Submitted:
17 April 2024
Posted:
18 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
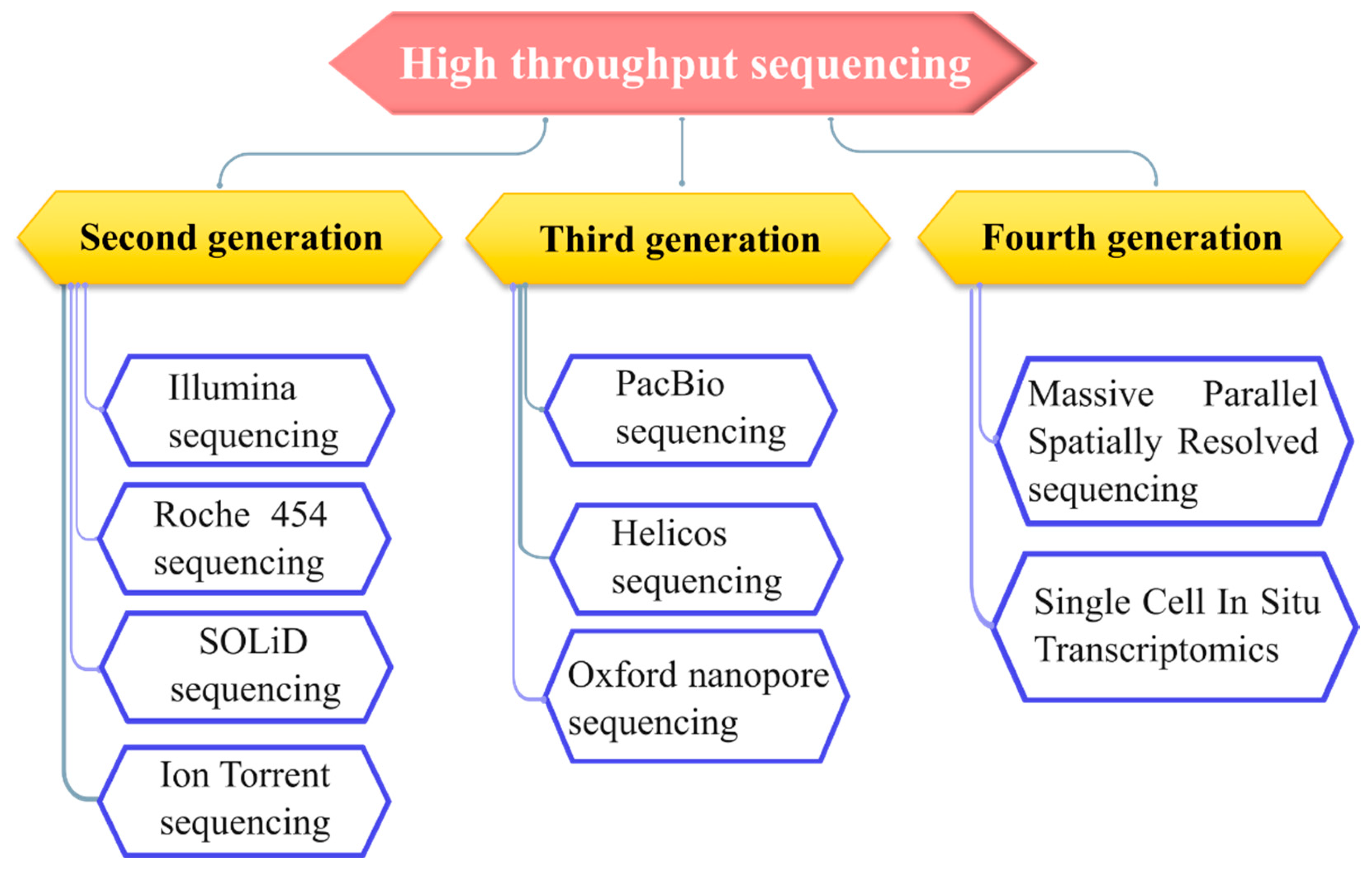
2. High Throughput Sequencing
2.1. Second Generation Sequencing
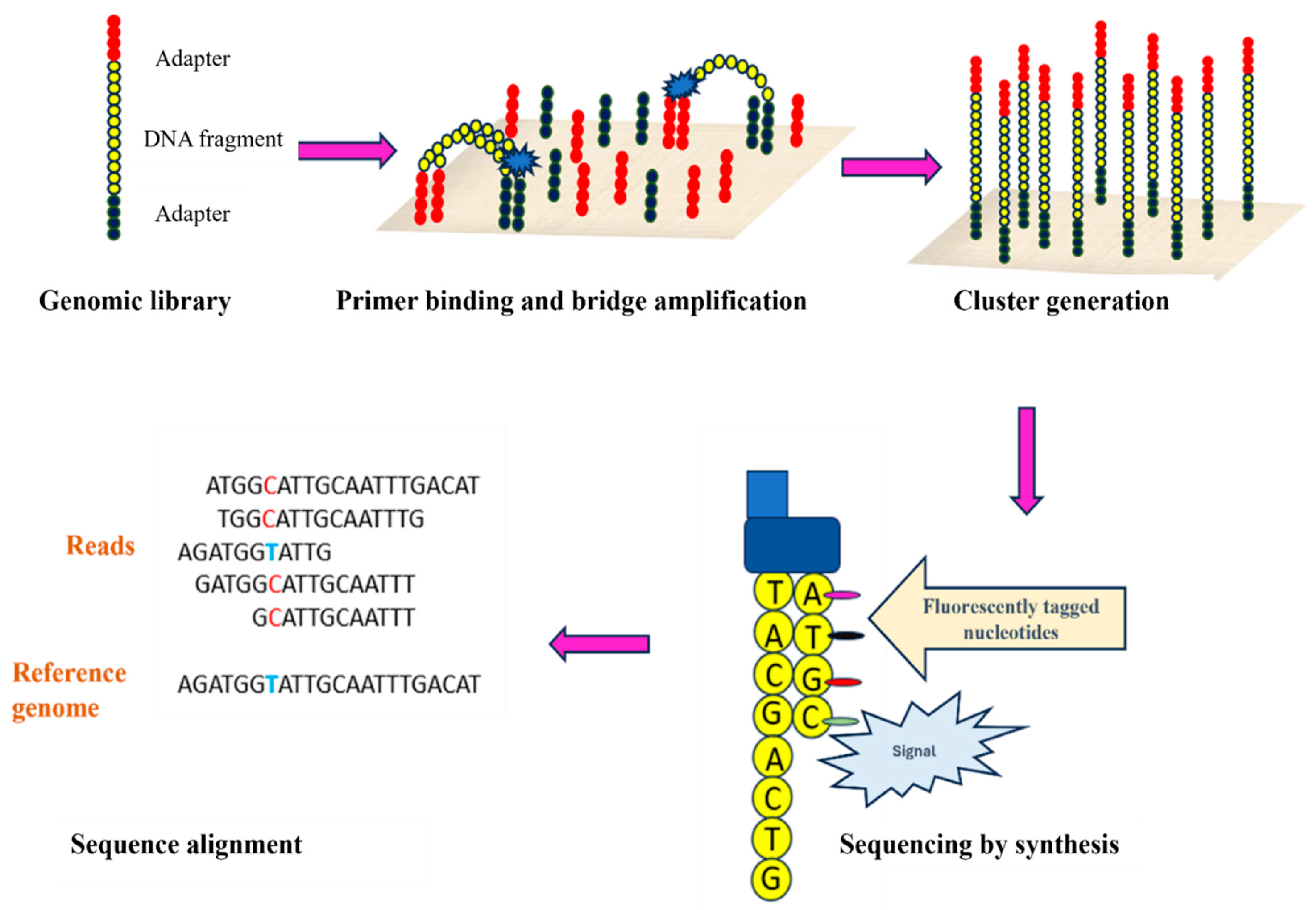
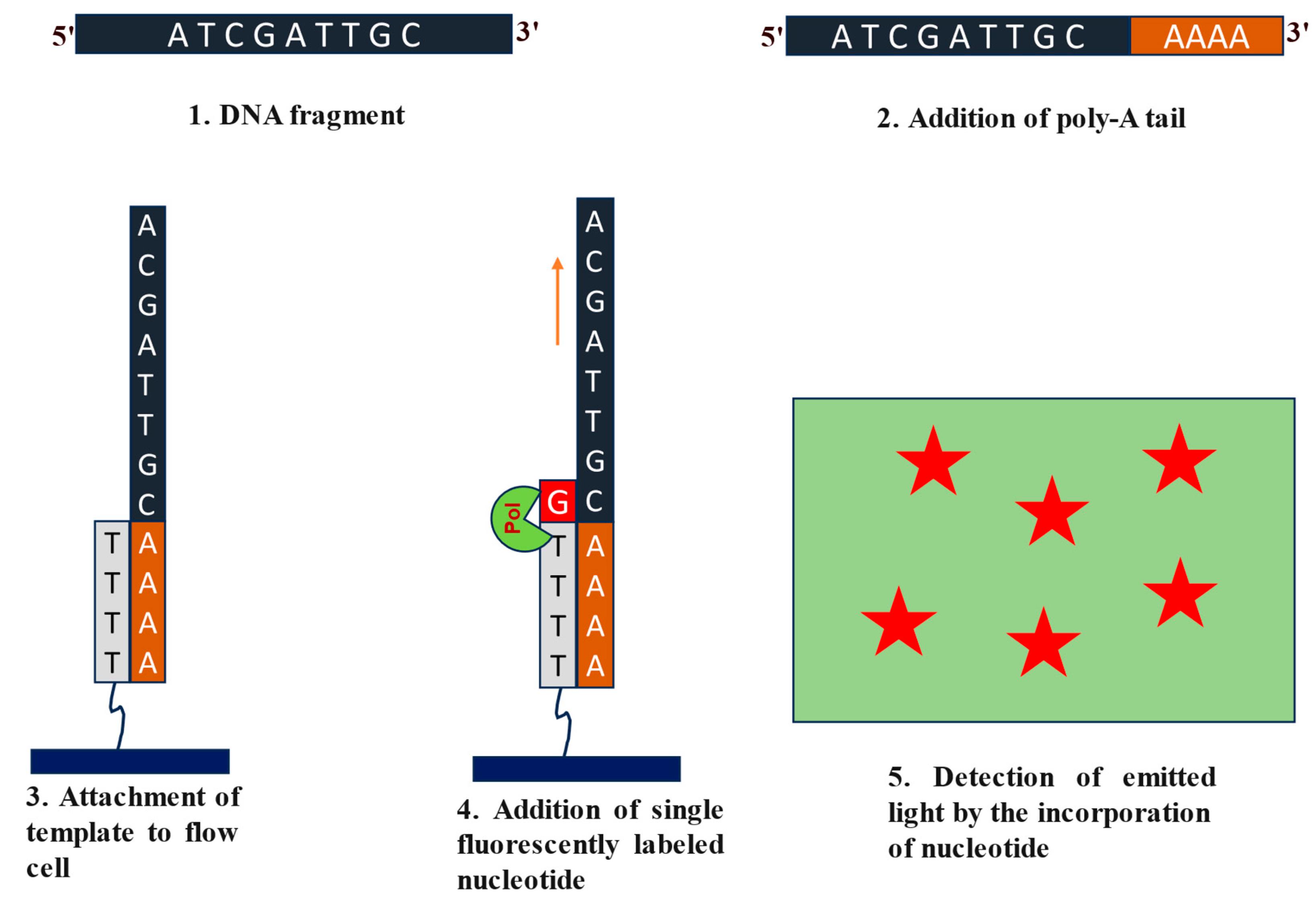
2.1.1. Illumina Sequencing
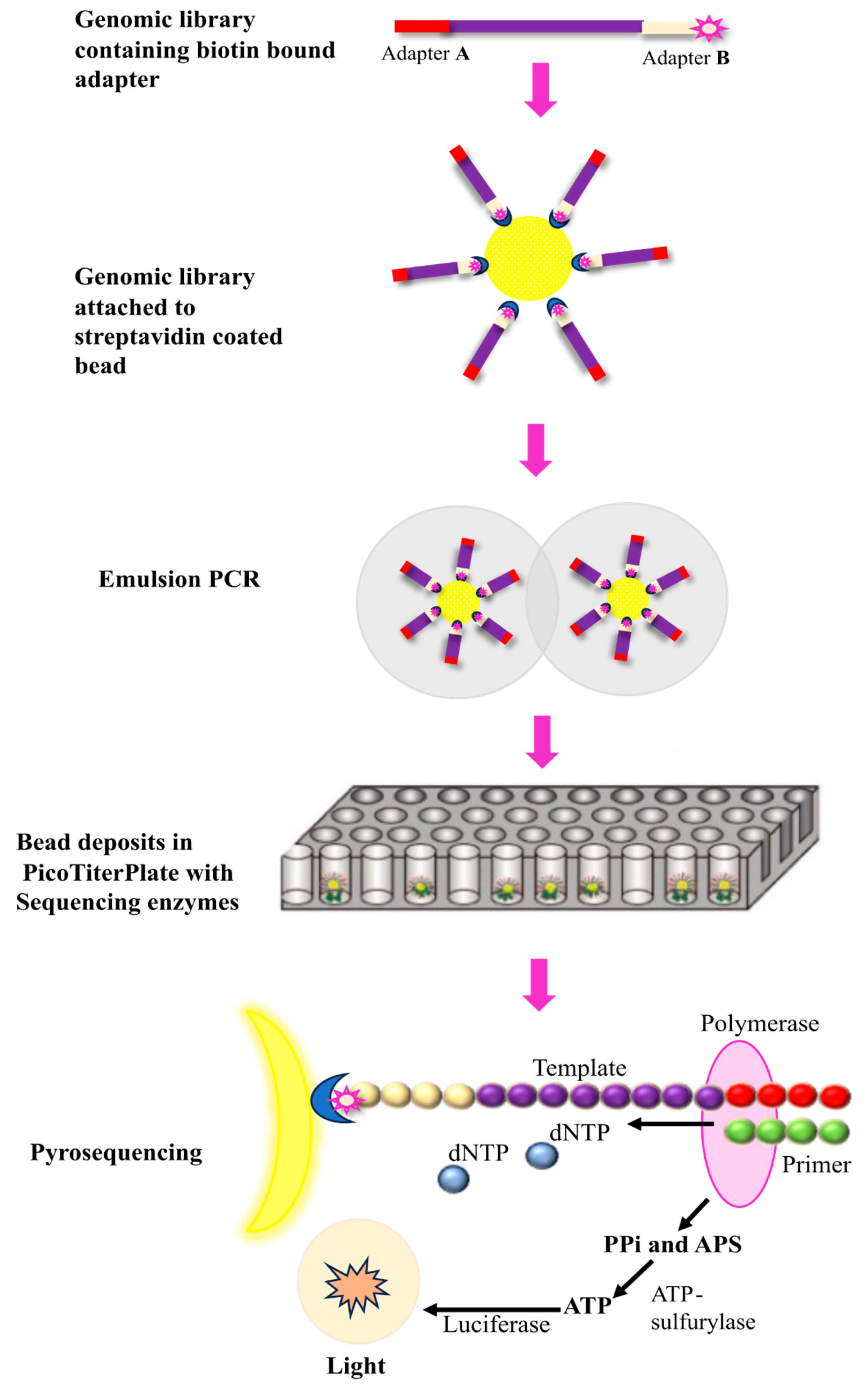
2.1.2. Roche 454 Sequencing
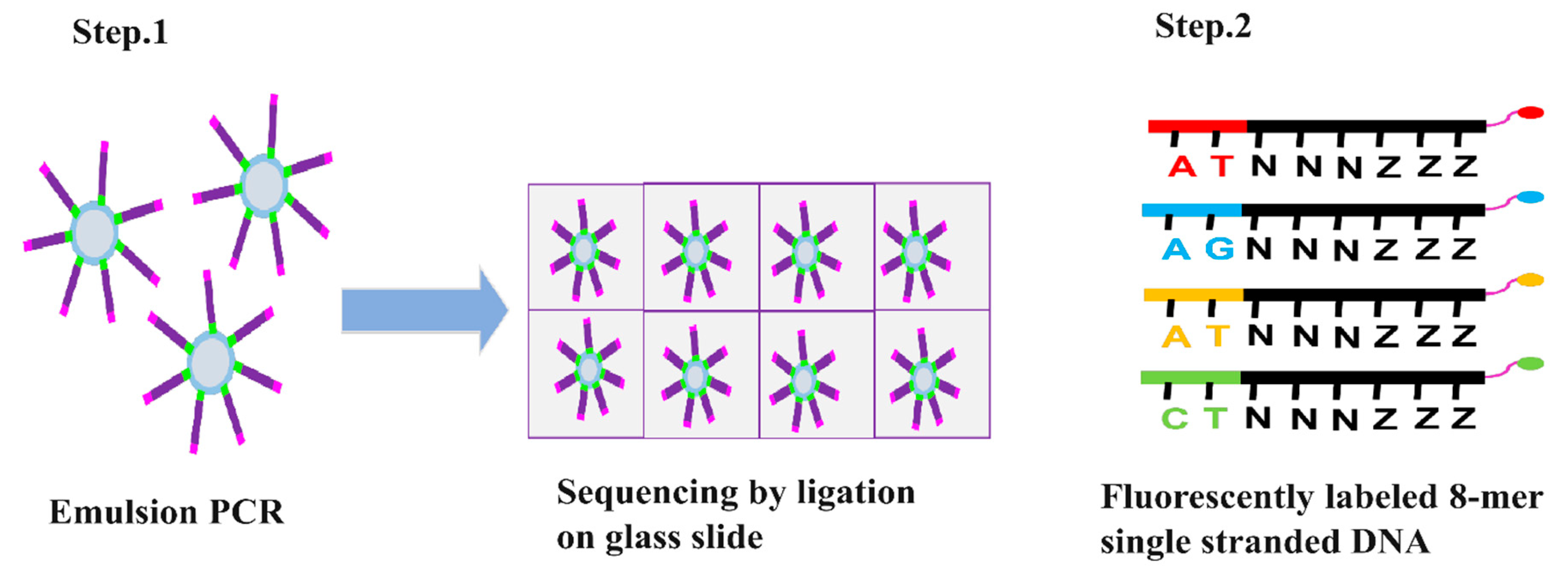
