Submitted:
28 March 2024
Posted:
28 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
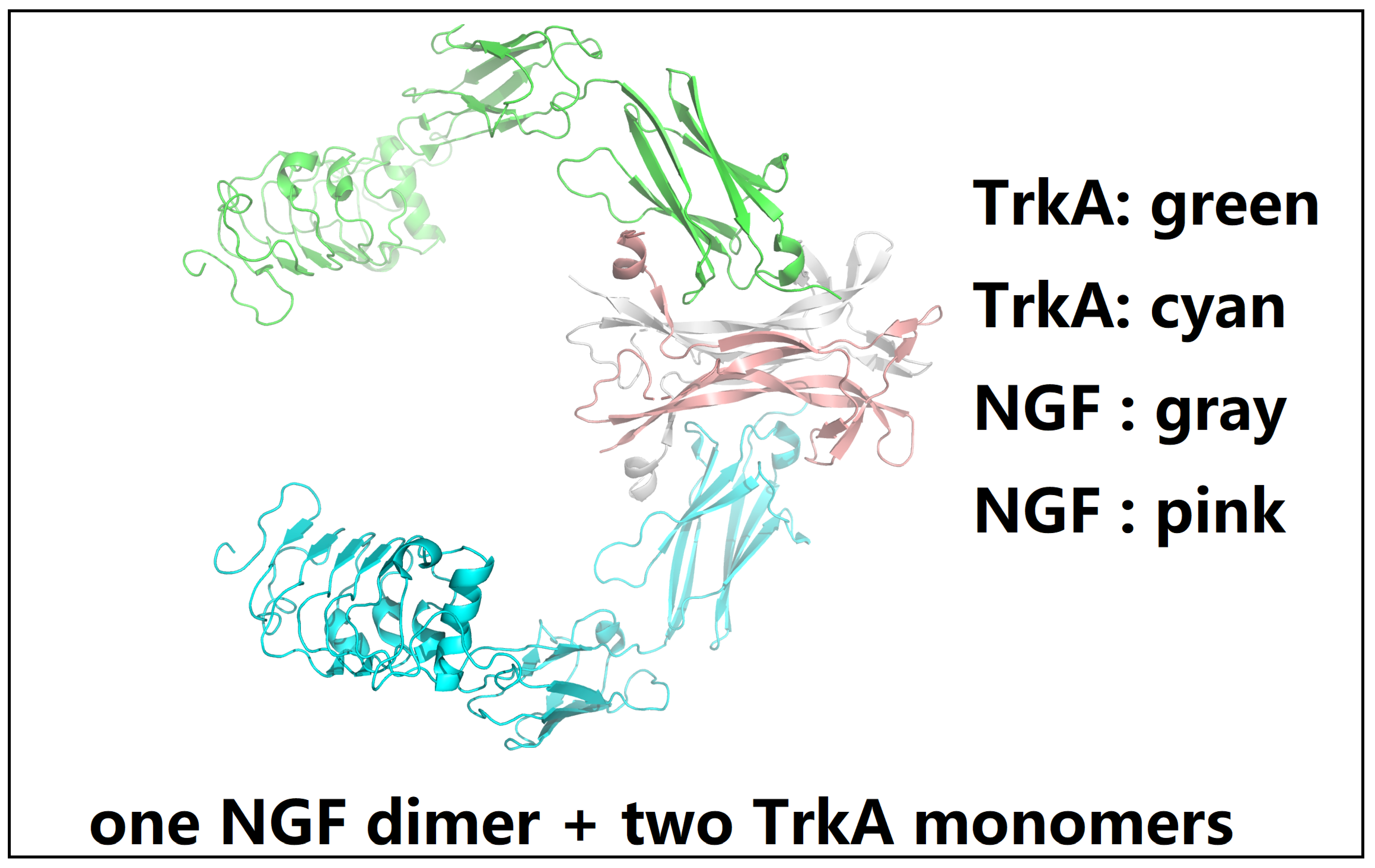
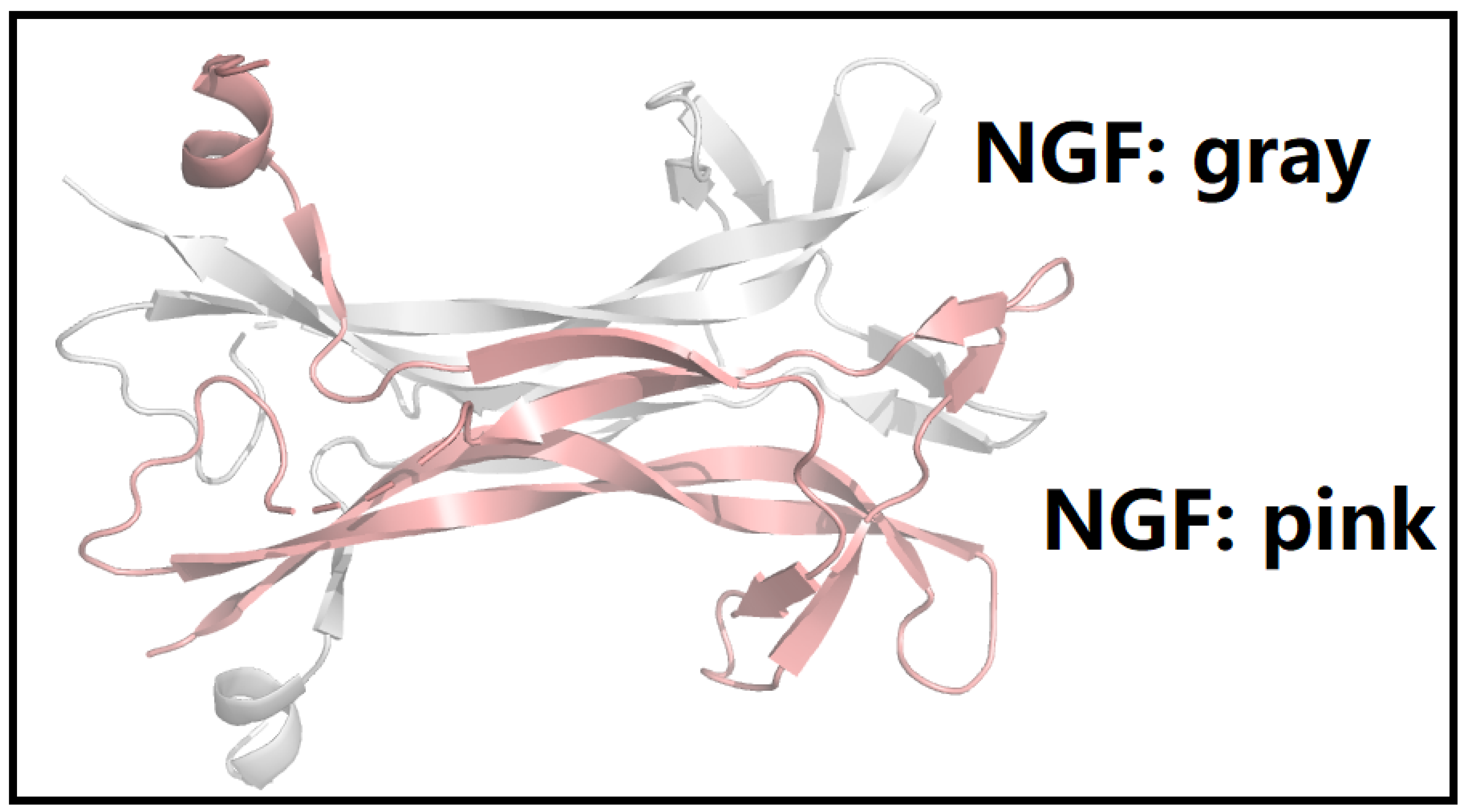
2.1. A Comprehensive Structural and Biophysical Analysis of the p75NTR-NGF-TrkA Complex
2.2. The Design of NGF Analogues to Suppress Pain Signal Transduction Mediated by the p75NTR-NGF-TrkA Complex
3. Results
- 1.
