Submitted:
09 April 2024
Posted:
10 April 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Therapy Resistance in GBM
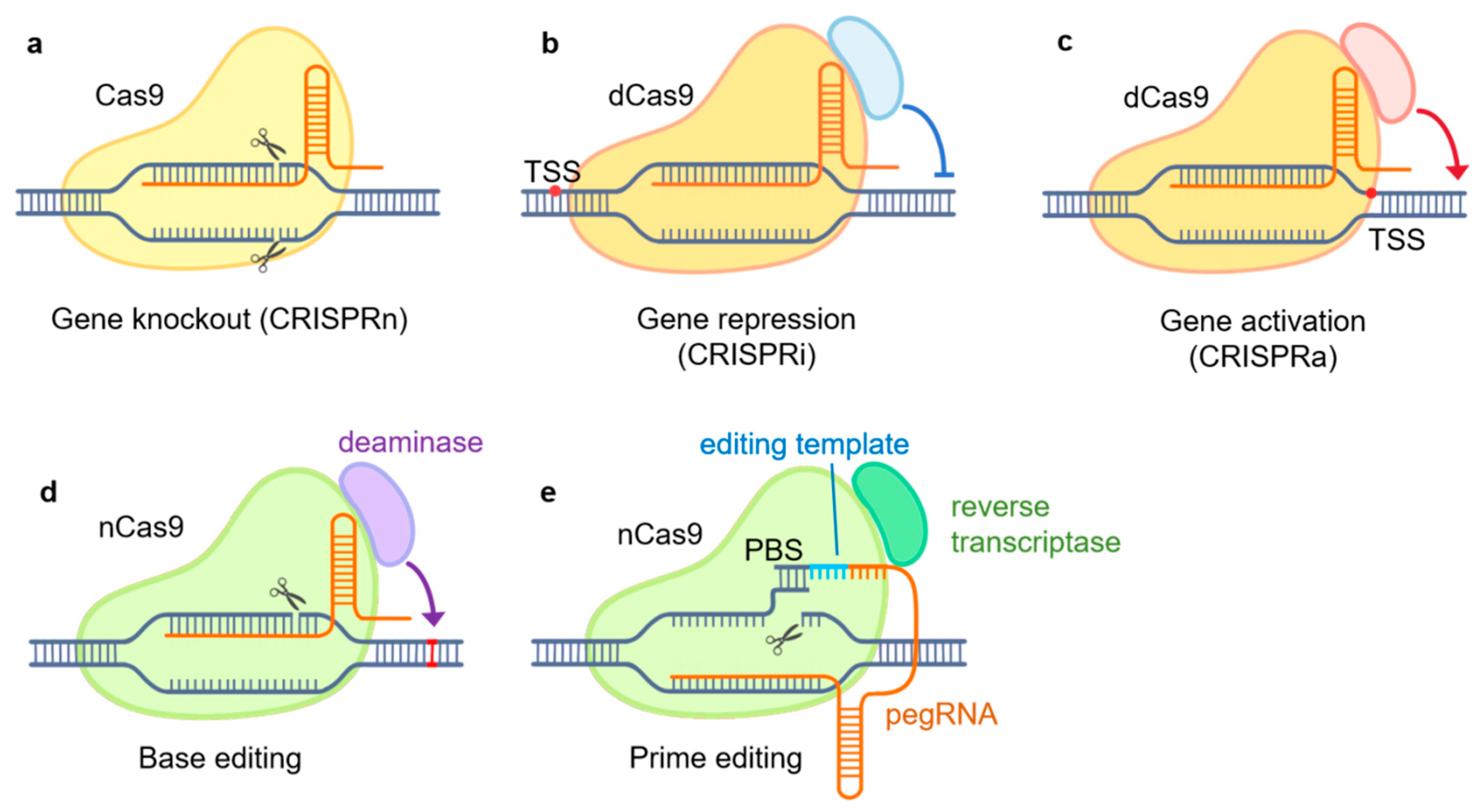
CRISPR/Cas9-Based Genome Engineering Tools
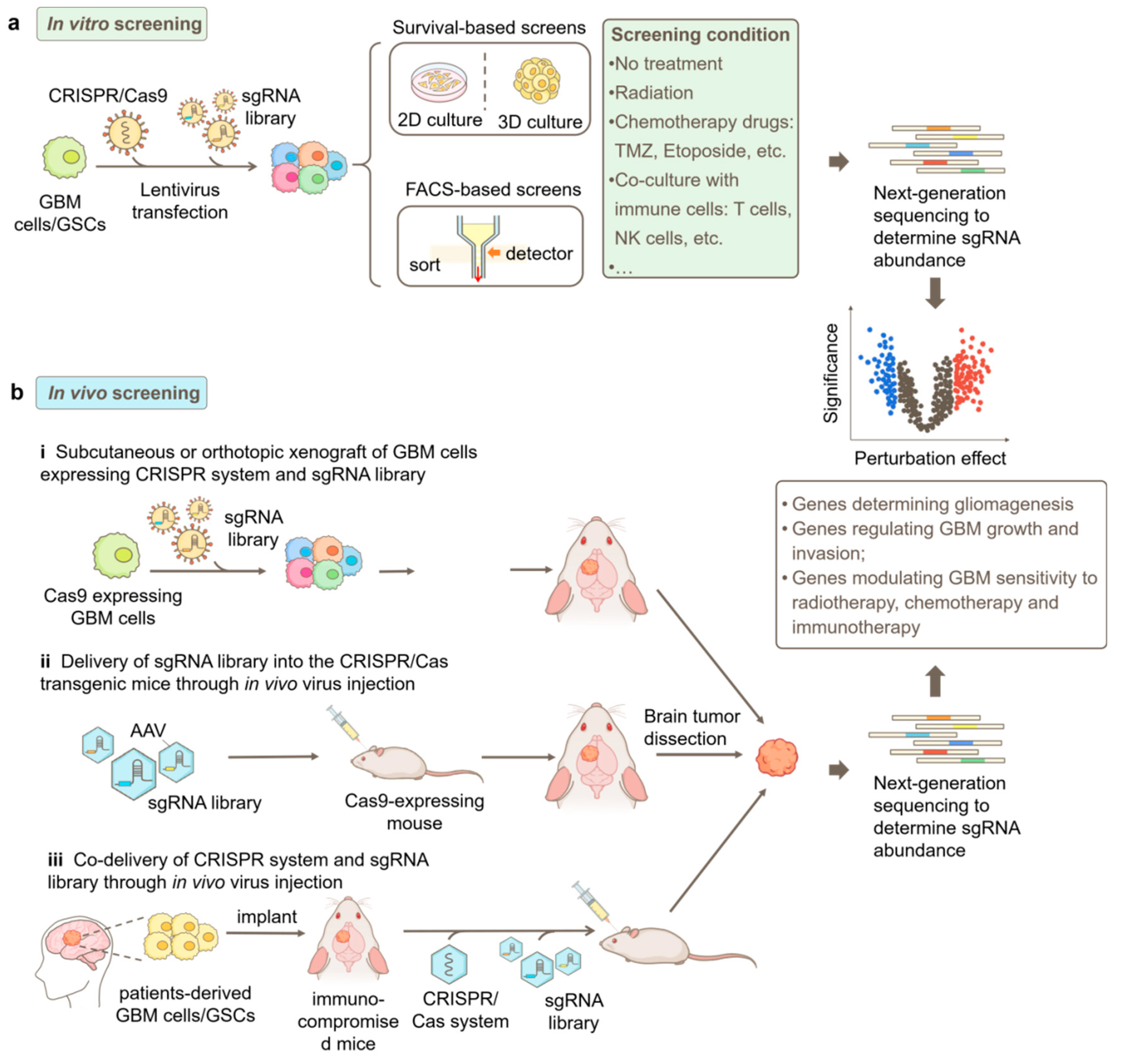
CRISPR/Cas9-Based Functional Genomics Screening Strategies in GBM
CRISPR/Cas9-Based Genetic Screening in GBM
- GBM progression
- 2.
- Treatment susceptibility
Susceptibility to Chemotherapy
Susceptibility to Immunotherapy
| Phenotype | Cell model | CRISPR system | Screening condition | sgRNA library | Main findings | Ref. | ||
|---|---|---|---|---|---|---|---|---|
| GBM progression | Tumori-genesis | astrocytes | CRISPRn | AAV delivery of sgRNA library into the brain of LSL-Cas9 transgenic mice | mTSG library: 286 sgRNAs targeting 56 tumor suppressor genes | determined mutational profiles in GBM tumorigenesis; identified co-occurring driver combinations like B2m-Nf1 and Zc3h13-Rb1 | [41] | |
| Growth | U87 | CRISPRn | normal condition, without treatment | Custom library: 557 sgRNA targeting 557 E3 ligase genes | identified RNF185 as a tumor suppressor regulated by miR-587 | [76] | ||
| U87 | CRISPRi | implanted in the brain of NU/J mice | CRinCL—Unique to U87: 23,317 sgRNAs targeting 2307 lncRNAs | identified 17 lncRNA essential for GBM growth in vivo | [47] | |||
| U87, U251 | CRISPRi | normal condition, without treatment | Custom library: 9,083 sgRNAs targeting 1,209 lncRNAs dysregulated in GBM cells | DARS1-AS1 promotes GBM growth through interaction with YBX1 | [48] | |||
| T98G-TERT-ON GBM cells | CRISPRn | normal condition, without treatment | Custom Library: sgRNAs targeting AAVS1, TERT and GABPB1L | did not detect genetic vulnerabilities specific to GBM carrying TERT promoter mutations (TPMs) | [53] | |||
| GBM progression | Growth | T98G, U373 | CRISPRn | normal condition, without treatment | EpiDoKOL: 1,628 sgRNA targeting 251 chromatin modifiers | ASH2L is essential for GBM cell survival by regulating cell cycle, transcription and histone methylation through interactions with histone methyltransferases | [45] | |
| patient-derived GSCs, NSCs | CRISPRn | normal condition, without treatment | GeCKO library: 64,751 sgRNAs targeting 18,080 genes |
knockout of PKMYT1 specifically inhibits the growth of GSCs by impairing cell division | [42] | |||
| patient-derived GSCs | CRISPRn | normal condition, without treatment | Custom library: targeting 160 chromatin regulator genes from ChIP-seq profiling | knockout of YY1 inhibits the proliferation and self-renewal of GSCs by controlling transcription and m6A modification | [23] | |||
| patient-derived GSCs, NSCs | CRISPRn | normal condition, without treatment | TKOv1/TKOv3 library: 70,948 sgRNAs targeting 18,053 genes | identified transcription factors SOX2 and SOX9, histone methyltransferase DOT1L, and cytokine signaling suppressor SOCS3 as important regulators of GSC stemness and fitness | [43] | |||
| patient-derived GSCs | CRISPRn | cultured as spheres or in the 3D bioprinted tissue model | Brunello library: 76,441 sgRNAs targeting 19,114 genes | identified PAG1, ZNF830, ATP5H, RNF19A as essential genes of GSCs | [44] | |||
