Submitted:
16 April 2024
Posted:
17 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
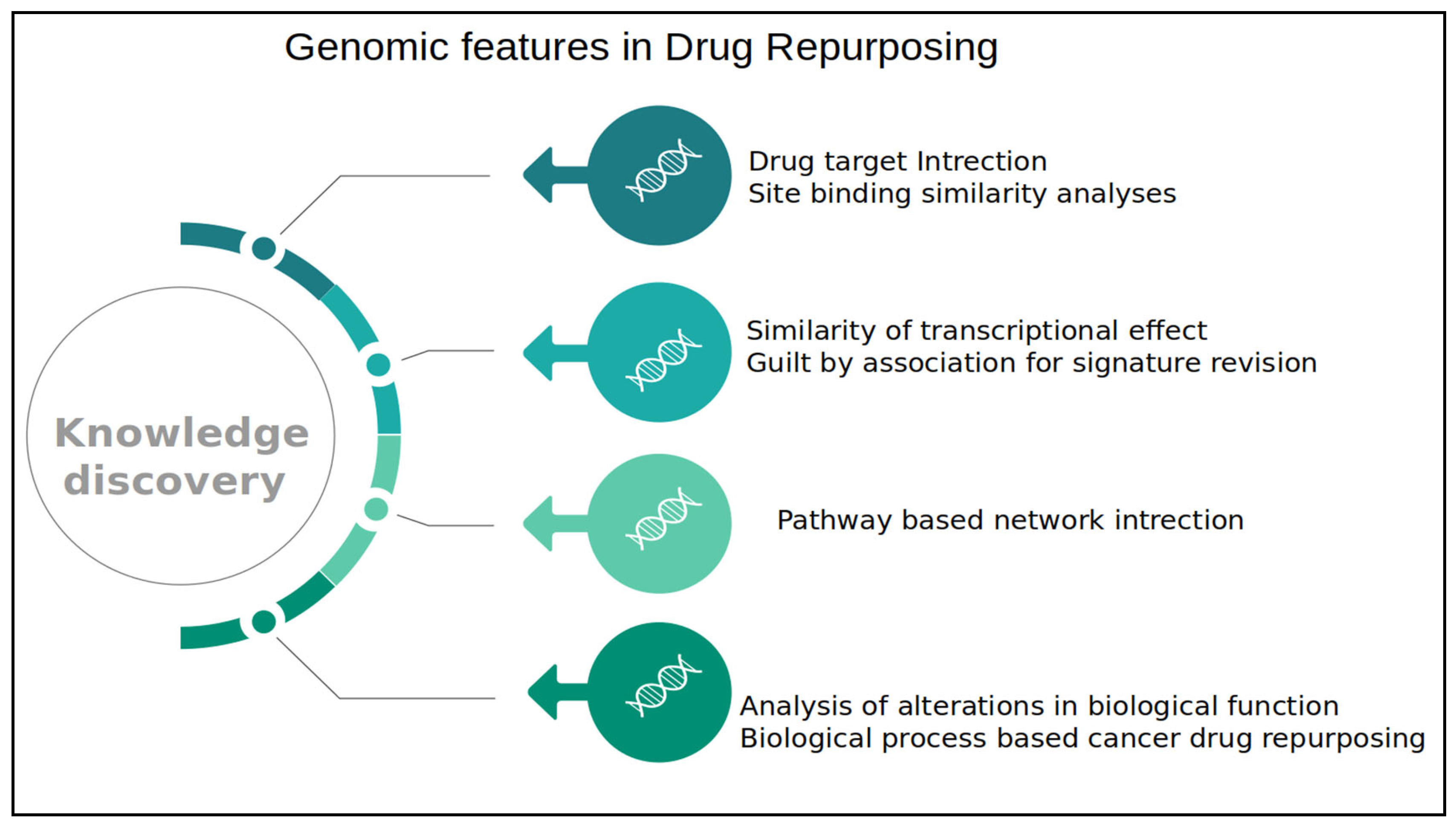
1.1. Functional Genomics and Drug Repurposing
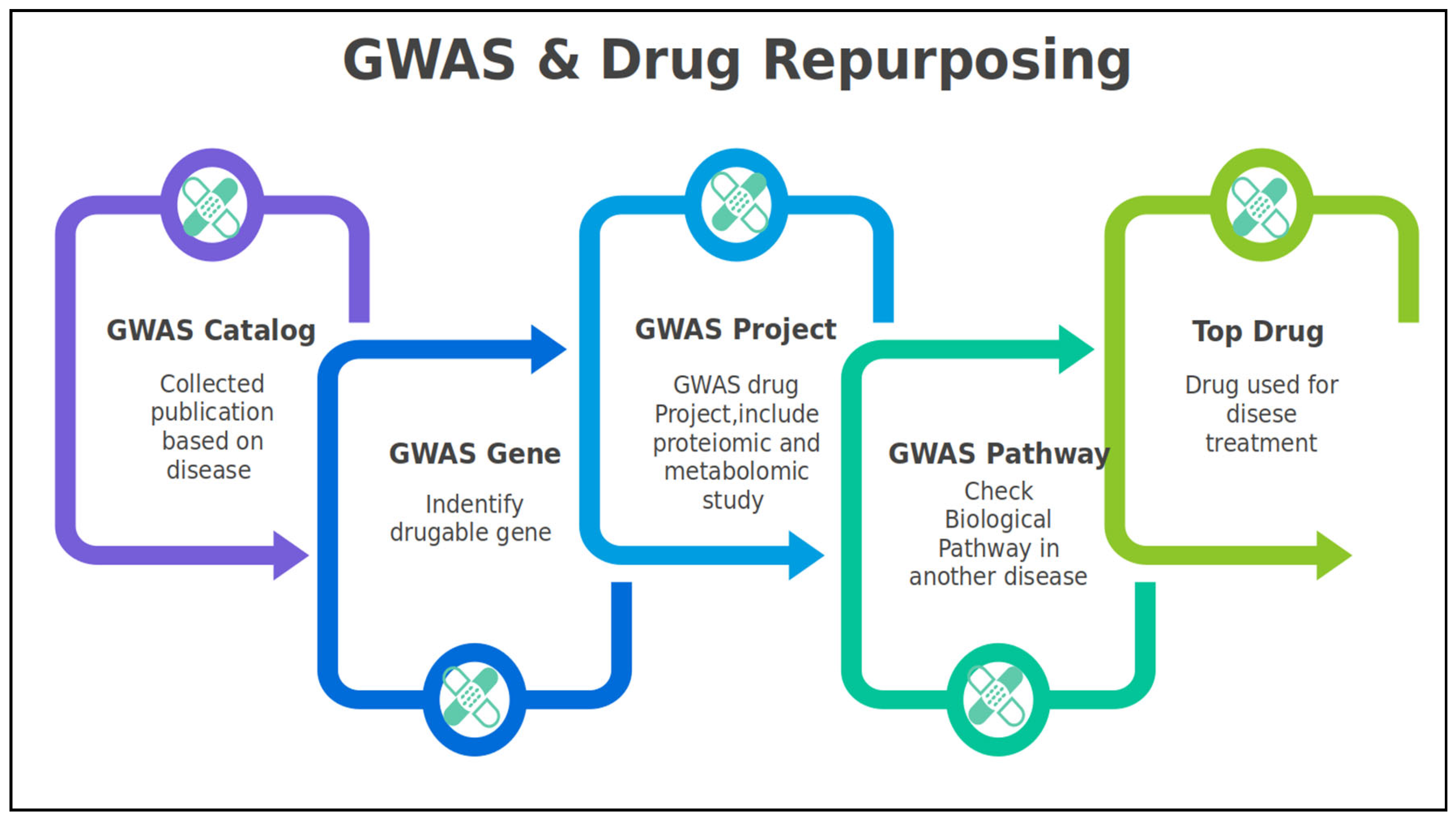
1.2. GWAS & Drug Repurposing
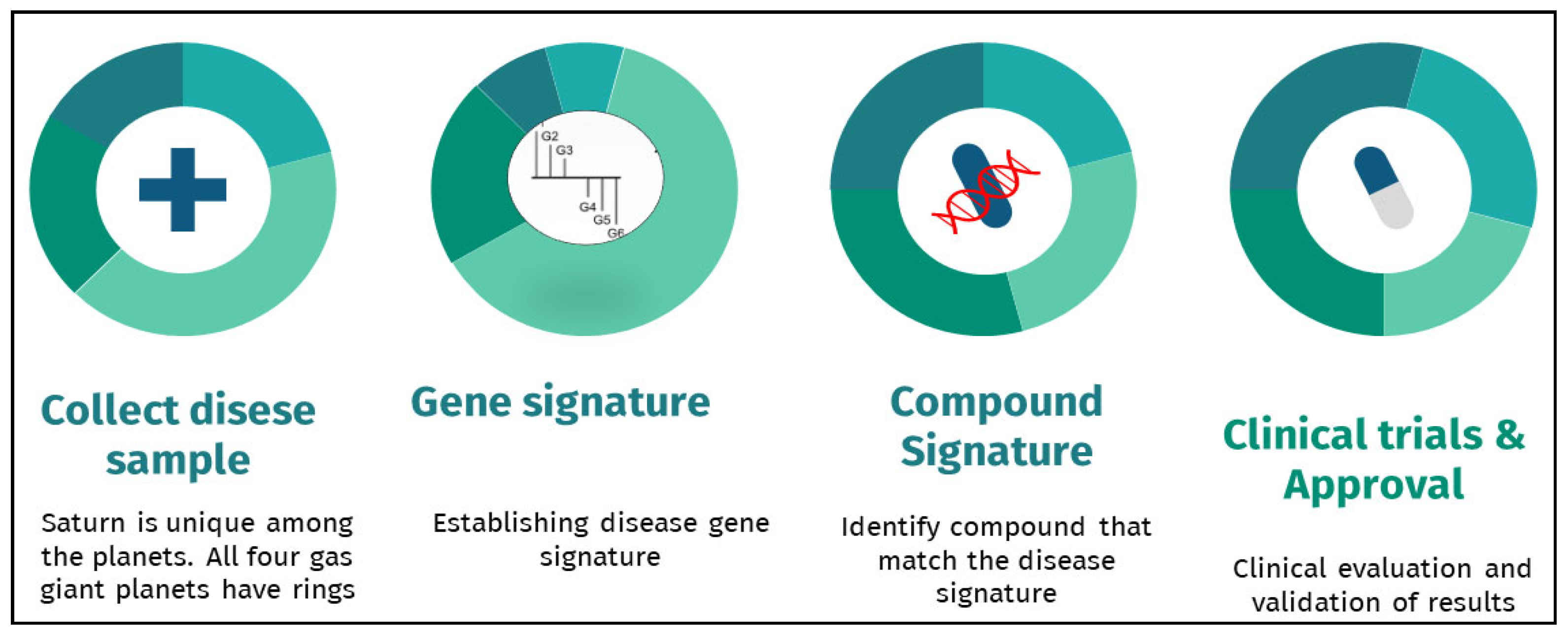
2. Gene Expression Signatures and Drug Repurposing
| Drug | Discovered | Repurposed | Ref. |
|---|---|---|---|
| Anastrazole | Ovulation induction | Breast cancer | [54] |
| Capecitabine | Colon cancer | Breast cancer | |
| Cyclophosphamide | As immuno-modulator in autoimmune | Breast cancer | |
| Everolimus (Votubia, Evertor) | Immunosuppressants during organ | Breast cancer | |
| Exemestane | Ovulation induction | Breast cancer | |
| Fluorouracil | Keratoacanthomas, actinic kerato- | Breast cancer | |
| Fulvestrant | Antiestrogen | Breast cancer | |
| Gemcitabine | Anti-viral drug | Breast cancer | |
| Goserelin | Prostate cancer, uterine fibroids, precursor cleavage | Breast cancer, a variety of cancers | |
