli, T.B.K.; Datta, S.; Oarabile, L.; Kemolatlhe, O.; Pebe, K.T.; et al. Emergence of High Pathogenicity Avian Influenza Virus H5N1 Clade 2.3.4.4b in Wild Birds and Poultry in Botswana. Viruses 2022, 14, 2601. [Google Scholar] [CrossRef]
- Grant, M. et al. Highly Pathogenic Avian Influenza (HPAI H5Nx, Clade 2.3.4.4.b) in Poultry and Wild Birds in Sweden: Synopsis of the 2020-2021 Season. Veterinary Sciences 9 (2022).
- King, J.; Schulze, C.; Engelhardt, A.; Hlinak, A.; Lennermann, S.-L.; Rigbers, K.; Skuballa, J.; Staubach, C.; Mettenleiter, T.C.; Harder, T.; et al. Novel HPAIV H5N8 Reassortant (Clade 2.3.4.4b) Detected in Germany. Viruses 2020, 12, 281. [Google Scholar] [CrossRef]
- Fereidouni, S.; Starick, E.; Grund, C.; Globig, A.; Mettenleiter, T.; Beer, M.; Harder, T. Rapid molecular subtyping by reverse transcription polymerase chain reaction of the neuraminidase gene of avian influenza A viruses. Veter- Microbiol. 2009, 135, 253–260. [Google Scholar] [CrossRef]
- Sagong, M.; Lee, Y.; Song, S.; Cha, R.M.; Lee, E.; Kang, Y.; Cho, H.; Kang, H.; Lee, Y.; Lee, K. Emergence of clade 2.3.4.4b novel reassortant H5N1 high pathogenicity avian influenza virus in South Korea during late 2021. Transbound. Emerg. Dis. 2022, 69, E3255–E3260. [Google Scholar] [CrossRef]
| Consideration | Reason | What to do |
|---|---|---|
| Assay is sensitive | Assay can detect influenza A viruses from samples adequately. | > Select an assay that is well validated > Select an assay that is frequently assessed against new strains containing mutations within primer and probe binding regions > Known well-characterised positive controls (should fall into known Ct value range) |
| Assay is specific to target (clade 2.3.4.4b HPAI H5N1) | Both LPAI H5 and HPAI H5 co-circulate, so imperative to distinguish as risk, response, notification pathway differs depending on the result. | > Select an assay that is well validated > Use both LPAI H5 and HPAI H5 controls > Sequencing of either the HA PCR products, or whole genome sequencing |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