| GBM progression | Invasion | U138 | CRISPRn | cultured in a transwell system | Custom library: 45,740 sgRNAs targeting 4,574 genes relative to cell motility and drug targets | knockout of MAP4K4 reduces invasion and inhibits mesenchymal transition of GBM cells | [50] | |
| patient-derived GSCs | CRISPRn | cultured in 3D hydrogel invasion devices | Custom library: 29790 sgRNAs targeting 2981 metabolic genes | knockdown or inhibition of CTH impaired GBM invasion in vitro and in vivo, and caused cysteine deficiency and ROS accumulation | [51] | |||
| Radiotherapy and chemotherapy |
Susceptibility to radiotherapy | U87 | CRISPRi | treated with radiation | CRiNC-U87 & HEK293T and CRiNCL-Unique to U87:38,011 sgRNAs targeting 3750 lncRNAs | knockdown of lncGRS-1(CTC-338 M12.4) selectively inhibits GBM growth and enhances GBM sensitivity to radiation | [35] | |
| U87, U251 | CRISPRa | treated with radiation | SAM library: 70, 290 sgRNAs targeting 23, 430 genes | CARHSP1 enhances radiation resistance in GBM via TNF-α/NF-kβ pathway | [55] | |||
| Susceptibility to TMZ | U138 | CRISPRn | treated with TMZ | GeCKO v2 library: 123,411 sgRNAs targeting 19,050 genes | knockout of MSH2, MSH6, CLCA2 or PTCH2 enhances TMZ resistance | [56] | ||
| U138 | CRISPRa | treated with TMZ | SAM library: 70, 290 sgRNAs targeting 23, 430 genes | NRF2 enhances TMZ resistance by controlling the expression of enzymes in GSH synthesis | [56] | |||
| Radiotherapy and chemotherapy |
Susceptibility to TMZ | RAD18+/+ and RAD18-/- U373 cells | CIRSPRn | treated with TMZ | DDR-CRISPR lentivirus library: 5040 sgRNAs targeting 504 DDR genes | knockout of POLD3 leads to greater TMZ sensitivity in RAD18-deficient GBM cells | [59] | |
| WT and EGFRvIII U87 cells | CRISPRn | treated with TMZ | GeCKO v2: library123,411 sgRNAs targeting 19,050 genes | E2F6 enhances TMZ resistance by promoting DNA repair | [57] | |||
| WT and EGFRvIII U87 cells | CRISPRn | treated with TMZ | GeCKO v2 library: 123,411 sgRNAs targeting 19,050 genes | MUC1 enhances TMZ resistance by regulating DSB repair and autophagy | [58] | |||
| patient-derived GSCs | CRISPRn | treated with TMZ |
TKOv1/TKOv3 library: 70,948 sgRNAs targeting 18,053 genes | knockout of genes in the MMR pathway including MLH1, MSH2, MSH6 and PMS2 leads to TMZ resistance in GSCs; knockout of genes in the FA and HR pathways (such as FANCA, MCM8 and MCM9) sensitizes GSCs to TMZ | [43] | |||
| Susceptibility to RSL3 | LN229 | CRISPRn | treated with RSL3 | GeCKO v2 library: 123,411 sgRNAs targeting 19,050 genes | identified ALOX15 as an essential driver of ferroptosis in GBM | [62] | ||
|
Radiotherapy and chemotherapy |
Susceptibility to etoposide | SNB19, U251, patient-derived GBM cells |
CRISPRn | treated with etoposide | Brunello Library: 76,441 sgRNAs targeting 19,114 genes | knockout of RPS11 reduces GBM susceptibility to etoposide by impairing the induction of the pro-apoptotic gene APAF1 | [61] | |
| Susceptibility to Gambogic amide (GA-amide) | patient-derived GSCs | CRISPRn | treated with GA-amide | Brunello Library: 76,441 sgRNAs targeting 19,114 genes | WDR1 is the direct binding target of GA-amide, a potential new chemotherapy drug for GBM | [63] | ||