| Methotrexate | Leukemia | Breast cancer | |
| Paclitaxel | Ovarian cancer, atrial restenosis | Breast cancer | |
| Raloxifene | Osteoporosis in postmenopausal | Breast cancer | |
| Thiotepa | Immunosuppressant | Breast cancer | |
| Letrozole | Ovulation induction | Breast cancer | |
| Toremifene | Infertility with an ovulatory disorder | Breast cancer | |
| Vinblastine | Hodgkin lymphoma, non-Hod- | Breast cancer | |
| Docetaxel | Hormone-refractory prostate cancer | Breast cancer and active against fungal biofilms | |
| Clarithromycin, pioglitazone, | Antibiotic | Non-small cell lung cancer | [55] |
| Digoxin | Treatment for cardiac diseases | Anticancer | |
| Disulfiram (Antabuse) | Reduces ethanol tolerance in alco- | Metastatic breast cancer & Alzheimer’s | [56] , |
| Mibefradil (Posicor) | Antihypertensive, calcium channel | Short term use as an adjuvant in cancer | |
| Metformin | Diabetes | Anti-nonsmall cell lung cancer, and aug- cancer | |
| Itraconazole | Antifungal | Anticancer | [57] |
| Mebendazole | Antiparasitic/Helminthiasis/Anti- infective | Brain cancer (i.e., medulloblastoma and | |
| Mycophenolic acid | Immunosuppressant | Anticancer |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- 1. Zheng, W., N. Thorne and J. C. McKew. "Phenotypic screens as a renewed approach for drug discovery." Drug Discovery Today 18 (2013): 1067-73. https://www.sciencedirect.com/science/article/pii/S135964461300202X. [CrossRef]
- Schroeder, H. W., Jr. "Mixing the old with the new: Drug repurposing for immune deficiency in the era of precision medicine and pediatric genomics." J Allergy Clin Immunol Pract 6 (2018): 2168-69. http://www.ncbi.nlm.nih.gov/pubmed/30390908. [CrossRef]
- Dudley, J. "The promise of genomics-based drug repurposing." Clin Adv Hematol Oncol 12 (2014): 601-3. http://www.ncbi.nlm.nih.gov/pubmed/25654481.