- 2.
- 3.
| Mutation | Energy mean | Energy std | Kd mean | Kd std |
|---|---|---|---|---|
| S4F | -10.270 | 0.671 | 8.953000000000001e-08 | 6.715793400634061e-08 |
| S4A | -10.310 | 0.342 | 6.23e-08 | 3.199703111227665e-08 |
| S4P | -10.320 | 0.389 | 6.8e-08 | 6.065311203887233e-08 |
| S4V | -10.370 | 0.335 | 5.59e-08 | 2.6135990511170603e-08 |
| S4I | -10.370 | 0.377 | 6.05e-08 | 4.102499238269277e-08 |
| S4M | -10.450 | 0.478 | 5.9399999999999996e-08 | 4.9218289283558e-08 |
| S4L | -10.560 | 0.578 | 5.859e-08 | 7.30497973987608e-08 |
| S4Y | -10.650 | 0.301 | 3.52e-08 | 1.70926884953772e-08 |
| S4G | -10.660 | 0.258 | 3.24e-08 | 1.3162066707018317e-08 |
| S4C | -10.850 | 0.461 | 2.911e-08 | 1.89068479657504e-08 |
| A98S | -10.860 | 0.595 | 3.832e-08 | 5.483358095182185e-08 |
| S4K | -11.020 | 0.328 | 1.958e-08 | 1.4229111005259603e-08 |
| A98Q | -11.070 | 0.454 | 2.023e-08 | 1.62038297942184e-08 |
| A98T | -11.150 | 0.364 | 1.6529999999999996e-08 | 1.0195494102788742e-08 |
| S4E | -11.180 | 0.464 | 1.726e-08 | 1.4366293885341482e-08 |
| S104I | -11.180 | 0.447 | 1.645e-08 | 1.0609736094738644e-08 |
| S4D | -11.200 | 0.410 | 1.5650000000000004e-08 | 1.0420772524146184e-08 |
| S104L | -11.260 | 0.250 | 1.24e-08 | 3.7812696280482305e-09 |
| N36A | -11.260 | 0.242 | 1.24e-08 | 4.991192242340502e-09 |
| S4W | -11.280 | 0.440 | 1.394e-08 | 8.486247698482527e-09 |
| N36P | -11.310 | 0.262 | 1.1369999999999999e-08 | 4.242652472215937e-09 |
| I35S | -11.370 | 0.618 | 1.4880000000000002e-08 | 1.4351431984300383e-08 |
| S104P | -11.420 | 0.392 | 1.025e-08 | 5.6498230060772695e-09 |
| S104F | -11.420 | 0.421 | 1.1350000000000002e-08 | 8.690368231553827e-09 |
| S104G | -11.430 | 0.473 | 1.126e-08 | 7.755282070950095e-09 |
| S4H | -11.440 | 0.472 | 1.229e-08 | 1.466243158551814e-08 |
| S104A | -11.460 | 0.361 | 9.48e-09 | 4.582313826005372e-09 |
| N36G | -11.470 | 0.323 | 9.34e-09 | 5.523078851510269e-09 |
| I35T | -11.500 | 0.490 | 1.1449999999999998e-08 | 1.2621509418449125e-08 |
| S104V | -11.540 | 0.508 | 1.091e-08 | 1.3590912404985913e-08 |
| Nddm | -12.700 | 0.000 | 1.2e-09 | 0.0 |
| Mutation | Energy mean | Energy std | Kd mean | Kd std |
|---|---|---|---|---|
| S4F_A98G | -8.600 | 0.000 | 9.1e-07 | 0.0 |
| S4A_L81D | -8.600 | 0.000 | 8.3e-07 | 0.0 |
| S4Y_L81A | -8.700 | 0.000 | 7.8e-07 | 0.0 |
| S4A_I22G | -8.800 | 0.000 | 6.3e-07 | 0.0 |
| S4A_D63G | -8.800 | 0.000 | 6e-07 | 0.0 |
| S4G_A98Q | -8.900 | 0.000 | 5.5e-07 | 0.0 |
| S4G_A88E | -8.900 | 0.000 | 5.2e-07 | 0.0 |
| S4A_V33A | -8.900 | 0.000 | 5.1e-07 | 0.0 |
| S4F_D63I | -9.000 | 0.000 | 4.6e-07 | 0.0 |
| S4A_A98Q | -9.000 | 0.000 | 4.4e-07 | 0.0 |
| S4Y_G1Q | -9.100 | 0.000 | 3.8e-07 | 0.0 |
| S4A_I22F | -9.100 | 0.000 | 3.9e-07 | 0.0 |