| Immunotherapy | Susceptibility to CAR T cell cytotoxicity | U87 | CRISPRn | co-cultured with EGFR-targeting CAR T cells | Brunello library: 76,441 sgRNAs targeting 19,114 genes | knockout of genes in the IFNγ signaling pathway, including IFNGR1, JAK1 and JAK2, induces GBM resistance to CAR T cell cytotoxicity | [65] | |
| patient-derived GSCs | CRISPRn | co-cultured with IL13Rα2-targeting CAR T cells | Brunello library: 76,441 sgRNAs targeting 19,114 genes | knockout of RELA or NPLOC4 sensitizes GBM to CAR T cell-mediated killing | [22] | |||
| Susceptibility to CAR T cell cytotoxicity | U87, U251 and T98G | CRISPRi | co-cultured with B7-H3 targeting CAR T cells | H1 library: 13,025 sgRNAs targeting 2,318 genes of kinases, phosphatases and drug targe | knockdown of ARPC4 or NDUFV1 in GBM cells enhances their killing by CAR T cells by activating TNFSF15-mediated cytokine signaling pathways. | [72] | ||
| Immunotherapy | Susceptibility to CD8+ T cell cytotoxicity | GL261 | CRISPRn | implanted in WT and CD8+ KO mice | Brie kinome KO library: 2856 sgRNA targeting 714 kinases | identified Chek2 as the most important kinase mediating GBM resistance to CD8+ T cell killing | [70] | |
| Susceptibility to NK cell cytotoxicity | patient-derived GSCs | CRISPRn | co-cultured with NK cells | Brunello library: 76,441 sgRNAs targeting 19,114 genes | knockout of CHMP2A in GSCs sensitizes them to NK cells by activating NF-κB signaling and increasing secretion of chemokines like CXCL10 and CXCL12 | [68] | ||
Conclusions
Abbreviations
| AAV | adeno-associated virus |
| CAR T | chimeric antigen receptor T |
| Chek2 | checkpoint kinase 2 |
| CRiNCL | CRISPRi Non-Coding Library |
| CRISPR activation, or CRISPRa | |
| CRISPR interference, or CRISPRi | |
| CRISPR | Regularly Interspaced Short Palindromic Repeats |
| CTH | cystathionine gamma lyase |
| dCas9 | inactive Cas9, dead Cas9 |
| DDR | DNA damage response |
| DSBs | double-strand breaks |
| EpiDoKOL | Epigenetic Domain-specific Knockout Library |
| FACS | fluorescence-activated cell sorting |
| GA-amide | gambogic amide |
| GBM | Glioblastoma, Glioblastoma multiforme |
| GeCKO | genome-scale CRISPR-Cas9 knockout |
| GSCs | Glioblastoma stem cells, GBM stem-like cells |
| HR | homologous recombination |
| KD | knockdown |
| KO | knockout |
| lncGRS | lncRNA Glioma Radiation Sensitizers |
| lncRNAs | long non-coding RNAs |
| MGMT | methyl guanine methyl transferase |
| mTSG | mouse homolog tumor suppressor gene |
| nCas9 | Cas9 nickase |
| NGS | Next-generation sequencing |
| NHEJ | non-homologous end joining |
| NSCs | neural stem cells |
| PDX | patient-derived xenograft |
| pegRNA | prime editing guide RNA |
| RNAi | RNA interference |
| saRNASmall | activating RNA |
| sgRNA | single guide RNA |
| shRNA | short hairpin RNA |
| siRNA | small interfering RNA |
| TERT | telomerase reverse transcriptase |
| TMZ | temozolomide |
| TPMs | telomerase reverse transcriptase promoter mutations |
| TRAIL | tumor necrosis factor-related apoptosis-inducing ligand |
| WDR1 | WD repeat domain 1 |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yabo, Y.A.; Niclou, S.P.; Golebiewska, A. Cancer cell heterogeneity and plasticity: A paradigm shift in glioblastoma. Neuro-Oncology 2021, 24, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Vollmann-Zwerenz, A.; Leidgens, V.; Feliciello, G.; Klein, C.A.; Hau, P. Tumor Cell Invasion in Glioblastoma. Int. J. Mol. Sci. 2020, 21, 1932. [Google Scholar] [CrossRef]
