1. Introduction
Antibodies are an interesting resource for studies of membrane proteins once they can be used for different purposes. Antibodies provide high specificity for their target antigens, allowing for the selective labeling and visualization of specific proteins or domains within complex biological samples. In cell biology assays and biophysical studies, antibodies can be useful for exploring conformational changes, localization, inhibition, and other applications [
1,
2]. In cryo-electron microscopy (Cryo-EM), the addition of a fragment-antigen binding region (Fab region) directed to the protein of interest or to a protein belonging to a complex have many advantages. It can stabilize the protein in particular conformation, increase the size of the target and, consequently, enhance the contrast and signal-to-noise ratio, making it easier to analyze images, facilitate alignment of particles and improve three-dimensional reconstruction [
3]. In X-ray crystallography, the use of antibodies, and Fab fragments, can be used to stabilize targets, drive novel crystal forms, and solve the problem of phase resolution in structure determination [
4].
The development of antibodies against membrane proteins, however, is hard due the difficulties to obtain these proteins in purified form. The RAD-Display system was developed by Rossman and collaborators [
5] for the presentation of small peptides in a protein to enable the study of interactions between proteins. The system was based on the thermostable recombinase RadA from
Pyrococcus furiosus, engineered to contain only the ATPase domain with a replacement of so-called L2 loop with a cloning site to facilitate insertion of displayed epitopes. In this system, the coding sequences for peptides of interest are inserted with a ligation-independent cloning (LIC) into pRAD plasmid. By expressing peptides of interest in a highly stable and easily folded anchor protein, tethered at both ends, the aim is to maintain at least partially their structure (folded state) and reduce risks of proteolysis. Affinity tags on the scaffold itself facilitate large scale production and purification of the proteins.
In previous studies, we showed that this system was suitable for production of antibodies against extracellular loops of permeases from ABC transporters of
Escherichia coli and
Staphylococcus aureus without the need for expression and purification of complete membrane proteins [
6]. The system also was applied to evaluate the immunogenicity of sequences from SARS-CoV-2 Spike protein, production of antibodies and selection of antibodies with ability to neutralize the virus infection in human cells [
7].
In this study, we have evaluated the use of the RAD display system to produce antibodies against the
Mycobacterium tuberculosis Rv1819c ATP-Binding Cassette (ABC) transporter. The ABC transporter Rv1819c has been characterized as a pivotal component in the transport of essential molecules, including vitamin B12 and hydrophilic compounds [
8,
9]. As a member of the ABC transporter superfamily, Rv1819c facilitates the translocation of these substrates across cellular membranes, utilizing energy derived from ATP hydrolysis.
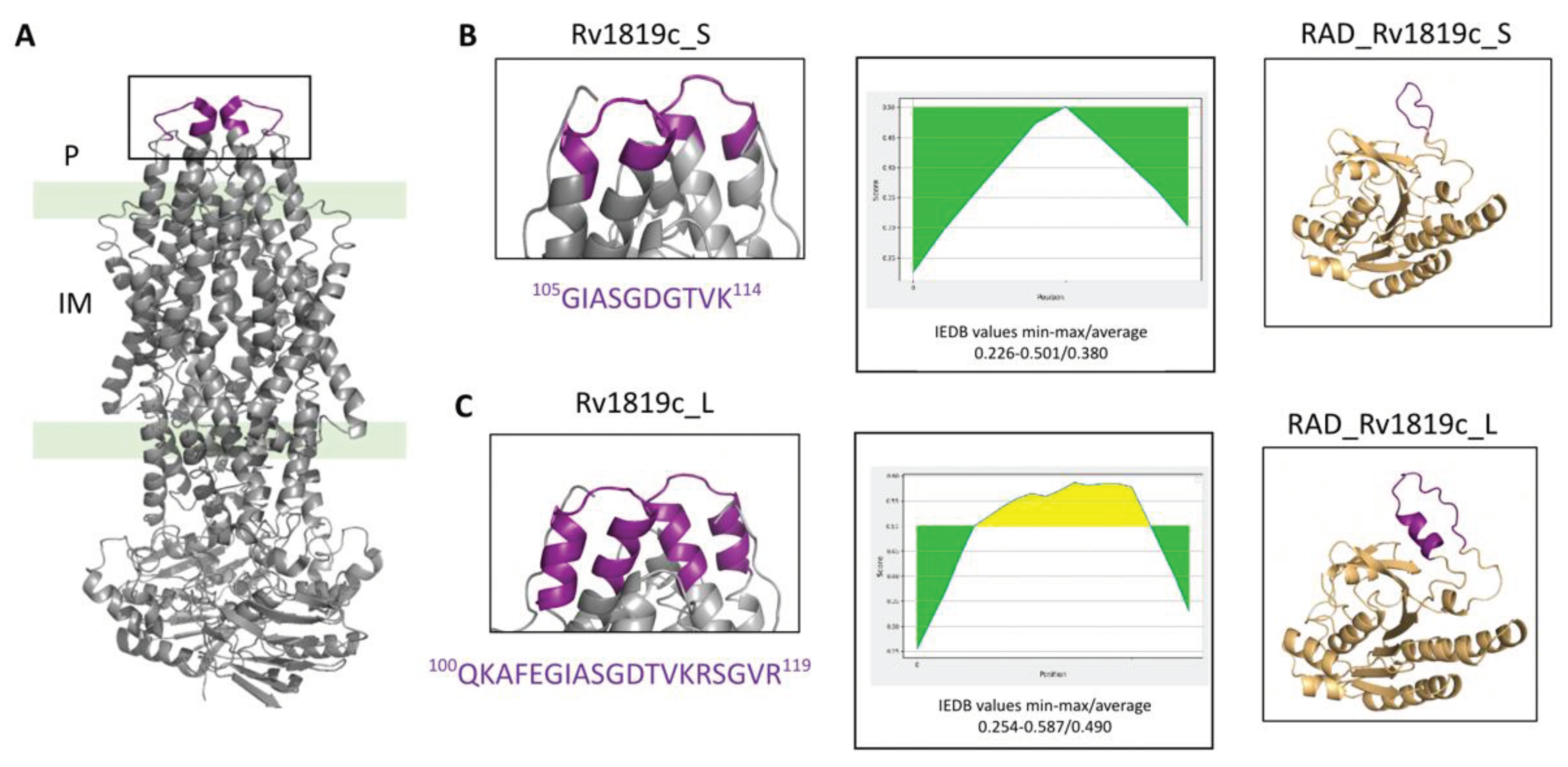
Rv1819c functions as a homodimeric transport complex composed of two identical half-transporters. The TMDs are responsible for recognizing and binding substrates while the NBDs act as the energy-transducing units, utilizing ATP to power the transport process. Based on the three-dimensional structure of the transporter, we selected most prominent extracellular loop of the permease to be presented by the RAD-display system. We designed two versions of this loop, different in their length, based on their predicted antigenicity. Rv1819c-derived peptides expressed in the RAD display system were used as antigens for immunization of C57BL/6 mice and production of polyclonal antibodies. We tested the ability of the antibodies for recognition of the Rv1819c transporter using different approaches. We found that the antibodies identified the purified Rv1819c transporter in vitro, in M. tuberculosis cell extracts and on the surface of the bacterium.