- Bisson, W. H. "Drug repurposing in chemical genomics: Can we learn from the past to improve the future?" Curr Top Med Chem 12 (2012): 1883-8. http://www.ncbi.nlm.nih.gov/pubmed/23116467. [CrossRef]
- Mirza, N., G. J. Sills, M. Pirmohamed and A. G. Marson. "Identifying new antiepileptic drugs through genomics-based drug repurposing." Hum Mol Genet 26 (2017): 527-37. http://www.ncbi.nlm.nih.gov/pubmed/28053048. [CrossRef]
- Power, A., A. C. Berger and G. S. Ginsburg. "Genomics-enabled drug repositioning and repurposing: Insights from an iom roundtable activity." JAMA 311 (2014): 2063-4. http://www.ncbi.nlm.nih.gov/pubmed/24867009. [CrossRef]
- Bateman, J. R. and C. T. Wu. "A genomewide survey argues that every zygotic gene product is dispensable for the initiation of somatic homolog pairing in drosophila." Genetics 180 (2008): 1329-42. http://www.ncbi.nlm.nih.gov/pubmed/18791221. [CrossRef]
- Ultsch, A., D. Kringel, E. Kalso, J. S. Mogil and J. Lotsch. "A data science approach to candidate gene selection of pain regarded as a process of learning and neural plasticity." Pain 157 (2016): 2747-57. http://www.ncbi.nlm.nih.gov/pubmed/27548044. [CrossRef]
- Moffat, J. G., F. Vincent, J. A. Lee, J. Eder and M. Prunotto. "Opportunities and challenges in phenotypic drug discovery: An industry perspective." Nat Rev Drug Discov 16 (2017): 531-43. http://www.ncbi.nlm.nih.gov/pubmed/28685762. [CrossRef]
- Kulkarni, V. S., V. Alagarsamy, V. R. Solomon, P. A. Jose and S. Murugesan. "Drug repurposing: An effective tool in modern drug discovery." Russ J Bioorg Chem 49 (2023): 157-66. http://www.ncbi.nlm.nih.gov/pubmed/36852389. [CrossRef]
- Giridhara Prema, S., J. Chandrasekaran, S. Kanekar, M. George, T. S. K. Prasad, R. Raju, S. Dagamajalu and R. D. A. Balaya. "Cisplatin and procaterol combination in gastric cancer? Targeting checkpoint kinase 1 for cancer drug discovery and repurposing by an integrated computational and experimental approach." OMICS 28 (2024): 8-23. http://www.ncbi.nlm.nih.gov/pubmed/38190280. [CrossRef]
- Botella, L. M. "Drug repurposing as a current strategy in medicine discovery." Semergen 48 (2022): 101790. http://www.ncbi.nlm.nih.gov/pubmed/35545490. [CrossRef]
- Yang, H. T., J. H. Ju, Y. T. Wong, I. Shmulevich and J. H. Chiang. "Literature-based discovery of new candidates for drug repurposing." Brief Bioinform 18 (2017): 488-97. http://www.ncbi.nlm.nih.gov/pubmed/27113728. [CrossRef]
- Deplanque, D., C. Fetro, A. Ferry, P. Lechat, T. Beghyn, C. Bernard, A. Bernasconi, H. Bienayme, C. Cougoule, J. Del Bano, et al. "Drug repurposing: From the discovery of a useful pharmacological effect to making the treatment available to the patient." Therapie 78 (2023): 10-18. http://www.ncbi.nlm.nih.gov/pubmed/36528417. [CrossRef]
- Boguski, M. S., K. D. Mandl and V. P. Sukhatme. "Drug discovery. Repurposing with a difference." Science 324 (2009): 1394-5. 10.1126/science.1169920. http://www.ncbi.nlm.nih.gov/pubmed/19520944. [CrossRef]
- Wishart, D. S., Y. D. Feunang, A. C. Guo, E. J. Lo, A. Marcu, J. R. Grant, T. Sajed, D. Johnson, C. Li, Z. Sayeeda, et al. "Drugbank 5.0: A major update to the drugbank database for 2018." Nucleic Acids Res 46 (2018): D1074-D82. http://www.ncbi.nlm.nih.gov/pubmed/29126136. [CrossRef]