- Zhao, T.; Li, C.; Ge, H.; Lin, Y.; Kang, D. Glioblastoma vaccine tumor therapy research progress. Chin. Neurosurg. J. 2022, 8, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Bahadur, S.; Sahu, A.K.; Baghel, P.; Saha, S. Current promising treatment strategy for glioblastoma multiform: A review. Oncol. Rev. 2019, 13, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.C.; Ashley, D.M.; López, G.Y.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef]
- Kim, Y.Z.; Kim, C.-Y.; Lim, D.H. The Overview of Practical Guidelines for Gliomas by KSNO, NCCN, and EANO. Brain Tumor Res. Treat. 2022, 10, 83–93. [Google Scholar] [CrossRef]
- Immanuel, S.R.C.; Ghanate, A.D.; Parmar, D.S.; Yadav, R.; Uthup, R.; Panchagnula, V.; Raghunathan, A. Integrated genetic and metabolic landscapes predict vulnerabilities of temozolomide resistant glioblastoma cells. npj Syst. Biol. Appl. 2021, 7, 1–10. [Google Scholar] [CrossRef]
- Jackson, C.M.; Choi, J.; Lim, M. Mechanisms of immunotherapy resistance: Lessons from glioblastoma. Nat. Immunol. 2019, 20, 1100–1109. [Google Scholar] [CrossRef]
- Przybyla, L.; Gilbert, L.A. A new era in functional genomics screens. Nat. Rev. Genet. 2021, 23, 89–103. [Google Scholar] [CrossRef]
- Kulkarni, S.; Goel-Bhattacharya, S.; Sengupta, S.; Cochran, B.H. A Large-Scale RNAi Screen Identifies SGK1 as a Key Survival Kinase for GBM Stem Cells. Mol. Cancer Res. 2018, 16, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Sa, J.K.; Yoon, Y.; Kim, M.; Kim, Y.; Cho, H.J.; Lee, J.-K.; Kim, G.-S.; Han, S.; Kim, W.J.; Shin, Y.J.; et al. In vivoRNAi screen identifies NLK as a negative regulator of mesenchymal activity in glioblastoma. Oncotarget 2015, 6, 20145–20159. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Mao, A.; Xu, M.; Weng, Q.; Mao, J.; Ji, J. CRISPR-Cas9 for cancer therapy: Opportunities and challenges. Cancer Lett. 2019, 447, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat Rev Dis Primers. 2015, 1. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E. Glioblastoma: Overview of Disease and Treatment. Clin. J. Oncol. Nurs. 2016, 20 (Suppl. S5), S2–S8. [Google Scholar] [CrossRef]
- Annovazzi, L.; Mellai, M.; Schiffer, D. Chemotherapeutic Drugs: DNA Damage and Repair in Glioblastoma. Cancers 2017, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Klockow, J.L.; Zhang, M.; Lafortune, F.; Chang, E.; Jin, L.; Wu, Y.; Daldrup-Link, H.E. Glioblastoma multiforme (GBM): An overview of current therapies and mechanisms of resistance. Pharmacol. Res. 2021, 171, 105780. [Google Scholar] [CrossRef] [PubMed]
- Oldrini, B.; Vaquero-Siguero, N.; Mu, Q.; Kroon, P.; Zhang, Y.; Galán-Ganga, M.; Bao, Z.; Wang, Z.; Liu, H.; Sa, J.K.; et al. MGMT genomic rearrangements contribute to chemotherapy resistance in gliomas. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, M.; Gan, H.; Wang, H.; Lee, J.-H.; Fang, D.; Kitange, G.J.; He, L.; Hu, Z.; Parney, I.F.; et al. A novel enhancer regulates MGMT expression and promotes temozolomide resistance in glioblastoma. Nat. Commun. 2018, 9, 2949. [Google Scholar] [CrossRef]