| S4G_E46D | -9.100 | 0.000 | 4.1e-07 | 0.0 |
| S4G_K79E | -9.100 | 0.000 | 3.7e-07 | 0.0 |
| S4G_H66A | -9.100 | 0.000 | 3.9e-07 | 0.0 |
| S4A_K106L | -9.100 | 0.000 | 3.9e-07 | 0.0 |
| S4Y_A89L | -9.200 | 0.000 | 3.4e-07 | 0.0 |
| S4G_W90P | -9.200 | 0.000 | 3.4e-07 | 0.0 |
| S4A_K65R | -9.200 | 0.000 | 3.2e-07 | 0.0 |
| S4G_L81E | -9.300 | 0.000 | 2.6e-07 | 0.0 |
| S4A_I95A | -9.300 | 0.000 | 2.9e-07 | 0.0 |
| S4G_L30M | -9.300 | 0.000 | 2.9e-07 | 0.0 |
| S4G_W90G | -9.300 | 0.000 | 2.7e-07 | 0.0 |
| S4L_H75I | -9.300 | 0.000 | 3e-07 | 0.0 |
| Nddm | -12.700 | 0.000 | 1.2e-09 | 0.0 |
| Mutation | Energy mean | Energy std | Kd mean | Kd std |
|---|---|---|---|---|
| S4A_A98Q_V33A | -7.400 | 0.000 | 5.8e-06 | 0.0 |
| S4A_A98Q_V33G | -7.400 | 0.000 | 6.1e-06 | 0.0 |
| S4G_A98Q_K79P | -7.500 | 0.000 | 4.7e-06 | 0.0 |
| S4G_A98Q_W90A | -7.500 | 0.000 | 4.8e-06 | 0.0 |
| S4Y_A98Q_R105I | -7.500 | 0.000 | 5.5e-06 | 0.0 |
| S4Y_A98Q_K41P | -7.600 | 0.000 | 4.2e-06 | 0.0 |
| S4F_A98Q_Y70L | -7.700 | 0.000 | 3.5e-06 | 0.0 |
| S4G_A98Q_W90V | -7.700 | 0.000 | 4e-06 | 0.0 |
| S4F_A98Q_K41V | -7.700 | 0.000 | 3.5e-06 | 0.0 |
| S4G_A98Q_K79M | -7.700 | 0.000 | 3.5e-06 | 0.0 |
| S4A_A98Q_L81A | -7.700 | 0.000 | 3.5e-06 | 0.0 |
| S4Y_A98Q_K41V | -7.800 | 0.000 | 3.2e-06 | 0.0 |
| S4A_A98Q_W12G | -7.800 | 0.000 | 3.1e-06 | 0.0 |
| S4F_A98Q_L81K | -7.800 | 0.000 | 3e-06 | 0.0 |
| S4G_A98Q_Q42G | -7.800 | 0.000 | 2.9e-06 | 0.0 |
| S4L_L81G_I35Q | -7.800 | 0.000 | 3e-06 | 0.0 |
| S4G_A98Q_K41P | -7.800 | 0.000 | 3.4e-06 | 0.0 |
| S4G_A98Q_D63G | -7.900 | 0.000 | 2.9e-06 | 0.0 |
| S4F_A98Q_K79G | -7.900 | 0.000 | 2.5e-06 | 0.0 |
| S4G_A98Q_R105G | -7.900 | 0.000 | 2.5e-06 | 0.0 |
| Nddm | -12.700 | 0.000 | 1.2e-09 | 0.0 |
4. Conclusions and Discussion
5. Ethical Statement
6. Declaration of Generative AI and AI-Assisted Technologies in the Writing Process
Author Contributions
Funding
Conflicts of Interest
References
- Rankin, S.L.; Guy, C.S.; Mearow, K.M. TrkA NGF receptor plays a role in the modulation of p75NTR expression. Neuroscience Letters 2005, 383, 305–310. [Google Scholar] [CrossRef]
- White, D.M.; Mansfield, K.; Kelleher, K. Increased neurite outgrowth of cultured rat dorsal root ganglion cells following transection or inhibition of axonal transport of the sciatic nerve. Neuroscience Letters 1996, 208, 93–96. [Google Scholar] [CrossRef]
- Xu, Z.L.; Chen, G.; Liu, X.; Xie, D.; Zhang, J.; Ying, Y. Effects of ginsenosides on memory impairment in propofol-anesthetized rats. Bioengineered 2021, 13, 617–623. [Google Scholar] [CrossRef]
- Genty, J.; Tetsi Nomigni, M.; Anton, F.; Hanesch, U. The combination of postnatal maternal separation and social stress in young adulthood does not lead to enhanced inflammatory pain sensitivity and depression-related behavior in rats. PLOS ONE 2018, 13, e0202599. [Google Scholar] [CrossRef]