- Southan, C., M. Sitzmann and S. Muresan. "Comparing the chemical structure and protein content of chembl, drugbank, human metabolome database and the therapeutic target database." Mol Inform 32 (2013): 881-97. http://www.ncbi.nlm.nih.gov/pubmed/24533037. [CrossRef]
- Tao, X. and L. Tong. "Crystal structure of the map kinase binding domain and the catalytic domain of human mkp5." Protein Sci 16 (2007): 880-6. http://www.ncbi.nlm.nih.gov/pubmed/17400920. [CrossRef]
- Fan, H. W., G. Y. Liu, C. F. Zhao, X. F. Li and X. Y. Yang. "Differential expression of cox-2 in osteoarthritis and rheumatoid arthritis." Genet Mol Res 14 (2015): 12872-9. [CrossRef]
- Bai, J. P., A. V. Alekseyenko, A. Statnikov, I. M. Wang and P. H. Wong. "Strategic applications of gene expression: From drug discovery/development to bedside." AAPS J 15 (2013): 427-37. http://www.ncbi.nlm.nih.gov/pubmed/23319288. [CrossRef]
- Clough, E. and T. Barrett. "The gene expression omnibus database." Methods Mol Biol 1418 (2016): 93-110. http://www.ncbi.nlm.nih.gov/pubmed/27008011. [CrossRef]
- Barrett, T. and R. Edgar. "Mining microarray data at ncbi's gene expression omnibus (geo)*." Methods Mol Biol 338 (2006): 175-90. http://www.ncbi.nlm.nih.gov/pubmed/16888359. [CrossRef]
- Ortega, S. S., L. C. Cara and M. K. Salvador. "In silico pharmacology for a multidisciplinary drug discovery process." Drug Metabol Drug Interact 27 (2012): 199-207. http://www.ncbi.nlm.nih.gov/pubmed/23152402. [CrossRef]
- Chang, B., R. Kustra and W. Tian. "Functional-network-based gene set analysis using gene-ontology." PLoS One 8 (2013): e55635. http://www.ncbi.nlm.nih.gov/pubmed/23418449. [CrossRef]
- Kang, H., S. Pan, S. Lin, Y. Y. Wang, N. Yuan and P. Jia. "Pharmgwas: A gwas-based knowledgebase for drug repurposing." Nucleic Acids Res 52 (2024): D972-D79. http://www.ncbi.nlm.nih.gov/pubmed/37831083. [CrossRef]
- Khosravi, A., B. Jayaram, B. Goliaei and A. Masoudi-Nejad. "Active repurposing of drug candidates for melanoma based on gwas, phewas and a wide range of omics data." Mol Med 25 (2019): 30. http://www.ncbi.nlm.nih.gov/pubmed/31221082. [CrossRef]
- Lin, W. Z., Y. C. Liu, M. C. Lee, C. T. Tang, G. J. Wu, Y. T. Chang, C. M. Chu and C. Y. Shiau. "From gwas to drug screening: Repurposing antipsychotics for glioblastoma." J Transl Med 20 (2022): 70. http://www.ncbi.nlm.nih.gov/pubmed/35120529. [CrossRef]
- Sabik, O. L. and C. R. Farber. "Using gwas to identify novel therapeutic targets for osteoporosis." Transl Res 181 (2017): 15-26. http://www.ncbi.nlm.nih.gov/pubmed/27837649. [CrossRef]
- Nanda, H., N. Ponnusamy, R. Odumpatta, J. Jeyakanthan and A. Mohanapriya. "Exploring genetic targets of psoriasis using genome wide association studies (gwas) for drug repurposing." 3 Biotech 10 (2020): 43. http://www.ncbi.nlm.nih.gov/pubmed/31988837. [CrossRef]
- Kaya, S., C. A. Schurman, N. S. Dole, D. S. Evans and T. Alliston. "Prioritization of genes relevant to bone fragility through the unbiased integration of aging mouse bone transcriptomics and human gwas analyses." J Bone Miner Res 37 (2022): 804-17. http://www.ncbi.nlm.nih.gov/pubmed/35094432. [CrossRef]
- Dand, N., P. E. Stuart, J. Bowes, D. Ellinghaus, J. Nititham, J. R. Saklatvala, M. Teder-Laving, L. F. Thomas, T. Traks, S. Uebe, et al. "Gwas meta-analysis of psoriasis identifies new susceptibility alleles impacting disease mechanisms and therapeutic targets." medRxiv (2023): 10.1101/2023.10.04.23296543. http://www.ncbi.nlm.nih.gov/pubmed/37873414.