- Mansouri, A.; Hachem, L.D.; Mansouri, S.; Nassiri, F.; Laperriere, N.J.; Xia, D.; Lindeman, N.I.; Wen, P.Y.; Chakravarti, A.; Mehta, M.P.; et al. MGMT promoter methylation status testing to guide therapy for glioblastoma: Refining the approach based on emerging evidence and current challenges. Neuro-Oncol. 2018, 21, 167–178. [Google Scholar] [CrossRef]
- Immanuel, S.R.C.; Ghanate, A.D.; Parmar, D.S.; Marriage, F.; Panchagnula, V.; Day, P.J.; Raghunathan, A. Integrative analysis of rewired central metabolism in temozolomide resistant cells. Biochem. Biophys. Res. Commun. 2018, 495, 2010–2016. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Prager, B.C.; Gimple, R.C.; Aguilar, B.; Alizadeh, D.; Tang, H.; Lv, D.; Starr, R.; Brito, A.; Wu, Q.; et al. CRISPR Screening of CAR T Cells and Cancer Stem Cells Reveals Critical Dependencies for Cell-Based Therapies. Cancer Discov. 2021, 11, 1192–1211. [Google Scholar] [CrossRef]
- Qiu, Z.; Zhao, L.; Shen, J.Z.; Liang, Z.; Wu, Q.; Yang, K.; Min, L.; Gimple, R.C.; Yang, Q.; Bhargava, S.; et al. Transcription Elongation Machinery Is a Druggable Dependency and Potentiates Immunotherapy in Glioblastoma Stem Cells. Cancer Discov. 2022, 12, 502–521. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.X.; Prager, B.C.; Wu, Q.L.; Kim, L.J.Y.; Gimple, R.C.; Shi, Y.; Yang, K.; Morton, A.R.; Zhou, W.; Zhu, Z.; et al. Reciprocal Signaling between Glioblastoma Stem Cells and Differentiated Tumor Cells Promotes Malignant Progression. Cell Stem Cell 2018, 22, 514. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Qi, L.S. A CRISPR-dCas Toolbox for Genetic Engineering and Synthetic Biology. J Mol Biol. 2019, 431, 34–47. [Google Scholar] [CrossRef]
- Knott, G.J.; Doudna, J.A. CRISPR-Cas guides the future of genetic engineering. Science 2018, 361, 866–869. [Google Scholar] [CrossRef]
- Kampmann, M. CRISPRi and CRISPRa Screens in Mammalian Cells for Precision Biology and Medicine. ACS Chem. Biol. 2017, 13, 406–416. [Google Scholar] [CrossRef]
- Weber, J.; Braun, C.J.; Saur, D.; Rad, R. In vivo functional screening for systems-level integrative cancer genomics. Nat. Rev. Cancer 2020, 20, 573–593. [Google Scholar] [CrossRef]
- Bak, R.O.; Gomez-Ospina, N.; Porteus, M.H. Gene Editing on Center Stage. Trends Genet. 2018, 34, 600–611. [Google Scholar] [CrossRef]
- Makarova, K.S.; Zhang, F.; Koonin, E.V. SnapShot: Class 2 CRISPR-Cas Systems. Cell 2017, 168, 328-e1. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Li, K.; Ouyang, M.; Zhan, J.; Tian, R. CRISPR-based functional genomics screening in human-pluripotent-stem-cell-derived cell types. Cell Genom. 2023, 3, 100300. [Google Scholar] [CrossRef]
- Kampmann, M. CRISPR-based functional genomics for neurological disease. Nat. Rev. Neurol. 2020, 16, 465–480. [Google Scholar] [CrossRef]
- Tian, R.; Gachechiladze, M.A.; Ludwig, C.H.; Laurie, M.T.; Hong, J.Y.; Nathaniel, D.; Prabhu, A.V.; Fernandopulle, M.S.; Patel, R.; Abshari, M.; et al. CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron 2019, 104, 239–255.e12. [Google Scholar] [CrossRef]
- Liu, S.J.; Malatesta, M.; Lien, B.V.; Saha, P.; Thombare, S.S.; Hong, S.J.; Pedraza, L.; Koontz, M.; Seo, K.; Horlbeck, M.A.; et al. CRISPRi-based radiation modifier screen identifies long non-coding RNA therapeutic targets in glioma. Genome Biol. 2020;21(1):83.