2.1.3. SOLiD DNA Sequencing
2.1.4. Ion Torrent Sequencing
2.2. Third Generation Sequencing
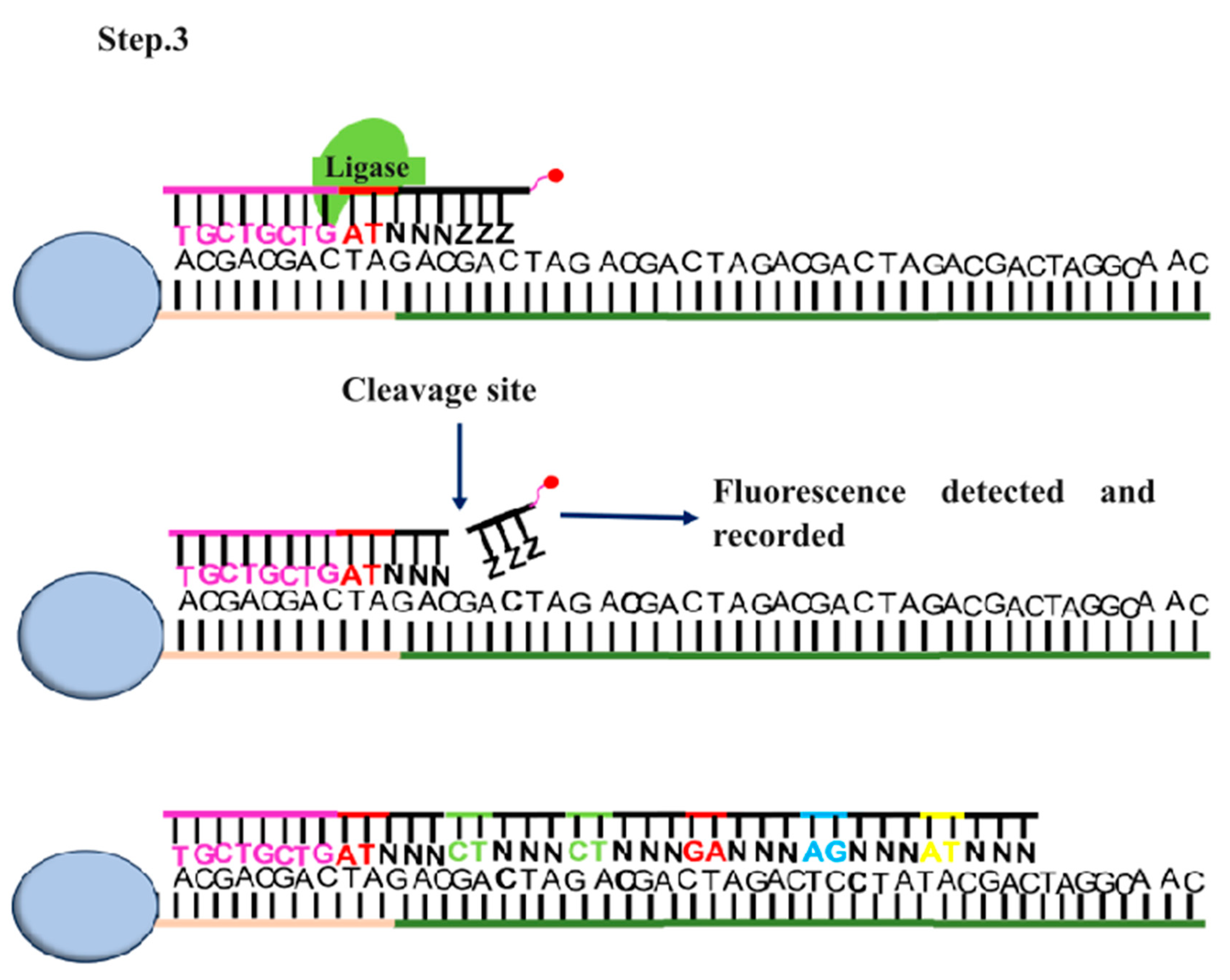
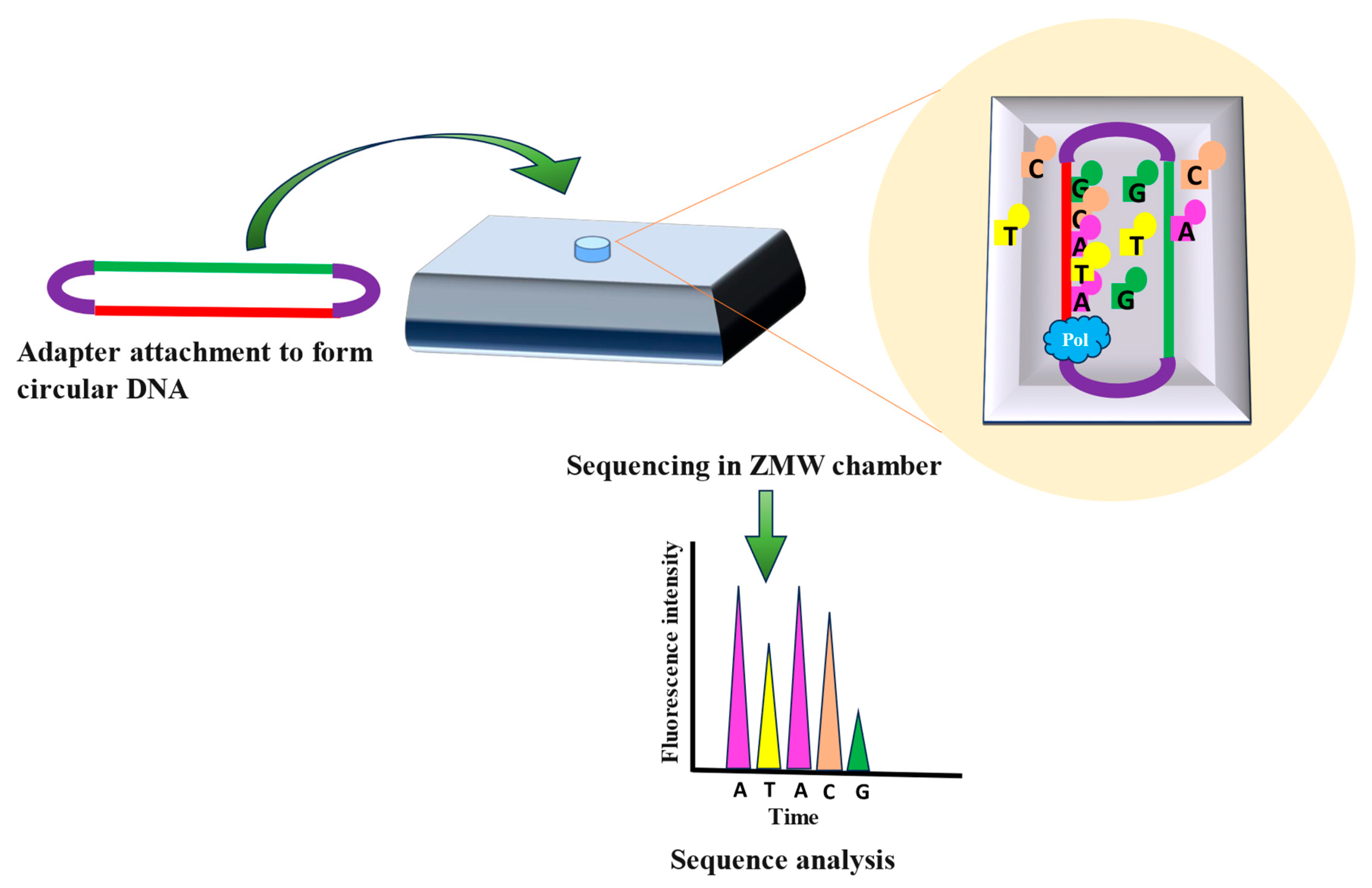
2.2.1. PacBio Sequencing
2.2.2. Helicos Sequencing
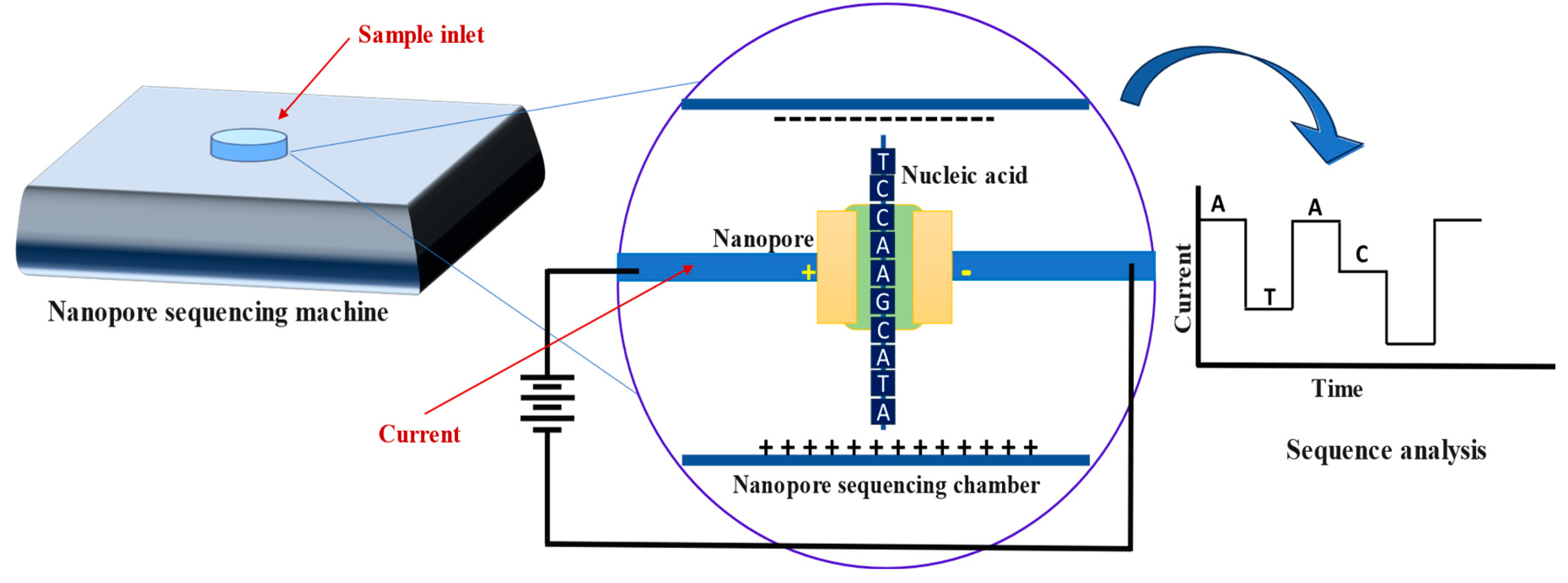
2.2.3. Nanopore-Based Sequencing
2.3. Fourth Generation Sequencing
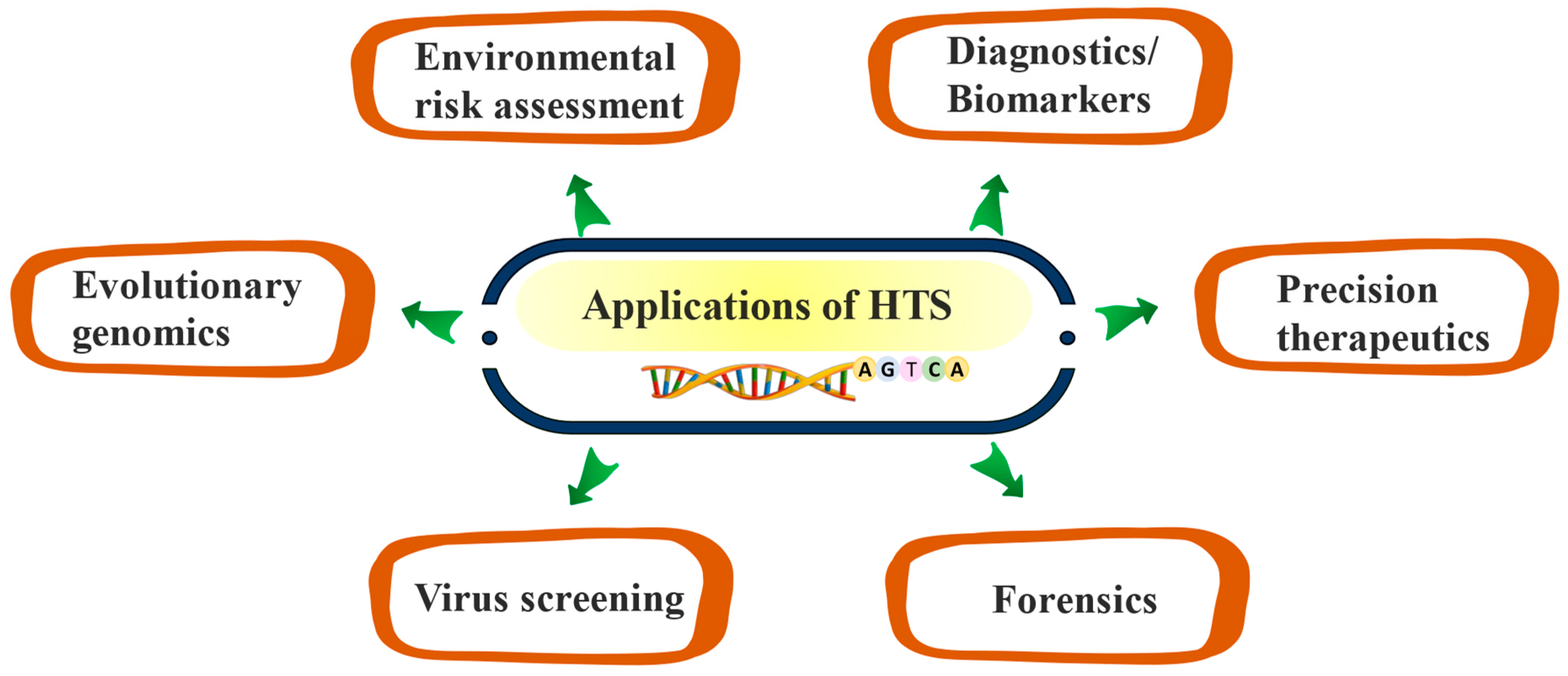
3. Applications
4. Conclusions
5. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgements
Conflicts of Interest
References
- Aganezov, S.; et al. A complete reference genome improves analysis of human genetic variation. Science 2022, 376, eabl3533. [Google Scholar] [CrossRef]
- Shendure, J.; et al. DNA sequencing at 40: past, present and future. Nature 2017, 550, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Johnston, H.R.; Keats, B.J.; Sherman, S.L. Population genetics, in Emery and Rimoin’s Principles and Practice of Medical Genetics and Genomics; Elsevier, 2019; pp. 359–373. [Google Scholar]
- Scacheri, C.A.; Scacheri, P.C. Mutations in the non-coding genome. Current opinion in pediatrics 2015, 27, 659. [Google Scholar] [CrossRef]
- Loewe, L.; Hill, W.G. The population genetics of mutations: good, bad and indifferent; The Royal Society, 2010; pp. 1153–1167. [Google Scholar]
- Hershberg, R. Mutation—the engine of evolution: studying mutation and its role in the evolution of bacteria. Cold Spring Harbor perspectives in biology 2015, 7, a018077. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.; et al. The genetic basis of disease. Essays in biochemistry 2018, 62, 643–723. [Google Scholar] [CrossRef]
- Bossuyt, P.M.; et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. 2003.