- Shu, L., M. Blencowe and X. Yang. "Translating gwas findings to novel therapeutic targets for coronary artery disease." Front Cardiovasc Med 5 (2018): 56. http://www.ncbi.nlm.nih.gov/pubmed/29900175. [CrossRef]
- Xu, Y., J. Kong and P. Hu. "Computational drug repurposing for alzheimer's disease using risk genes from gwas and single-cell rna sequencing studies." Front Pharmacol 12 (2021): 617537. http://www.ncbi.nlm.nih.gov/pubmed/34276354. [CrossRef]
- Lippmann, C., D. Kringel, A. Ultsch and J. Lotsch. "Computational functional genomics-based approaches in analgesic drug discovery and repurposing." Pharmacogenomics 19 (2018): 783-97. http://www.ncbi.nlm.nih.gov/pubmed/29792109. [CrossRef]
- Breen, G., Q. Li, B. L. Roth, P. O'Donnell, M. Didriksen, R. Dolmetsch, P. F. O'Reilly, H. A. Gaspar, H. Manji, C. Huebel, et al. "Translating genome-wide association findings into new therapeutics for psychiatry." Nat Neurosci 19 (2016): 1392-96. [CrossRef]
- Uffelmann, E., Q. Q. Huang, N. S. Munung, J. de Vries, Y. Okada, A. R. Martin, H. C. Martin, T. Lappalainen and D. Posthuma. "Genome-wide association studies." Nature Reviews Methods Primers 1 (2021): 59. https://doi.org/10.1038/s43586-021-00056-9. [CrossRef]
- Wishart, D. S., C. Knox, A. C. Guo, D. Cheng, S. Shrivastava, D. Tzur, B. Gautam and M. Hassanali. "Drugbank: A knowledgebase for drugs, drug actions and drug targets." Nucleic Acids Res 36 (2008): D901-6. [CrossRef]
- Klein, T. E. and R. B. Altman. "Pharmgkb: The pharmacogenetics and pharmacogenomics knowledge base." The Pharmacogenomics Journal 4 (2004): 1-1. 10.1038/sj.tpj.6500230. [CrossRef]
- Bovijn, J., J. C. Censin, C. M. Lindgren and M. V. Holmes. "Commentary: Using human genetics to guide the repurposing of medicines." Int J Epidemiol 49 (2020): 1140-46. http://www.ncbi.nlm.nih.gov/pubmed/32097451. [CrossRef]
- Truong, V. Q., J. A. Woerner, T. A. Cherlin, Y. Bradford, A. M. Lucas, C. C. Okeh, M. K. Shivakumar, D. H. Hui, R. Kumar, M. Pividori, et al. "Quality control procedures for genome-wide association studies." Curr Protoc 2 (2022): e603. http://www.ncbi.nlm.nih.gov/pubmed/36441943. [CrossRef]
- Reay, W. R. and M. J. Cairns. "Advancing the use of genome-wide association studies for drug repurposing." Nat Rev Genet 22 (2021): 658-71. http://www.ncbi.nlm.nih.gov/pubmed/34302145. [CrossRef]
- Satish, M., K. Sandhya, K. Nitin, N. Yashas Kiran, B. Aleena, A. Satish Kumar, K. G and E. Rajakumara. "Computational, biochemical and ex vivo evaluation of xanthine derivatives against phosphodiesterases to enhance the sperm motility." J Biomol Struct Dyn 41 (2023): 5317-27. http://www.ncbi.nlm.nih.gov/pubmed/35696450. [CrossRef]
- Wu, P., Q. Feng, V. E. Kerchberger, S. D. Nelson, Q. Chen, B. Li, T. L. Edwards, N. J. Cox, E. J. Phillips, C. M. Stein, et al. "Integrating gene expression and clinical data to identify drug repurposing candidates for hyperlipidemia and hypertension." Nat Commun 13 (2022): 46. [CrossRef]
- Koudijs, K. K. M., S. Böhringer and H. J. Guchelaar. "Validation of transcriptome signature reversion for drug repurposing in oncology." Brief Bioinform 24 (2023). [CrossRef]