- Katti, A.; Diaz, B.J.; Caragine, C.M.; Sanjana, N.E.; Dow, L.E. CRISPR in cancer biology and therapy. Nat. Rev. Cancer 2022, 22, 259–279. [Google Scholar] [CrossRef]
- Tong, H.; Wang, X.; Liu, Y.; Liu, N.; Li, Y.; Luo, J.; Ma, Q.; Wu, D.; Li, J.; Xu, C.; et al. Programmable A-to-Y base editing by fusing an adenine base editor with an N-methylpurine DNA glycosylase. Nat. Biotechnol. 2023, 41, 1080–1084. [Google Scholar] [CrossRef]
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef]
- Li, C.; Kasinski, A.L. In Vivo Cancer-Based Functional Genomics. Trends Cancer 2020, 6, 1002–1017. [Google Scholar] [CrossRef]
- Chow, R.D.; Chen, S. Cancer CRISPR Screens In Vivo. Trends Cancer 2018, 4, 349–358. [Google Scholar] [CrossRef]
- Chow, R.D.; Guzman, C.D.; Wang, G.; Schmidt, F.; Youngblood, M.W.; Ye, L.; Errami, Y.; Dong, M.B.; Martinez, M.A.; Zhang, S.; et al. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat. Neurosci. 2017, 20, 1329–1341. [Google Scholar] [CrossRef]
- Toledo, C.M.; Ding, Y.; Hoellerbauer, P.; Davis, R.J.; Basom, R.; Girard, E.J.; Lee, E.; Corrin, P.; Hart, T.; Bolouri, H.; et al. Genome-wide CRISPR-Cas9 Screens Reveal Loss of Redundancy between PKMYT1 and WEE1 in Glioblastoma Stem-like Cells. Cell Rep. 2015, 13, 2425–2439. [Google Scholar] [CrossRef]
- MacLeod, G.; Bozek, D.A.; Rajakulendran, N.; Monteiro, V.; Ahmadi, M.; Steinhart, Z.; Kushida, M.M.; Yu, H.; Coutinho, F.J.; Cavalli, F.M.G.; et al. Genome-Wide CRISPR-Cas9 Screens Expose Genetic Vulnerabilities and Mechanisms of Temozolomide Sensitivity in Glioblastoma Stem Cells. Cell Rep. 2019, 27, 971–986.e9. [Google Scholar] [CrossRef]
- Tang, M.; Xie, Q.; Gimple, R.C.; Zhong, Z.; Tam, T.; Tian, J.; Kidwell, R.L.; Wu, Q.; Prager, B.C.; Qiu, Z.; et al. Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions. Cell Res. 2020, 30, 833–853. [Google Scholar] [CrossRef]
- Ozyerli-Goknar, E.; Kala, E.Y.; Aksu, A.C.; Bulut, I.; Cingöz, A.; Nizamuddin, S.; Biniossek, M.; Seker-Polat, F.; Morova, T.; Aztekin, C.; et al. Epigenetic-focused CRISPR/Cas9 screen identifies (absent, small, or homeotic)2-like protein (ASH2L) as a regulator of glioblastoma cell survival. Cell Commun. Signal. 2023, 21, 1–16. [Google Scholar] [CrossRef]
- Humphreys, L.M.; Smith, P.; Chen, Z.; Fouad, S.; D’angiolella, V. The role of E3 ubiquitin ligases in the development and progression of glioblastoma. Cell Death Differ. 2021, 28, 522–537. [Google Scholar] [CrossRef]
- Attenello, F.J.; Tsung, K.; Bishara, I.; Loh, Y.E.; Chen, T.C. In vivo CRISPR screening for novel noncoding RNA functional targets in glioblastoma models. J. Neurosci. Res. 2021, 99, 2029–2045. [Google Scholar] [CrossRef]
- Zheng, C.; Wei, Y.; Zhang, Q.; Sun, M.; Wang, Y.; Hou, J.; Zhang, P.; Lv, X.; Su, D.; Jiang, Y.; et al. Multiomics analyses reveal DARS1-AS1/YBX1–controlled posttranscriptional circuits promoting glioblastoma tumorigenesis/radioresistance. Sci. Adv. 2023, 9, eadf3984. [Google Scholar] [CrossRef]
- Verdugo, E.; Puerto, I.; Medina, M. An update on the molecular biology of glioblastoma, with clinical implications and progress in its treatment. Cancer Commun. 2022, 42, 1083–1111. [Google Scholar] [CrossRef]