2. Material and Methods
Bioinformatics analyses. The published structures of Rv1819c were used in the analysis (PDB codes: 6TQF and 6TQE). The sequence was obtained from NCBI (
https://www.ncbi.nlm.nih.gov/). The structure was used to identify the extracellular loops of transmembrane helices that extend to the membrane and are exposed to the periplasm (ECLs), identification of residues, cloning design, and experimental strategy. The transporter and the ECLs were analyzed in the PyMOL viewer (Schrondinger, LLC. The PyMOL Molecular Graphics System, Version 1.2.7). After the choice of the sequences, they were submitted to the IEDB server [
10] for prediction of linear B cell epitopes. All results were analyzed together with data from the literature to design cloning strategies.
Cloning, protein expression and purification. The nucleotide sequences corresponding to the short and long ECLs of Rv1819c transporter were cloned into a phRAD vector to present the peptides of interest in the RAD-Display system, using the Ligase Independent Cloning (LIC) methodology [
11] following the instructions described in Rossmann and collaborators [
5]. The constructs were incubated on ice for 30 minutes and then transformed into chemocompetent
E. coli DH5α cells. Cloning was confirmed by digestion of the vectors with
HindIII and sequencing. The vectors were then transformed into chemocompetent
E. coli BL21 (DE3) cells containing the pUBS24 plasmid that encodes a tRNA with a rare anticodon for Arginine present in the RadA sequence, allowing the expression of chimeric proteins. BL21 (DE3)/pUBS520 cells transformed with the vector constructed for the expression of RAD-peps proteins grown overnight were inoculated at a ratio of 1/100 in 500 mL of 2xYT medium supplemented with 100 μg/mL of ampicillin and 25 μg/mL of kanamycin at 37ºC under stirring at 200 rpm until reaching an optical density (OD 600 nm) of 0.8, when they were induced with IPTG (isopropyl β-D-1-thiogalactopyranoside) at a final concentration of 0.4 mM. Expression occurred for 3 hours at 37ºC under shaking at 200 rpm. Aliquots of the culture were collected before induction (T0) and after 3 hours of expression (T2), for analysis of expression on SDS-polyacrylamide gel. After expression, cells were recovered by centrifugation at 5520
g for 15 minutes. The pellet was resuspended in 25 mL of equilibrium buffer with 1mM PMSF, 1 mg/mL DNAse and 1 mg/mL lysozyme at the pH appropriate to the pI of each protein, followed by incubation at 4ºC in a homogenizer for 30 minutes. Cell lysis was performed by sonication for 10 minutes (15 second pulse, 20 second pause) and the soluble fraction of the culture was obtained by centrifugation of the cell lysate at 30,500 g for 40 min at 4ºC. Purification of the proteins was carried out in 50 mM Tris HCl, 150 NaCl using the nickel affinity chromatography technique with Ni Sepharose 6 Fast Flow resin (Cytiva Life Sciences). The elution was obtained after two washes of the column with 20 mM and 50 mM and then elution with 400 mM imidazole. After IMAC, samples were purified with size exclusion chromatography (SEC) using the Superdex 200 HiLoad 16/60 column in AKTA start system (GE Healthcare) in 50 mM Tris HCl, 150 NaCl. Proteins were concentrated and quantified by Bradford.
Immunization and antibody production. Polyclonal antibodies against the RAD-peps proteins and against the RAD protein without peptide insertion were produced in female C57BL/6 mice immunized subcutaneously with 3 doses and 15-day intervals between doses. The immunization was performed combining 10 μg of the target protein with 10 μg of Aluminum hydroxide (Alum) as an adjuvant in 100 µL of saline. 15 days after the third immunization, approximately 200 µL of blood was collected from each animal via submandibular vein puncture. Serum was obtained after incubating the collected blood for 30 minutes at room temperature and 30 minutes at 4ºC, followed by centrifugation at 425 g for 30 minutes. Serum was then collected, aliquoted and stored at -20ºC.