- Ehrlich, G.D.; Hiller, N.L.; Hu, F.Z. What makes pathogens pathogenic. Genome biology 2008, 9, 1–7. [Google Scholar] [CrossRef]
- Goetz, L.H.; Schork, N.J. Personalized medicine: motivation, challenges, and progress. Fertility and sterility 2018, 109, 952–963. [Google Scholar] [CrossRef] [PubMed]
- Heather, J.M.; Chain, B. The sequence of sequencers: The history of sequencing DNA. Genomics 2016, 107, 1–8. [Google Scholar] [CrossRef]
- Mooney, S.D. Progress towards the integration of pharmacogenomics in practice. Human genetics 2015, 134, 459–465. [Google Scholar] [CrossRef]
- Perrier, L.; et al. Cost of genome analysis: the sanger sequencing method. Value in Health 2015, 18, A353. [Google Scholar] [CrossRef]
- Normand, R.; Yanai, I. An introduction to high-throughput sequencing experiments: design and bioinformatics analysis. Deep Sequencing Data Analysis 2013, 1–26. [Google Scholar]
- Genomics, C. Sanger sequencing: Introduction, principle, and protocol. 2020, CD Genomics Blog. Recuperado el.
- Del Vecchio, F.; et al. Next-generation sequencing: recent applications to the analysis of colorectal cancer. J Transl Med 2017, 15, 246. [Google Scholar] [CrossRef] [PubMed]
- Coleman, S.J.; et al. Structural annotation of equine protein-coding genes determined by mRNA sequencing. Anim Genet 2010, 41 (Suppl 2), 121–130. [Google Scholar] [CrossRef] [PubMed]
- Donkor, E.S. Sequencing of bacterial genomes: principles and insights into pathogenesis and development of antibiotics. Genes 2013, 4, 556–572. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.; Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc 2010, 2010, pdb-prot5448. [Google Scholar] [CrossRef] [PubMed]
- Bronner, I.F. Improved Protocols for Illumina Sequencing. Current Protocols in Human Genetics. Current Protocols in Human Genetics 2014, 79. [Google Scholar]
- Glenn, T.C.; et al. Adapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext). PeerJ 2019, 7, e7755. [Google Scholar]
- Sinha, R.; et al. Index switching causes “spreading-of-signal” among multiplexed samples in Illumina HiSeq 4000 DNA sequencing. BioRxiv 2017, 125724. [Google Scholar]
- Yang, X.; et al. HTQC: a fast quality control toolkit for Illumina sequencing data. BMC bioinformatics 2013, 14, 1–4. [Google Scholar] [CrossRef]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next-Generation Sequencing Technologies. Curr Protoc Mol Biol 2018, 122, e59. [Google Scholar] [CrossRef]
- Liu, L.; et al. Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012, 2012, 251364. [Google Scholar] [CrossRef]
- Klein, H.-U.; et al. R453Plus1Toolbox: an R/Bioconductor package for analyzing Roche 454 Sequencing data. Bioinformatics 2011, 27, 1162–1163. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; et al. ART: a next-generation sequencing read simulator. Bioinformatics 2012, 28, 593–594. [Google Scholar] [CrossRef]
- Luo, C.; et al. Direct comparisons of Illumina vs. Roche 454 sequencing technologies on the same microbial community DNA sample. PLoS One 2012, 7, e30087. [Google Scholar]
- Hedges, D.J.; et al. Comparison of three targeted enrichment strategies on the SOLiD sequencing platform. PloS one 2011, 6, e18595. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Verma, V.K. Next-Generation Sequencing and Its Application: Empowering in Public Health Beyond Reality, in Microbial Technology for the Welfare of Society. 2019. p. 313-341.
- Ari, Ş.; Arikan, M. Next-Generation Sequencing: Advantages, Disadvantages, and Future, in Plant Omics: Trends and Applications. 2016. p. 109-135.
- Shendure, J.; et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 2005, 309, 1728–1732. [Google Scholar] [CrossRef]
- Garrido-Cardenas, J.A.; et al. DNA Sequencing Sensors: An Overview. Sensors (Basel) 2017, 17. [Google Scholar] [CrossRef]
- Pickrell, W.O.; Rees, M.I.; Chung, S.-K. Next generation sequencing methodologies-an overview. Advances in protein chemistry and structural biology 2012, 89, 1–26. [Google Scholar] [PubMed]
- Malapelle, U.; et al. Ion Torrent next-generation sequencing for routine identification of clinically relevant mutations in colorectal cancer patients. Journal of clinical pathology 2015, 68, 64–68. [Google Scholar] [CrossRef]
- Merriman, B.; R&, *!!! REPLACE !!!*; D Team, I.T.; Rothberg, J.M. T.; Rothberg, J.M. Progress in ion torrent semiconductor chip based sequencing. Electrophoresis 2012, 33, 3397–3417. [Google Scholar] [CrossRef]
- Lahens, N.F.; et al. A comparison of Illumina and Ion Torrent sequencing platforms in the context of differential gene expression. BMC Genomics 2017, 18, 602. [Google Scholar] [CrossRef] [PubMed]
- Malapelle, U.; et al. Ion Torrent next-generation sequencing for routine identification of clinically relevant mutations in colorectal cancer patients. J Clin Pathol 2015, 68, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Davies, K. It’s “Watson Meets Moore” as Ion Torrent Introduces Semiconductor Sequencing. Bio-IT World 2010. [Google Scholar]
- Bleidorn, C. Third generation sequencing: technology and its potential impact on evolutionary biodiversity research. Systematics and biodiversity 2016, 14, 1–8. [Google Scholar] [CrossRef]
- Zhang, W.; Jia, B.; Wei, C. PaSS: a sequencing simulator for PacBio sequencing. BMC Bioinformatics 2019, 20, 352. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; et al. Advantages of genome sequencing by long-read sequencer using SMRT technology in medical area. Hum Cell 2017, 30, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genomics Proteomics Bioinformatics 2015, 13, 278–289. [Google Scholar] [CrossRef]
- Wang, Y.; et al. Direct Pacbio sequencing methods and applications for different types of DNA sequences. bioRxiv 2023. [Google Scholar] [CrossRef]
- Coupland, P.; et al. Direct sequencing of small genomes on the Pacific Biosciences RS without library preparation. Biotechniques 2012, 53, 365–372. [Google Scholar] [CrossRef]
- Milos, P.M. Helicos single molecule sequencing: unique capabilities and importance for molecular diagnostics. Genome Biology 2010, 11 (Suppl 1), I14. [Google Scholar] [CrossRef]
- Thompson, J.F.; Raz, T.; Milos, P.M. Helicos Single-Molecule Sequencing for Accurate Tag-Based RNA Quantitation. Tag-Based Next Generation Sequencing 2011, 353–365. [Google Scholar]
- Thompson, J.F.; Steinmann, K.E. Single molecule sequencing with a HeliScope genetic analysis system. Curr Protoc Mol Biol 2010, 92, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.F.; Milos, P.M. The properties and applications of single-molecule DNA sequencing. Genome biology 2011, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.K. Single-molecule DNA sequencing technologies for future genomics research. Trends in biotechnology 2008, 26, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Nanopore, O. Oxford Nanopore announcement sets sequencing sector abuzz. Nature biotechnology 2012, 30, 295. [Google Scholar]