- Jha, A., M. Quesnel-Vallières, D. Wang, A. Thomas-Tikhonenko, K. W. Lynch and Y. Barash. "Identifying common transcriptome signatures of cancer by interpreting deep learning models." Genome Biology 23 (2022): 117. [CrossRef]
- Abdelhafiz, A. S., M. A. Fouda, N. A. Elzefzafy, Taha, II, O. M. Mohemmed, N. H. Alieldin, I. Toony, A. A. Abdel Wahab and I. G. Farahat. "Gene expression analysis of invasive breast carcinoma yields differential patterns in luminal subtypes of breast cancer." Ann Diagn Pathol 55 (2021): 151814. http://www.ncbi.nlm.nih.gov/pubmed/34517157. [CrossRef]
- Nguyen, H. T. N., H. Xue, V. Firlej, Y. Ponty, M. Gallopin and D. Gautheret. "Reference-free transcriptome signatures for prostate cancer prognosis." BMC Cancer 21 (2021): 394. http://www.ncbi.nlm.nih.gov/pubmed/33845808. [CrossRef]
- Slebioda, T. J., M. Stanislawowski, M. Cyman, P. M. Wierzbicki, D. Zurawa-Janicka, J. Kobiela, W. Makarewicz, M. Guzek and Z. Kmiec. "Distinct expression patterns of two tumor necrosis factor superfamily member 15 gene isoforms in human colon cancer." Dig Dis Sci 64 (2019): 1857-67. http://www.ncbi.nlm.nih.gov/pubmed/30788683. [CrossRef]
- Koufos, N., J. Syrios, D. Michailidou, I. D. Xynos, A. Lazaris, N. Kavantzas, P. Tomos, S. Kakaris, C. Kosmas and N. Tsavaris. "Distinct patterns of angiogenic factor expression as a predictive factor of response to chemotherapy in stage iiia non-small-cell lung cancer patients." Mol Clin Oncol 5 (2016): 440-46. http://www.ncbi.nlm.nih.gov/pubmed/27699040. [CrossRef]
- Kutay, M., D. Gozuacik and T. Cakir. "Cancer recurrence and omics: Metabolic signatures of cancer dormancy revealed by transcriptome mapping of genome-scale networks." OMICS 26 (2022): 270-79. http://www.ncbi.nlm.nih.gov/pubmed/35394340. [CrossRef]
- Li, H. R., J. Wang-Rodriguez, T. M. Nair, J. M. Yeakley, Y. S. Kwon, M. Bibikova, C. Zheng, L. Zhou, K. Zhang, T. Downs, et al. "Two-dimensional transcriptome profiling: Identification of messenger rna isoform signatures in prostate cancer from archived paraffin-embedded cancer specimens." Cancer Res 66 (2006): 4079-88. http://www.ncbi.nlm.nih.gov/pubmed/16618727. [CrossRef]
- Deng, Z., T. Guo, J. Bi, G. Wang, Y. Hu, H. Du, Y. Zhou, S. Jia, X. Xing and J. Ji. "Transcriptome profiling of patient-derived tumor xenografts suggests novel extracellular matrix-related signatures for gastric cancer prognosis prediction." J Transl Med 21 (2023): 638. http://www.ncbi.nlm.nih.gov/pubmed/37726803. [CrossRef]
- Koudijs, K. K. M., A. G. T. Terwisscha van Scheltinga, S. Böhringer, K. J. M. Schimmel and H. J. Guchelaar. "Transcriptome signature reversion as a method to reposition drugs against cancer for precision oncology." Cancer J 25 (2019): 116-20. [CrossRef]
- Aggarwal, S., S. S. Verma and S. C. Gupta. "Drug repurposing for breast cancer therapy: Old weapon for new battle." Semin Cancer Biol 68 (2021): 8-20. [CrossRef]
- Hernandez, J. J., M. Pryszlak, L. Smith, C. Yanchus, N. Kurji, V. M. Shahani and S. V. Molinski. "Giving drugs a second chance: Overcoming regulatory and financial hurdles in repurposing approved drugs as cancer therapeutics." Front Oncol 7 (2017): 273. 10.3389/fonc.2017.00273.