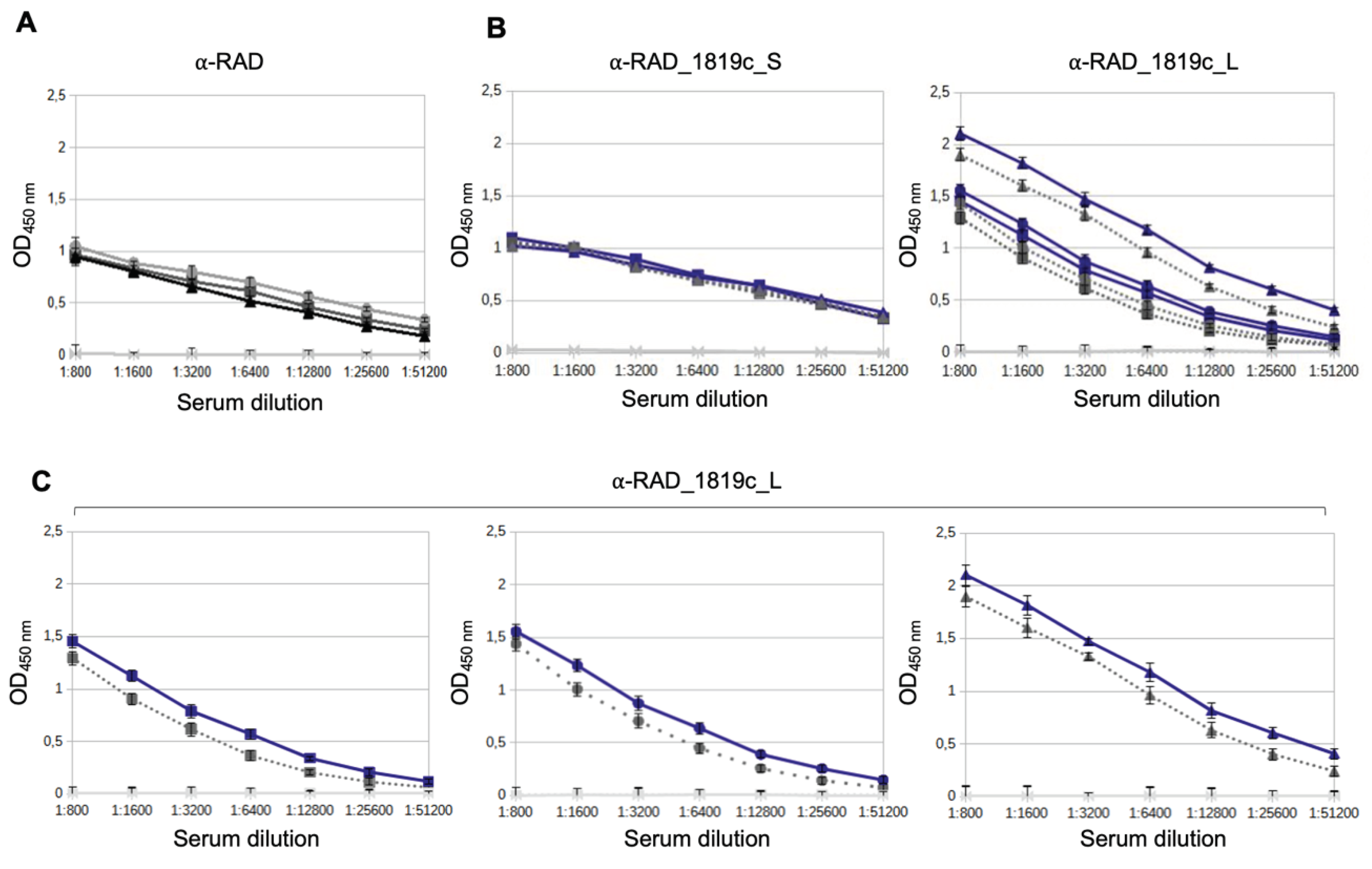
Enzyme-linked immunosorbent assay analysis of the sera samples. Nunc high-protein binding polystyrene plates (Thermo Fisher Scientific) were sensitized with RAD, RAD_Rv1819c_S and RAD_Rv1819c_L proteins (400 ng/well) diluted in carbonate buffer pH 9.6 (100 µL/well) for 14 hours at 4ºC. A cycle of 3 washes was then carried out with PBS buffer containing 0.05% Tween-20. Blocking was carried out for 3 hours at 37ºC with 300 µL/well of buffer (5% skimmed milk, 0.5% BSA in PBS containing 0.05% Tween-20) and after a washing cycle, sera containing antibodies relating to each RAD-pep protein in dilutions starting from 1:400 in 1X PBS buffer, 5% BSA. The plate was incubated for 1 h at 37ºC and then washed. Next, the secondary antibody Anti-Mouse IgG conjugated to peroxidase (Sigma-Aldrich) was added at a dilution of 1:5000. The plate was incubated for 1 h at 37ºC and after washing, 50 µL of TMB (3.3’, 5.5’-Tetramethylbenzidine) was added per well to react with peroxidase and reveal the binding of primary antibodies to their respective target antigen. The reaction was stopped by adding 50 µL/well of 0.2 N sulfuric acid. The optical density reading was performed at a wavelength of 450 nm. The last dilution value in which the OD450nm was greater than twice the value obtained in the blank was considered as the titer value.