- Prolo, L.M.; Li, A.; Owen, S.F.; Parker, J.J.; Foshay, K.; Nitta, R.T.; Morgens, D.W.; Bolin, S.; Wilson, C.M.; Vega, L.J.C.; et al. Targeted genomic CRISPR-Cas9 screen identifies MAP4K4 as essential for glioblastoma invasion. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Garcia, J.H.; Akins, E.A.; Jain, S.; Wolf, K.J.; Zhang, J.; Choudhary, N.; Aghi, M.K. Multi-omic screening of invasive GBM cells reveals targetable transsulfuration pathway alterations. J. Clin. Investig. 2023, 134. [Google Scholar]
- Dresser, L.; Wlodarski, R.; Rezania, K.; Soliven, B. Myasthenia Gravis: Epidemiology, Pathophysiology and Clinical Manifestations. J Clin Med. 2021, 10. [Google Scholar] [CrossRef]
- Tu, K.J.; Stewart, C.E.; Hendrickson, P.G.; Regal, J.A.; Kim, S.Y.; Ashley, D.M.; Waitkus, M.S.; Reitman, Z.J. Pooled genetic screens to identify vulnerabilities in TERT-promoter-mutant glioblastoma. Oncogene 2023, 42, 3274–3286. [Google Scholar] [CrossRef]
- Matt, S.; Hofmann, T.G. The DNA damage-induced cell death response: A roadmap to kill cancer cells. Cell. Mol. Life Sci. 2016, 73, 2829–2850. [Google Scholar] [CrossRef]
- Zhu, G.-D.; Yu, J.; Sun, Z.-Y.; Chen, Y.; Zheng, H.-M.; Lin, M.-L.; Ou-Yang, S.; Liu, G.-L.; Zhang, J.-W.; Shao, F.-M. Genome-wide CRISPR/Cas9 screening identifies CARHSP1 responsible for radiation resistance in glioblastoma. Cell Death Dis. 2021, 12, 1–9. [Google Scholar] [CrossRef]
- Rocha, C.R.R.; Reily Rocha, A.; Molina Silva, M.; Rodrigues Gomes, L.; Teatin Latancia, M.; Andrade Tomaz, M.; de Souza, I.; Karolynne Seregni Monteiro, L.; Menck, C.F.M. Revealing Temozolomide Resistance Mechanisms via Genome-Wide CRISPR Libraries. Cells 2020, 9, 2573. [Google Scholar] [CrossRef]
- Huang, K.; Liu, X.; Li, Y.; Wang, Q.; Zhou, J.; Wang, Y.; Dong, F.; Yang, C.; Sun, Z.; Fang, C.; et al. Genome-Wide CRISPR-Cas9 Screening Identifies NF-kappaB/E2F6 Responsible for EGFRvIII-Associated Temozolomide Resistance in Glioblastoma. Adv Sci. 2019, 6, 1900782. [Google Scholar] [CrossRef]
- Tong, F.; Zhao, J.X.; Fang, Z.Y.; Cui, X.T.; Su, D.Y.; Liu, X.; Zhou, J.H.; Wang, G.X.; Qiu, Z.J.; Liu, S.Z.; et al. MUC1 promotes glioblastoma progression and TMZ resistance by stabilizing EGFRvIII. Pharmacol. Res. 2023, 187. [Google Scholar] [CrossRef]
- Cheng, X.; An, J.; Lou, J.; Gu, Q.; Ding, W.; Droby, G.N.; Wang, Y.; Wang, C.; Gao, Y.; Anand, J.R.; et al. Trans-lesion synthesis and mismatch repair pathway crosstalk defines chemoresistance and hypermutation mechanisms in glioblastoma. Nat. Commun. 2024, 15, 1–20. [Google Scholar] [CrossRef]
- Nitiss, J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 2009, 9, 327–337. [Google Scholar] [CrossRef]
- Awah, C.U.; Chen, L.; Bansal, M.; Mahajan, A.; Winter, J.; Lad, M.; Warnke, L.; Gonzalez-Buendia, E.; Park, C.; Zhang, D.; et al. Ribosomal protein S11 influences glioma response to TOP2 poisons. Oncogene 2020, 39, 5068–5081. [Google Scholar] [CrossRef]
- Cao, Z.; Liu, X.; Zhang, W.; Zhang, K.; Pan, L.; Zhu, M.; Qin, H.; Zou, C.; Wang, W.; Zhang, C.; et al. Biomimetic Macrophage Membrane-Camouflaged Nanoparticles Induce Ferroptosis by Promoting Mitochondrial Damage in Glioblastoma. ACS Nano 2023, 17, 23746–23760. [Google Scholar] [CrossRef]