- Schein, C. H. "Repurposing approved drugs on the pathway to novel therapies." Med Res Rev 40 (2020): 586-605. [CrossRef]
- Shim, J. S. and J. O. Liu. "Recent advances in drug repositioning for the discovery of new anticancer drugs." Int J Biol Sci 10 (2014): 654-63. [CrossRef]
- Wang, B., F. van der Kloet, M. Kes, J. Luirink and L. W. Hamoen. "Improving gene set enrichment analysis (gsea) by using regulation directionality." Microbiol Spectr (2024): e0345623. http://www.ncbi.nlm.nih.gov/pubmed/38294221. [CrossRef]
- Gns, H. S., S. Gr, M. Murahari and M. Krishnamurthy. "An update on drug repurposing: Re-written saga of the drug’s fate." Biomedicine & Pharmacotherapy 110 (2019): 700-16. https://www.sciencedirect.com/science/article/pii/S0753332218372871. [CrossRef]
| Gene names and functions | ||
|---|---|---|
| AmiGO (search utility for GO) | Search utility for Gene Ontology (GO) | http://amigo.geneontology.org/ |
| Gene Ontology (GO) | A standardized classification system for gene functions and products. | www.geneontology.org/ |
| HUGO Gene Nomenclature Committee | Assigns standardized nomenclature to human genes. | www.genenames.org/ |
| Database for Annotation, Visualization, and Integrated Discovery (DAVID) | Provides tools for functional annotation and enrichment analysis of gene sets. | https://david.ncifcrf.gov |
| NCBI gene index database | National Center for Biotechnology Information's database for gene information. | www.ncbi.nlm.nih.gov/gene/ |
| GeneCards | Comprehensive database for human genes, their products, and their associations. | www.genecards.org |
| Human diseases | ||
| Online Mendelian Inheritance in Man (OMIM) database | Catalogs human genes and genetic disorders with a focus on the relationships between phenotype and genotype. | www.ncbi.nlm.nih.gov/omim |
| MalaCards: The human disease database | Database providing comprehensive information on human diseases. | www.malacards.org |
| DiseaseGenes database | A database cataloging genes associated with various diseases. | www.jbldesign.com/jmogil/enter.html |
| The Human Disease Genetics Database (HPGDB) | Database focused on the genetics of human diseases. | https://humandiseasegenetics.org/hpgdb/ |
| Comparative Toxicogenomics Database (CTD) | Integrates toxicology and genomics to advance understanding of the effects of environmental exposures on human health | http://ctd.mdibl.org |
| Biological pathways | ||
| Pathway Commons | A resource for biological pathway analysis and integration. | www.pathwaycommons.org |
| Kyoto Encyclopedia of Genes and Genomes (KEGG) | Provides information on biological pathways, diseases, and drugs. | www.genome.jp/kegg/ |
| Drugs, small molecules, and/or (potential) targets | ||
| DrugBank database | Database containing information on drugs, their targets, and mechanisms of action. | www.drugbank.ca |
| Connectivity map (CMap) | Analyzes the effects of small molecules on gene expression and cellular phenotypes. | https://clue.io/cmap |
| DrugSig | Identifies gene expression signatures related to drug response. | http://biotechlab.fundan.edu.cn/database/drugsig/ |
| Thomson Reuters Integrity database (non-free) | Database providing information on drugs and their properties. | https://integrity.thomson-pharma.com |
| ChEMBL | Database of bioactive molecules with drug-like properties. | www.ebi.ac.uk/chembl/ |
| UniProtKB/Swiss-Prot | Comprehensive resource for protein information. | www.uniprot.org |
| Gene Expression Omnibus (GEO) | Repository for high-throughput gene expression and functional genomics data. | www.ncbi.nlm.nih.gov/geo |
| Software | ||
| R software | Programming language and software environment for statistical computing and graphics. | http://CRAN.R-project.org/ |
| Drug versus disease (DvD) | Analytical approach comparing drug and disease-related data. | www.ebi.ac.uk/saezrodriguez/DVD/ |
| GeneTrail | Software for the statistical evaluation of biological pathways. | http://genetrail.bioinf.uni-sb.de/ |
| Bioconductor | Open-source software for the analysis and comprehension of high-throughput genomic data. | http://bioconductor.org/ |
| R package ‘org.Hs.eg.db’ | R package providing gene annotation for the human genome. | https://bioconductor.org/packages/org.Hs.eg.db/ |
| R package ‘GO.db’ | R package providing Gene Ontology annotations. | http://bioconductor.org/packages/GO.db/ |
| R package ‘dbtORA’ | R package for over-representation analysis in genomics. | https://github.com/IME-TMP-FFM/dbtORA |
| Graphviz | Open-source graph visualization software. | www.graphviz.org |
| Literature | ||
| PubMed | A comprehensive database of biomedical literature, providing access to a vast collection of articles, research papers, and other scholarly materials. | www.ncbi.nlm.nih.gov/pubmed/ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