Western blot assays.Growth conditions. M. bovis BCG Moreau,
M. tuberculosis H27Ra cells (avirulent strains) and
M. tuberculosis H37Rv were grown in 100 ml of 7H9 medium with 43 mM sodium pyruvate, supplemented with 10% OADC until they reached an OD600 of 0.4. To induce the Rv1819c production, cultures were treated with ½ MIC of Isoniazid (0.015 µg /ml) for 24 hours [
12].
Cellular extract preparation. M. bovis BCG and M. tuberculosis H37Ra 50 mL cultures were centrifuged, resuspended in 5 mL of PBS, PMSF 1 mM and lysozyme 1 mg/mL [
13,
14], and homogenized for 15 minutes at 4ºC. Then, the cultures were sonicated for 10 minutes (10 second pulse, 20 second pause). The total cell lysates were centrifuged for 1 hour, 27.000 g before applying it to a 12%, 1 mm polyacrylamide gel.
M. tuberculosis H37Rv culture was centrifuged and resuspended in 10 ml of PBS, 1% PMSF and 1 mg/mL lysozyme and fractionated into 10 2 ml tubes containing ice-cold 0.1 mm silica beads. The tubes were positioned in the cell disruptor Tissue Lyser LT (QIAGEN) and cell lysis was performed with 2 sets of 2.5 min with an oscillation of 40. The lysate was transferred to clean tubes and centrifuged at 15,000 rpm to obtain the soluble fraction, which was filtered through a 0.22 µm syringe filter.
Membrane fractions were obtained according to Mot and Vanderleyden [