- Qu, J.; Qiu, B.; Zhang, Y.; Hu, Y.; Wang, Z.; Guan, Z.; Qin, Y.; Sui, T.; Wu, F.; Li, B.; et al. The tumor-enriched small molecule gambogic amide suppresses glioma by targeting WDR1-dependent cytoskeleton remodeling. Signal Transduct. Target. Ther. 2023, 8, 1–15. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Larson, R.C.; Kann, M.C.; Bailey, S.R.; Haradhvala, N.J.; Llopis, P.M.; Bouffard, A.A.; Scarfó, I.; Leick, M.B.; Grauwet, K.; Berger, T.R.; et al. CAR T cell killing requires the IFNγR pathway in solid but not liquid tumours. Nature 2022, 604, 563–570. [Google Scholar] [CrossRef]
- Tomaszewski, W.; Sanchez-Perez, L.; Gajewski, T.F.; Sampson, J.H. Brain Tumor Microenvironment and Host State: Implications for Immunotherapy. Clin. Cancer Res. 2019, 25, 4202–4210. [Google Scholar] [CrossRef]
- Shimasaki, N.; Jain, A.; Campana, D. NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 2020, 19, 200–218. [Google Scholar] [CrossRef]
- Bernareggi, D.; Xie, Q.; Prager, B.C.; Yun, J.; Cruz, L.S.; Pham, T.V.; Kim, W.; Lee, X.; Coffey, M.; Zalfa, C.; et al. CHMP2A regulates tumor sensitivity to natural killer cell-mediated cytotoxicity. Nat. Commun. 2022, 13, 1–14. [Google Scholar] [CrossRef]
- Mittrücker, H.-W.; Visekruna, A.; Huber, M. Heterogeneity in the Differentiation and Function of CD8+ T Cells. Arch. Immunol. et Ther. Exp. 2014, 62, 449–458. [Google Scholar] [CrossRef]
- Dmello, C.; Zhao, J.; Chen, L.; Gould, A.; Castro, B.; Arrieta, V.A.; Zhang, D.Y.; Kim, K.S.; Kanojia, D.; Zhang, P.; et al. Checkpoint kinase 1/2 inhibition potentiates anti-tumoral immune response and sensitizes gliomas to immune checkpoint blockade. Nat. Commun. 2023, 14. [Google Scholar] [CrossRef]
- Schmidts, A.; Maus, M.V. Making CAR T Cells a Solid Option for Solid Tumors. Front. Immunol. 2018, 9, 2593. [Google Scholar] [CrossRef]
- Li, X.; Sun, S.; Zhang, W.; Liang, Z.; Fang, Y.; Sun, T.; Wan, Y.; Ma, X.; Zhang, S.; Xu, Y.; et al. Identification of genetic modifiers enhancing B7-H3-targeting CAR T cell therapy against glioblastoma through large-scale CRISPRi screening. J. Exp. Clin. Cancer Res. 2024, 43, 1–16. [Google Scholar] [CrossRef]
- Serra, R.; Mangraviti, A.; Gorelick, N.L.; Shapira-Furman, T.; Alomari, S.; Cecia, A.; Darjee, N.; Brem, H.; Rottenberg, Y.; Domb, A.J.; et al. Combined intracranial Acriflavine, temozolomide and radiation extends survival in a rat glioma model. Eur. J. Pharm. Biopharm. 2021, 170, 179–186. [Google Scholar] [CrossRef]
- Zhu, S.; Li, W.; Liu, J.; Chen, C.-H.; Liao, Q.; Xu, P.; Xu, H.; Xiao, T.; Cao, Z.; Peng, J.; et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR–Cas9 library. Nat. Biotechnol. 2016, 34, 1279–1286. [Google Scholar] [CrossRef]
- Datlinger, P.; Rendeiro, A.F.; Schmidl, C.; Krausgruber, T.; Traxler, P.; Klughammer, J.; Schuster, L.C.; Kuchler, A.; Alpar, D.; Bock, C. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 2017, 14, 297–301. [Google Scholar] [CrossRef]
- Lin, K.; Shen, S.-H.; Lu, F.; Zheng, P.; Wu, S.; Liao, J.; Jiang, X.; Zeng, G.; Wei, D. CRISPR screening of E3 ubiquitin ligases reveals Ring Finger Protein 185 as a novel tumor suppressor in glioblastoma repressed by promoter hypermethylation and miR-587. J. Transl. Med. 2022, 20, 1–14. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