- Koo, H.M.; Yong, M.S.; Na, S.S. The effect of low-intensity laser therapy (LILT) on cutaneous wound healing and pain relief in rats. Journal of Physical Therapy Science 2015, 27, 3421–3423. [Google Scholar] [CrossRef]
- Bishop, T.; Hewson, D.W.; Yip, P.K.; Fahey, M.S.; Dawbarn, D.; Young, A.R.; McMahon, S.B. Characterisation of ultraviolet-B-induced inflammation as a model of hyperalgesia in the rat. Pain 2007, 131, 70–82. [Google Scholar] [CrossRef]
- Kashyap, M.P.; Roberts, C.; Waseem, M.; Tyagi, P. Drug Targets in Neurotrophin Signaling in the Central and Peripheral Nervous System. Molecular Neurobiology 2018, 55, 6939–6955. [Google Scholar] [CrossRef]
- Das, V.; Kc, R.; Li, X.; O-Sullivan, I.; van Wijnen, A.J.; Kroin, J.S.; Pytowski, B.; Applegate, D.T.; Votta-Velis, G.; Ripper, R.L.; et al. Blockade of vascular endothelial growth factor receptor-1 (Flt-1), reveals a novel analgesic for osteoarthritis-induced joint pain. Gene Reports 2018, 11, 94–100. [Google Scholar] [CrossRef]
- Gugliandolo, E.; Peritore, A.f.; Impellizzeri, D.; Cordaro, M.; Siracusa, R.; Fusco, R.; D’Amico, R.; Di Paola, R.; Schievano, C.; Cuzzocrea, S.; et al. Dietary Supplementation with Palmitoyl-Glucosamine Co-Micronized with Curcumin Relieves Osteoarthritis Pain and Benefits Joint Mobility. Animals 2020, 10, 1827. [Google Scholar] [CrossRef]
- Vellani, V.; Kinsey, A.M.; Prandini, M.; Hechtfischer, S.C.; Reeh, P.; Magherini, P.C.; Giacomoni, C.; McNaughton, P.A. Protease Activated Receptors 1 and 4 Sensitize TRPV1 in Nociceptive Neurones. Molecular Pain 2010, 6, 1744-8069-6-61. [Google Scholar] [CrossRef]
- Kingery, W.S. Role of Neuropeptide, Cytokine, and Growth Factor Signaling in Complex Regional Pain Syndrome. Pain Medicine 2010, 11, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Huang, Z.; Zhang, L.; Li, X.; Xu, B.; Xiao, Y.; Shi, X.; Zhang, H.; Liao, T.; Wang, P. Vanillic Acid Reduces Pain-Related Behavior in Knee Osteoarthritis Rats Through the Inhibition of NLRP3 Inflammasome-Related Synovitis. Frontiers in Pharmacology 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Day-Williams, A.; Southam, L.; Panoutsopoulou, K.; Rayner, N.; Esko, T.; Estrada, K.; Helgadottir, H.; Hofman, A.; Ingvarsson, T.; Jonsson, H.; et al. A Variant in MCF2L Is Associated with Osteoarthritis. The American Journal of Human Genetics 2011, 89, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Isa, I.L.M.; Srivastava, A.; Tiernan, D.; Owens, P.; Rooney, P.; Dockery, P.; Pandit, A. Hyaluronic Acid Based Hydrogels Attenuate Inflammatory Receptors and Neurotrophins in Interleukin-1β Induced Inflammation Model of Nucleus Pulposus Cells. Biomacromolecules 2015, 16, 1714–1725. [Google Scholar] [CrossRef]