15]. After centrifugation at 30.500 g for 1 hour at 4°C to eliminate the cell wall fraction, the supernatant was centrifuged at 100.000 g for 1 h. The supernatant was gently removed and the pellet, containing the membrane protein fraction, was taken with 1X PBS and centrifuged again at 100,000 g for 1 h. The supernatant was removed, and the pellet was homogenized in buffer containing 1% (w/v) sodium deoxycholate detergent for 1 hour at 15ºC. The solution was centrifuged again at 100.000 g for 1 h and the supernatant containing the solubilized proteins was gently collected and stored at -20ºC.
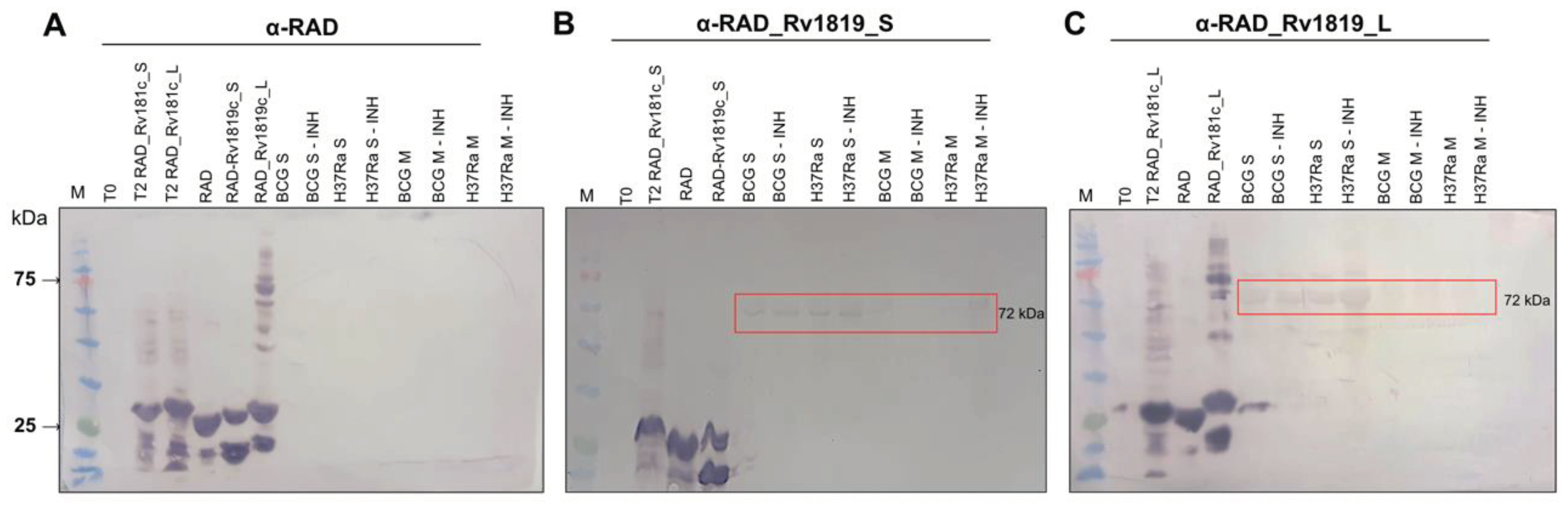
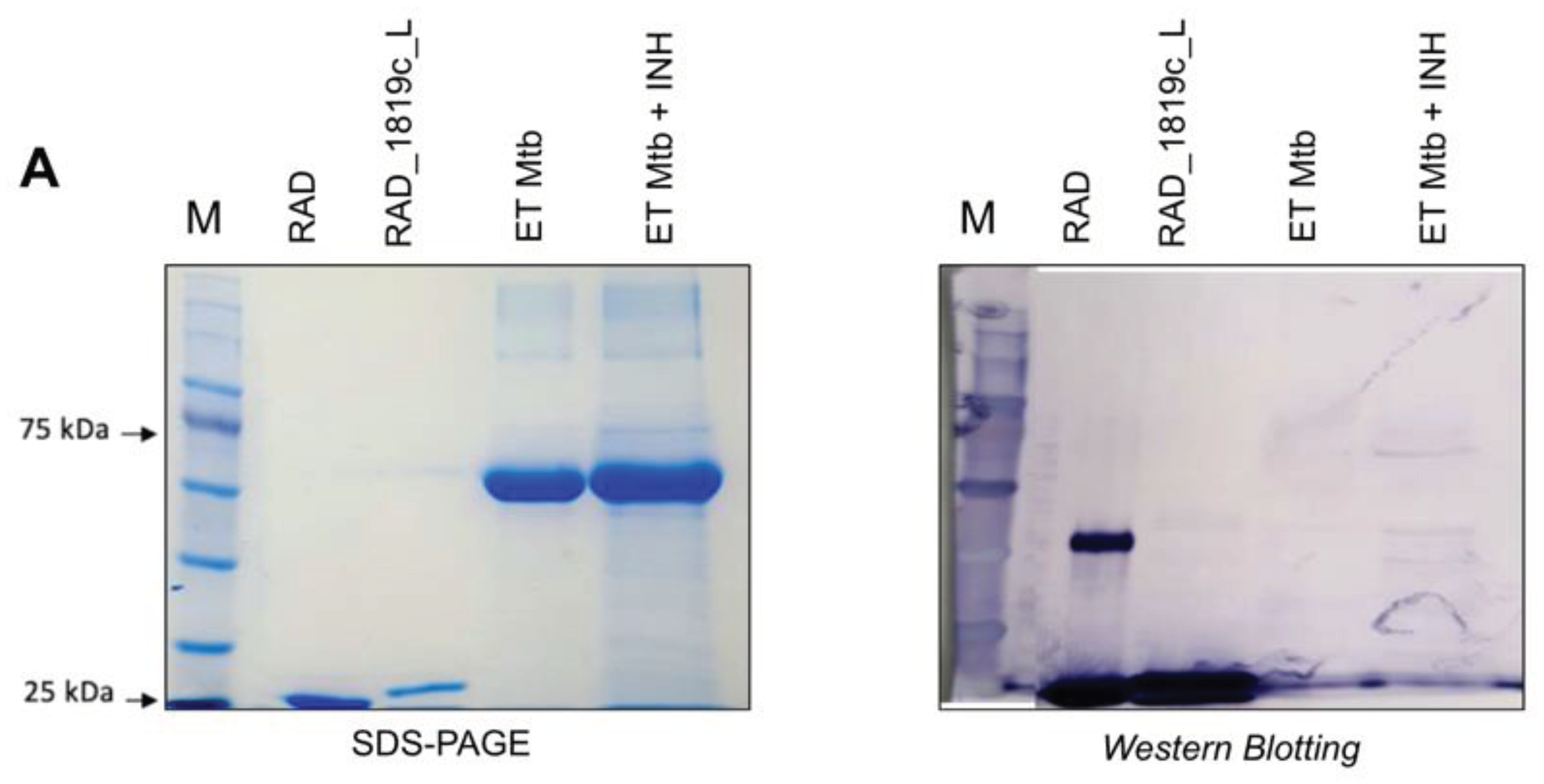
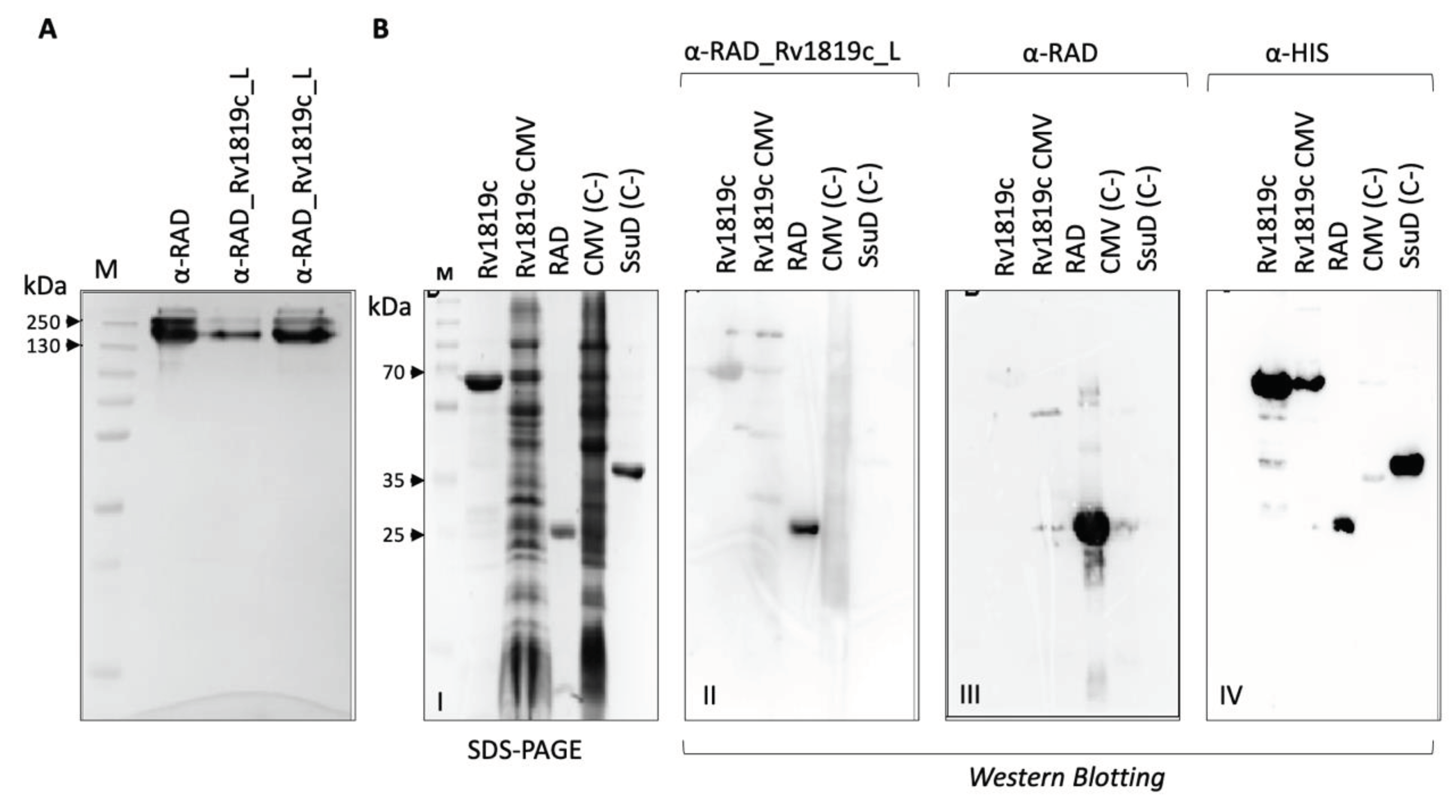
Western blot. For analysis of antibody binding to mycobacterial protein extracts, 35 µg/well of soluble and membrane fractions were transferred from the SDS-PAGE gel to the PVDF membrane. The transfer was carried out on a PVDF membrane at 15 V for 30 minutes in a Trans-Blot SD semi-dry transfer cell (BioRad). They were then incubated for 1 hour at room temperature under agitation in blocking buffer (TBST, Gelatin 3%). After a cycle of three 5-minute washes (TBST), the membranes were incubated with serum containing specific primary antibodies at a 1:400 dilution in buffer (TBST, Gelatin 1%) for 4 hours at room temperature. Then, the membranes were submitted to a new washing cycle and incubated with the anti-mouse IgG secondary antibody conjugated to alkaline phosphatase (A4312 – Sigma-Aldrich) at a dilution of 1:10,000 for 1 h in buffer (TBST, Gelatin 1%). Three 5-minute washes were performed again (TBST) and the membranes were developed with