- Gunter, H.M.; et al. Library adaptors with integrated reference controls improve the accuracy and reliability of nanopore sequencing. Nature Communications 2022, 13, 6437. [Google Scholar] [CrossRef]
- Dumschott, K.; et al. Oxford Nanopore sequencing: new opportunities for plant genomics? J Exp Bot 2020, 71, 5313–5322. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION Sequencing and Genome Assembly . Genomics Proteomics Bioinformatics 2016, 14, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol 2019, 20, 129. [Google Scholar] [CrossRef]
- Jain, M.; et al. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol 2016, 17, 239. [Google Scholar]
- Kumar, A.; Sharma, V.K.; Kumar, P. Nanopore sequencing: The fourthgeneration sequencing. J Entom Zool Stud 2019, 7, 1400–1403. [Google Scholar]
- Feng, Y.; et al. Nanopore-based fourth-generation DNA sequencing technology. Genomics, proteomics & bioinformatics 2015, 13, 4–16. [Google Scholar]
- Gaur, R.; et al. Sequencing Technologies: Introduction and Applications. Int. J. Hum. Genet 2019, 19, 123–133. [Google Scholar]
- Lin, Y.; et al. Application of nanopore adaptive sequencing in pathogen detection of a patient with Chlamydia psittaci infection. Frontiers in Cellular and Infection Microbiology 2023, 13, 1064317. [Google Scholar] [CrossRef] [PubMed]
- Ke, R.; et al. Fourth generation of next-generation sequencing technologies: Promise and consequences. Human mutation 2016, 37, 1363–1367. [Google Scholar] [CrossRef] [PubMed]
- Strell, C.; et al. Placing RNA in context and space–methods for spatially resolved transcriptomics. The FEBS journal 2019, 286, 1468–1481. [Google Scholar] [CrossRef] [PubMed]
- Alon, S.; et al. Expansion sequencing: Spatially precise in situ transcriptomics in intact biological systems. Science 2021, 371, eaax2656. [Google Scholar] [CrossRef]
- Mignardi, M.; Nilsson, M. Fourth-generation sequencing in the cell and the clinic. Genome medicine 2014, 6, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Ratan, A.; et al. Identification of indels in next-generation sequencing data. BMC bioinformatics 2015, 16, 1–8. [Google Scholar] [CrossRef]
- Fischer, A.; Hacein-Bey-Abina, S. Gene therapy for severe combined immunodeficiencies and beyond. J Exp Med 2020, 217. [Google Scholar] [CrossRef]
- Strand, J.; et al. Second-tier next generation sequencing integrated in nationwide newborn screening provides rapid molecular diagnostics of severe combined immunodeficiency. Frontiers in immunology 2020, 11, 545364. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Hacein-Bey-Abina, S. Gene therapy for severe combined immunodeficiencies and beyond. Journal of Experimental Medicine 2019, 217, e20190607. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.M.; et al. Clinical Implementation of Routine Whole-genome Sequencing for Hospital Infection Control of Multi-drug Resistant Pathogens. Clin Infect Dis 2023, 76, e1277–e1284. [Google Scholar] [CrossRef]
- Márquez, S.; et al. Genome sequencing of the first SARS-CoV-2 reported from patients with COVID-19 in Ecuador. MedRxiv 2020. [Google Scholar]
- Quer, J.; et al. Next-generation sequencing for confronting virus pandemics. Viruses 2022, 14, 600. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.J.; Daetwyler, H.D. 1000 bull genomes project to map simple and complex genetic traits in cattle: applications and outcomes. Annual review of animal biosciences 2019, 7, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Weldenegodguad, M.; et al. Whole-genome sequencing of three native cattle breeds originating from the northernmost cattle farming regions. Frontiers in genetics 2019, 9, 728. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, G.L.; et al. Pathogen genomics in public health. New England Journal of Medicine 2019, 381, 2569–2580. [Google Scholar] [CrossRef] [PubMed]
- Sintchenko, V.; Holmes, E.C. The role of pathogen genomics in assessing disease transmission. Bmj 2015, 350. [Google Scholar] [CrossRef]
- Lopez-Labrador, F.X.; et al. Recommendations for the introduction of metagenomic high-throughput sequencing in clinical virology, part I: Wet lab procedure. J Clin Virol 2021, 134, 104691. [Google Scholar] [CrossRef]
- Bik, H.M.; et al. Metagenetic community analysis of microbial eukaryotes illuminates biogeographic patterns in deep-sea and shallow water sediments. Molecular Ecology 2012, 21, 1048–1059. [Google Scholar] [CrossRef]
- Diaz-Lara, A.; et al. High-Throughput Sequencing of Grapevine in Mexico Reveals a High Incidence of Viruses including a New Member of the Genus Enamovirus. Viruses 2023, 15, 1561. [Google Scholar] [CrossRef]
- Massart, S.; et al. Virus Detection by High-Throughput Sequencing of Small RNAs: Large-Scale Performance Testing of Sequence Analysis Strategies. Phytopathology 2019, 109, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Semenova, E.M.; et al. Crude Oil Degradation in Temperatures Below the Freezing Point by Bacteria from Hydrocarbon-Contaminated Arctic Soils and the Genome Analysis of Sphingomonas sp. AR_OL41. Microorganisms 2023, 12, 79. [Google Scholar] [CrossRef]
- Waldrop, M.P.; et al. Permafrost microbial communities and functional genes are structured by latitudinal and soil geochemical gradients. The ISME Journal 2023, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, M.; et al. Bioremediation by oil degrading marine bacteria: An overview of supplements and pathways in key processes. Chemosphere 2022, 303, 134956. [Google Scholar] [CrossRef]
- Goel, N.; Khandnor, P. TCGA: A multi-genomics material repository for cancer research. Materials Today: Proceedings 2020, 28, 1492–1495. [Google Scholar]
- Bailey, M.H.; et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018, 173, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. Review The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemporary Oncology/Współczesna Onkologia 2015, 2015, 68–77. [Google Scholar] [CrossRef]
- Sengupta, D.; et al. Population stratification and underrepresentation of Indian subcontinent genetic diversity in the 1000 genomes project dataset. Genome biology and evolution 2016, 8, 3460–3470. [Google Scholar] [CrossRef]
- Dugger, S.A.; Platt, A.; Goldstein, D.B. Drug development in the era of precision medicine. Nature reviews Drug discovery 2018, 17, 183–196. [Google Scholar] [CrossRef]
- Pereira, M.A.; et al. Application of Next-Generation Sequencing in the Era of Precision Medicine, in Applications of RNA-Seq and Omics Strategies - From Microorganisms to Human Health. 2017.