- Facer, P.; Casula, M.A.; Smith, G.D.; Benham, C.D.; Chessell, I.P.; Bountra, C.; Sinisi, M.; Birch, R.; Anand, P. Differential expression of the capsaicin receptor TRPV1 and related novel receptors TRPV3, TRPV4 and TRPM8 in normal human tissues and changes in traumatic and diabetic neuropathy. BMC Neurology 2007, 7. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Ustinova, E.E.; Patnam, R.; Fraser, M.O.; Gutkin, D.W.; Pezzone, M.A. Enhanced expression of mast cell growth factor and mast cell activation in the bladder following the resolution of trinitrobenzenesulfonic acid (TNBS) colitis in female rats. Neurourology and Urodynamics 2007, 26, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.P.; Tendi, E.A.; Dib-Hajj, S.D.; Fields, R.D.; Waxman, S.G. Patterned electrical activity modulates sodium channel expression in sensory neurons. Journal of Neuroscience Research 2003, 74, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Wijaya, L.K.; Stumbles, P.A.; Drummond, P.D. Tumor necrosis factor α induces α1B-adrenergic receptor expression in keratinocytes. Cytokine 2020, 125, 154851. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, T.F.; Baracat, E.C.; Doumouchtsis, S.K.; Haddad, J.M. Biomarkers in the diagnosis and symptom assessment of patients with bladder pain syndrome: A systematic review. International Urogynecology Journal 2019, 30, 1785–1794. [Google Scholar] [CrossRef]
- Li, B.; Jing, L.; Jia, L.; Qian, T.; Jianyi, C.; Zhongsheng, H.; Xiaohong, Z.; Guowei, C. Acupuncture reduces pain in rats with osteoarthritis by inhibiting MCP2CCR2 signaling pathway. Experimental Biology and Medicine 2020, 245, 1722–1731. [Google Scholar] [CrossRef]
- D’Arcy, Y.; Mantyh, P.; Yaksh, T.; Donevan, S.; Hall, J.; Sadrarhami, M.; Viktrup, L. Treating osteoarthritis pain: Mechanisms of action of acetaminophen, nonsteroidal anti-inflammatory drugs, opioids, and nerve growth factor antibodies. Postgraduate Medicine 2021, 133, 879–894. [Google Scholar] [CrossRef] [PubMed]
- Borgonio Cuadra, V.M.; González-Huerta, N.C.; Romero-Córdoba, S.; Hidalgo-Miranda, A.; Miranda-Duarte, A. Altered Expression of Circulating MicroRNA in Plasma of Patients with Primary Osteoarthritis and In Silico Analysis of Their Pathways. PLoS ONE 2014, 9, e97690. [Google Scholar] [CrossRef] [PubMed]
- Schrenker, S.; Gao, L.; Cucchiarini, M.; Madry, H. Future Aspects of Clinical Osteoarthritis Therapies in the Continuum of Translational Research. Zeitschrift für Orthopädie und Unfallchirurgie 2019, 157, 629–643. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Zhang, M.; Jin, Y.; Di, X.; Liu, R.; Wang, Z. Safety, Tolerability, Pharmacokinetics, and Immunogenicity of a Novel Recombination Human Nerve Growth Factor in Healthy Chinese Subjects. CNS Drugs 2023, 37, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Kissel, C.; Kovács, K.; Larson, A. Evidence for the modulation of nociception in mice by central mast cells. European Journal of Pain 2017, 21, 1743–1755. [Google Scholar] [CrossRef] [PubMed]