BCIP/NBT. For analysis of antibody binding to the purified Rv1819c transporter, purified proteins and membrane fractions were transferred from the SDS-PAGE gel to nitrocellulose membranes at 25 V, 7 minutes in a Trans-blot Turbo (BioRad). The membranes were incubated overnight in blocking buffer (I-Block 0.2%, Thermo Fisher Scientific) at 4ºC. Each of the three membranes was incubated with a different primary antibody diluted in TBST, I-Block (0.1%): α-RAD_Rv1819c_L (I), dilution 1:375 (concentration 1.515 mg/mL) and α-RAD (II) (Negative control for Rv1819c recognition), dilution 1:400 (concentration 2.230 mg/mL) both using anti-mouse alkaline phosphatase as secondary antibody (A4312) at a dilution of 1:5000 and Anti-poly-his peroxidase antibody (III) (A7058), dilution 1:5000.
Purification of α-RAD_RV1819c_L and α-RAD antibodies. To purify InG antibodies from the sera obtained after immunization of mice with RAD and RAD_Rv1819c_L, the Serum Antibody Purification Kit (Protein G) (ABCAM) was used. 500 µL of animal serum was centrifuged for 10 min at 20.000 g at 4ºC. The sample was diluted to 5 ml in wash buffer with the addition of 500 µL of 10x binding buffer. The sample was decanted 4 times in a column containing 1 ml of protein G resin, which was then washed with 6 ml of washing buffer. Elutions were made with 6 fractions of 500 µL of elution buffer. Each elution was done in tubes containing 100 µL of neutralization buffer. Samples were analyzed on SDS-PAGE gel and on Nanodrop (ThermoScientific).