- Naithani, N.; et al. Precision medicine: Uses and challenges. Med J Armed Forces India 2021, 77, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Matrosova, V.Y.; et al. High-quality genome sequence of the radioresistant bacterium Deinococcus ficus KS 0460. Stand Genomic Sci 2017, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; et al. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev 2001, 65, 44–79. [Google Scholar] [CrossRef]
- Liu, F.; Li, N.; Zhang, Y. The radioresistant and survival mechanisms of Deinococcus radiodurans. Radiation Medicine and Protection 2023, 4, 70–79. [Google Scholar] [CrossRef]
- Lindsay, J.A.; Holden, M.T. Staphylococcus aureus: superbug, super genome? Trends in microbiology 2004, 12, 378–385. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiology and molecular biology reviews 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Shelenkov, A. Whole-Genome Sequencing of Pathogenic Bacteria-New Insights into Antibiotic Resistance Spreading. Microorganisms 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Erjavec, S.O.; et al. Whole exome sequencing in Alopecia Areata identifies rare variants in KRT82. Nat Commun 2022, 13, 800. [Google Scholar] [CrossRef]
- Wang, Z.; et al. Use of whole-exome sequencing to identify novel monogenic gene mutations and genotype–phenotype correlations in Chinese Han children with urolithiasis. Frontiers in Genetics 2023, 14, 1128884. [Google Scholar] [CrossRef]
- Rajagopal, A.; et al. Exome sequencing identifies a novel homozygous mutation in the phosphate transporter SLC34A1 in hypophosphatemia and nephrocalcinosis. The Journal of Clinical Endocrinology & Metabolism 2014, 99, E2451–E2456. [Google Scholar]
- Smola, M.J.; et al. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nature protocols 2015, 10, 1643–1669. [Google Scholar] [CrossRef]
- Spitale, R.C.; et al. RNA SHAPE analysis in living cells. Nature chemical biology 2013, 9, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; et al. In vivo architecture of the telomerase RNA catalytic core in Trypanosoma brucei. Nucleic Acids Research 2021, 49, 12445–12466. [Google Scholar] [CrossRef] [PubMed]
- Schlick, T.; et al. To knot or not to knot: multiple conformations of the SARS-CoV-2 frameshifting RNA element. Journal of the American Chemical Society 2021, 143, 11404–11422. [Google Scholar] [CrossRef]
- Dey, A.; et al. Abolished frameshifting for predicted structure-stabilizing SARS-CoV-2 mutants: Implications to alternative conformations and their statistical structural analyses. bioRxiv 2024. [Google Scholar]
- Boerneke, M.A.; et al. Structure-first identification of RNA elements that regulate dengue virus genome architecture and replication. Proceedings of the National Academy of Sciences 2023, 120, e2217053120. [Google Scholar] [CrossRef] [PubMed]
- Madden, E.A. Using SHAPE-MaP to model RNA secondary structure and identify 3′ UTR variation in chikungunya virus. Journal of virology 2020, 94, e00701-20. [Google Scholar] [CrossRef]
- Alvarez, D.R.; et al. The RNA structurome in the asexual blood stages of malaria pathogen plasmodium falciparum. RNA biology 2021, 18, 2480–2497. [Google Scholar] [CrossRef]
- Coria, A.; et al. Rotavirus RNA chaperone mediates global transcriptome-wide increase in RNA backbone flexibility. Nucleic Acids Research 2022, 50, 10078–10092. [Google Scholar] [CrossRef]
- Dey, A. , Structural Modifications and Novel Protein-Binding Sites in Pre-miR-675—Explaining Its Regulatory Mechanism in Carcinogenesis. Non-coding RNA 2023, 9, 45. [Google Scholar] [CrossRef]
- Monroy-Eklund, A.; et al. Structural analysis of MALAT1 long noncoding RNA in cells and in evolution. RNA 2023, 29, 691–704. [Google Scholar] [CrossRef]
- He, S.; Liu, S.; Zhu, H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC evolutionary biology 2011, 11, 1–14. [Google Scholar] [CrossRef]
- Turner, B.M. Epigenetic responses to environmental change and their evolutionary implications. Philosophical Transactions of the Royal Society B: Biological Sciences 2009, 364, 3403–3418. [Google Scholar] [CrossRef]
- Wu, D.; et al. Epigenetic mechanisms of immune remodeling in sepsis: targeting histone modification. Cell death & disease 2023, 14, 112. [Google Scholar]
- Kasuga, T.; Gijzen, M. Epigenetics and the evolution of virulence. Trends in microbiology 2013, 21, 575–582. [Google Scholar] [CrossRef]
- Bartlett, D.A.; et al. High-throughput single-cell epigenomic profiling by targeted insertion of promoters (TIP-seq). Journal of Cell Biology 2021, 220, e202103078. [Google Scholar] [CrossRef]
- Tuteja, R.; Tuteja, N. Serial analysis of gene expression: applications in human studies. BioMed Research International 2004, 2004, 113–120. [Google Scholar] [CrossRef]
- Li, G.; et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome biology 2010, 11, 1–13. [Google Scholar] [CrossRef]
- Steinhauser, S.; et al. A comprehensive comparison of tools for differential ChIP-seq analysis. Briefings in bioinformatics 2016, bbv110. [Google Scholar] [CrossRef]
- Halabian, R.; et al. Laboratory methods to decipher epigenetic signatures: a comparative review. Cellular & Molecular Biology Letters 2021, 26, 1–30. [Google Scholar]
- Grandi, F.C.; et al. Chromatin accessibility profiling by ATAC-seq. Nature protocols 2022, 17, 1518–1552. [Google Scholar] [CrossRef]
- Stephenson, W.; et al. Direct detection of RNA modifications and structure using single-molecule nanopore sequencing. Cell genomics 2022, 2. [Google Scholar] [CrossRef]
- Leger, A.; et al. RNA modifications detection by comparative Nanopore direct RNA sequencing. Nature communications 2021, 12, 7198. [Google Scholar] [CrossRef]
- Fleming, A.M.; et al. Direct nanopore sequencing for the 17 RNA modification types in 36 locations in the E. coli ribosome enables monitoring of stress-dependent changes. ACS Chemical Biology 2023, 18, 2211–2223. [Google Scholar] [CrossRef]
- Prezza, G.; et al. Improved bacterial RNA-seq by Cas9-based depletion of ribosomal RNA reads. Rna 2020, 26, 1069–1078. [Google Scholar] [CrossRef]
- Eberwine, J.; et al. The promise of single-cell sequencing. Nat Methods 2014, 11, 25–27. [Google Scholar] [CrossRef]
- Shen, X.; et al. Recent advances in high-throughput single-cell transcriptomics and spatial transcriptomics. Lab Chip 2022, 22, 4774–4791. [Google Scholar] [CrossRef]
- Alborelli, I.; et al. Cell-free DNA analysis in healthy individuals by next-generation sequencing: a proof of concept and technical validation study. Cell death & disease 2019, 10, 534. [Google Scholar]
- Esposito Abate, R.; et al. Next generation sequencing-based profiling of cell free DNA in patients with advanced non-small cell lung cancer: advantages and pitfalls. Cancers 2020, 12, 3804. [Google Scholar] [CrossRef]
- Bohers, E.; Viailly, P.J.; Jardin, F. , cfDNA Sequencing: Technological Approaches and Bioinformatic Issues. Pharmaceuticals (Basel) 2021, 14. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