Expression and purification of the M. tuberculosis Rv1819c transporter.Protein expression. E. coli MC1061 cells containing the pBAD24_Rv1819c_cHis8 vector were inoculated into 8 L of LB medium at a ratio of 1:100 containing ampicillin. Expression was performed in 5 L finned Erlenmeyer flasks containing 2 L of LB each. The culture was incubated at 37ºC, 200 rpm until reaching an OD600nm of 0.8 and expression was induced with 0.02% (w/v) L-arabinose. Expression occurred for 3 h under the same conditions and cells were recovered by centrifugation at 6000 g, 15 min, 4ºC. The pellet was resuspended in buffer with 50 mM Kpi pH 7.5, 4ºC to wash the cells, centrifuged again and resuspended in the same buffer added with 20% glycerol in a total volume of 100 ml. The suspension was quickly frozen in liquid nitrogen and stored at -80ºC. Preparation of membrane vesicles of Rv1819c transporter. The cell suspension containing the Rv1819c transporter was thawed on ice and added with a protease inhibitor tablet (Protease Inhibitor Cocktail, Roche), 0.2 mM PMSF, 1 mM MgSO4 and 20 µg/mL DNAseI. For cell lysis, the cells were subjected twice to the cell disruptor (HPL6, Maximator) previously cooled to 4ºC and equilibrated with the same buffer. The soluble fraction was obtained from centrifugation of the lysate at 16,000 g (JA-17 Fixed-Angle Rotor), 40 min, 4oC. To obtain the membrane fraction, the supernatant was subjected to ultracentrifugation at 40.000 rpm for 2.5 hours at 4ºC with the 45 TI rotor. The membrane fraction was resuspended in 50 mM Kpi buffer pH 7.5, 10% glycerol, 1 mM DTT with a “potter” homogenizer. The membranes were frozen in liquid nitrogen and stored at -80ºC. Purification of the ABC transporter Rv1819c. Purification of the transporter in 50 mM Tris pH 8.0. 300 mM NaCl, 15 mM Imidazole, 1% DDM/CHS, 10% Glycerol, 1 mM DTT was carried out using the nickel affinity chromatography technique with Ni Sepharose 6 Fast Flow resin (Cytiva Life Sciences) using 50 mM Tris pH 8.0. 300 mM NaCl, 15 mM Imidazole, 10% Glycerol, 1 mM DTT as equilibrium buffer, 50 mM Tris pH 8.0. 300 mM NaCl, 60 mM Imidazole, 0.05% DDM/CHS, 1 mM DTT as wash buffer and 50 mM Tris pH 8.0. 300 mM NaCl, 350 mM Imidazole, 0.05% DDM/CHS, 1 mM DTT as elution buffer. After IMAC, a SEC using the Superdex 200 Increase 10/300 column (Cytiva Life Sciences) in a Bio-Rad system was performed in buffer 25 mM Tris pH 8.0. 200 mM NaCl, 0.05% DDM/CHS, 1 mM DTT. A tube (500 µl) of membrane preparation was thawed on ice, solubilized in 10 ml of solubilization buffer and incubated for 1 hour at 4ºC under gentle agitation. The membrane solubilized in detergent was subjected to ultracentrifugation at 80,000 rpm for 25 min at 4ºC in the MLA80 rotor. Then, the supernatant was incubated with 1 ml of Ni-Sepharose 6 Fast Flow resin (50% v/v) in equilibration buffer for 1 hour at 4ºC under gentle agitation. Due to the DTT contained in the equilibration buffer, the resin must be pre-equilibrated with wash buffer. After incubation, the sample was decanted into a chromatography column. The resin is then washed with 20 CV of wash buffer. Elution is carried out first with 450 µl of buffer and then with 800 µl. The fractions were analyzed in Nanodrop (Thermo Fisher Scientific), centrifuged at 20.000 g, 5 min at 4ºC to remove aggregated proteins and then subjected to SEC on the column Superdex 200 increase 10/300 GL column equilibrated with buffer. The fractions referring to the chromatogram peaks were analyzed using the Nanodrop (Themo Fisher Scientific).
Statistical analysis. Statistical analyses were performed wih Graphpad Prism version 6. Specific statistical tests employed are addressed in figure legends.
4. Discussion
Antibodies are an interesting resource for studies of membrane proteins such as identification, exploring conformational changes, transport inhibition and localization [
2,
17]. In structural biology, antibodies can be used to increase the size of the targets, facilitating the reconstruction of the 3D structure in cryo-EM studies [
18,
19,
20], expanding the surface area available for the formation of crystal contacts in protein crystallization or still ensuring less flexibility and heterogeneity of samples for the formation of crystals [
1,
21,
22].
In clinical practice, detecting membrane proteins, is crucial as it helps identify mechanisms potentially linked to distinct diseases and increased drug resistance [
23,
24]. Efflux pumps are one of the most important drug resistance mechanisms involved in cancer treatment and have been considered important targets for modulation [
25]. Similarly, efflux pumps are upregulated in bacteria during treatment with antibiotics and could become candidate tools to treat infectious diseases [
26,
27]. However, the use of other targets like the transmembrane permeases for these means is sparsely explored, primarily due to the complications in their production, high hydrophobicity of the TM segments necessitating the use of detergents and uncertain immunogenicity of these proteins [
28,
29].
In this work, aiming to avoid the problems of membrane protein expression and purification, we present the RAD display as a quick and effective alternative for expressing small epitopes from membrane proteins as antigens for antibody production. As a model, we used a well characterized ABC transporter for vitamin B12 from
M. tuberculosis, Rv1819c. Rv1819c is needed for the maintenance of chronic infection in mice, mediates the uptake of antimicrobial peptides and other toxins, and is also essential for the import of vitamin B12 (cobalamin) in
M. tuberculosis [
8,
9,
30]. Once its three-dimensional structure is available in different conformations [
9], we could explore extracellular regions of permeases exposed to the periplasm. The choice of these specific regions was based on their localization (outside the membrane) and in the possibility that antibodies that target those regions could inhibit the transport. The constructs with the two potential immunogenic epitopes (short and long) were expressed in a soluble form and hence easily purified using Ni-IMAC followed by SEC.
Via in silico analysis the long epitope of Rv1819 TMD was predicted by IEDB to be more immunogenic than the short one and the ELISA results confirmed the superiority of a longer epitope. Fortunately, the RAD-display system easily tolerates the insertion of long peptides, without much restriction on their folding due to the flexibility of the system. In addition, our results confirm that the IEDB can be reliable tool for preliminary analysis of peptides, with greater agility and speed in the screening process for possible antigens of membrane proteins.
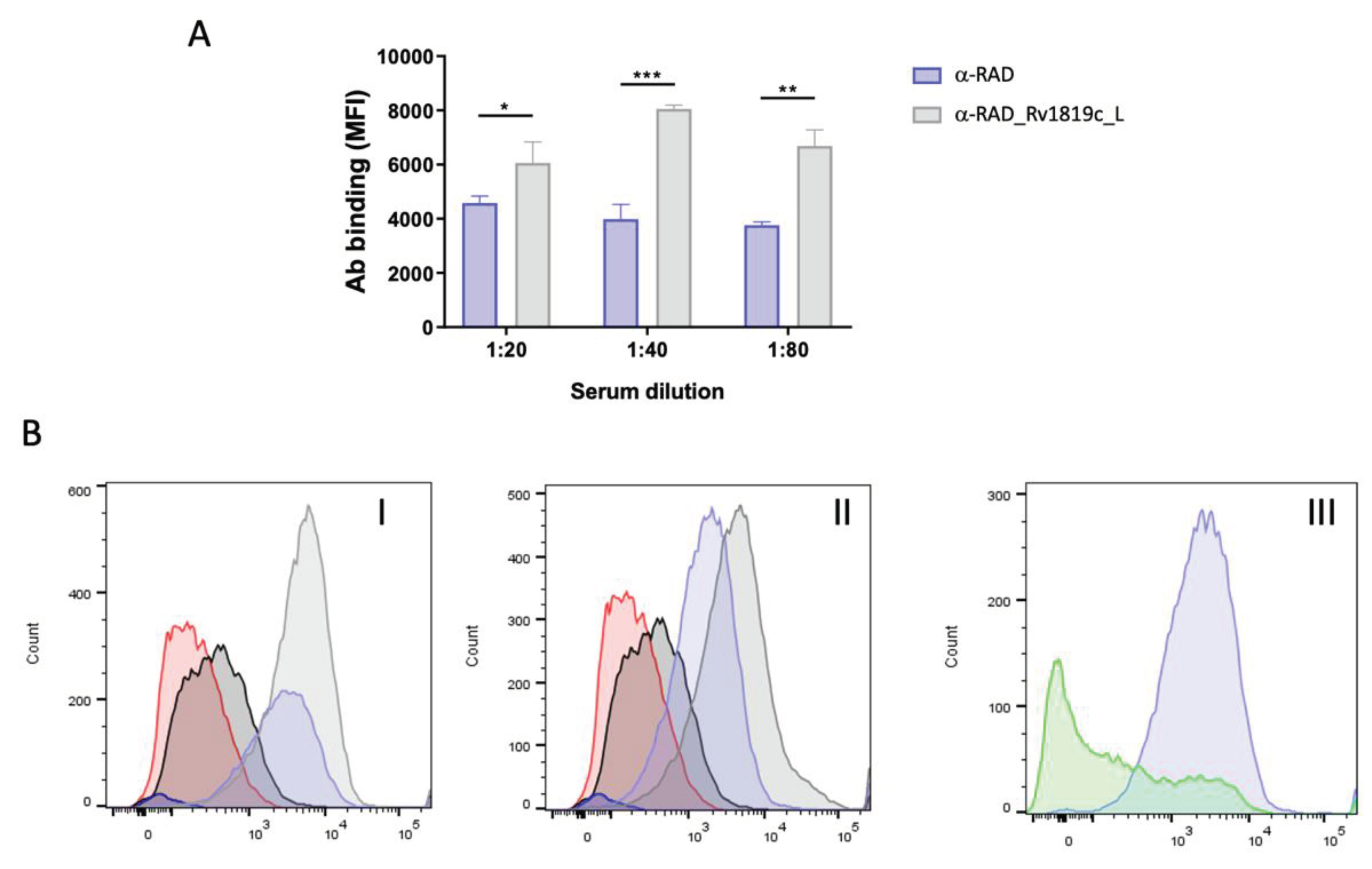
Interestingly, although the antibody titers produced against the short peptide were not as abundant as for the long peptide, when we take into account the recognition of the RAD scaffold, these antibodies also recognized the transporter in soluble fractions of
M. bovis BCG and
M. tuberculosis H37Ra suggesting both epitopes were able to induce antibodies’ production. Rv1819c shares 99.7% and 100% of sequence identity with its orthologs from
M. bovis and
M. tuberculosis H37Ra, respectively, and the sequences chose were identical. The Rv1819c corresponding gene in these lineages is assigned as probable drug transporter. The induction of the transporter from
M. bovis has been shown in isolates from different animals that show resistance not only to isoniazid but also to other first and second line antibiotics [
31]. However, it was not detected in the membrane fractions, where we anticipated stronger recognition. It is possible that the protocol used and treatment of cells to obtain membrane fractions was more harmful. Otherwise, in
M. tuberculosis H37Rv, the results show that INH has an effect in western blot assays and in flow cytometry. In flow cytometry, we observed that α-RAD antibodies have nonspecific interactions with the
M. tuberculosis cells, but not with the
Xanthomonas citri cells. Even so, the MFI observed for the α-RAD_Rv1819c_L antibodies in comparison with the control cells and exposed to α-RAD was significantly higher. Importantly, we must consider that we worked with a sample of polyclonal antibodies and that the target peptide corresponds to a small portion of the RAD display. In fact, it is not possible to quantify how much of peptide-specific antibodies are present in the sample. Therefore, the recognition of the purified Rv1819c corroborated the previous results and showed the strategy applied with RAD display can be a useful tool for screening of efflux pumps epitopes, with different perspectives.