- LA Binch, A.; Cole, A.A.; Breakwell, L.M.; Michael, A.L.; Chiverton, N.; Cross, A.K.; Le Maitre, C.L. Expression and regulation of neurotrophic and angiogenic factors during human intervertebral disc degeneration. Arthritis Research: Therapy 2014, 16. [Google Scholar] [CrossRef] [PubMed]
- Joseph, E.; Levine, J. Mu and delta opioid receptors on nociceptors attenuate mechanical hyperalgesia in rat. Neuroscience 2010, 171, 344–350. [Google Scholar] [CrossRef] [PubMed]
- FATANI, A.J.; AL-REJAIE, S.S.; ABUOHASHISH, H.M.; AL-ASSAF, A.; PARMAR, M.Y.; OLA, M.S.; AHMED, M.M. Neuroprotective effects of Gymnema sylvestre on streptozotocin-induced diabetic neuropathy in rats. Experimental and Therapeutic Medicine 2015, 9, 1670–1678. [Google Scholar] [CrossRef] [PubMed]
- Cruise, B.A.; Xu, P.; Hall, A.K. Wounds increase activin in skin and a vasoactive neuropeptide in sensory ganglia. Developmental Biology 2004, 271, 1–10. [Google Scholar] [CrossRef]
- Shaqura, M.; Khalefa, B.I.; Shakibaei, M.; Winkler, J.; Al-Khrasani, M.; Fürst, S.; Mousa, S.A.; Schäfer, M. Reduced Number, G Protein Coupling, and Antinociceptive Efficacy of Spinal Mu-Opioid Receptors in Diabetic Rats Are Reversed by Nerve Growth Factor. The Journal of Pain 2013, 14, 720–730. [Google Scholar] [CrossRef]
- Sharma, H.S.; F. Ali, S.; Patnaik, R.; Zimmermann-Meinzingen, S.; Sharma, A.; F. Muresanu, D. Cerebrolysin Attenuates Heat Shock Protein (HSP 72 KD) Expression in the Rat Spinal Cord Following Morphine Dependence and Withdrawal: Possible New Therapy for Pain Management. Current Neuropharmacology 2011, 9, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Tomalty, D.; Giovannetti, O.; Magliocchetti, S.; Williams, A.; Hannan, J.; Komisaruk, B.; Goldstein, S.; Goldstein, I.; Adams, M.A. Characterizing the innervation of the vulvar vestibule and the immunohistochemical features of neuroproliferative vestibulodynia. The Journal of Sexual Medicine 2023, 20, 716–731. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Muralidharan, A.; Smith, M.T. Attenuation of the Infiltration of Angiotensin II Expressing CD3+ T-Cells and the Modulation of Nerve Growth Factor in Lumbar Dorsal Root Ganglia - A Possible Mechanism Underpinning Analgesia Produced by EMA300, An Angiotensin II Type 2 (AT2) Receptor Antagonist. Frontiers in Molecular Neuroscience 2017, 10. [Google Scholar]
- Palma, J.A.; Yadav, R.; Gao, D.; Norcliffe-Kaufmann, L.; Slaugenhaupt, S.; Kaufmann, H. Expanding the Genotypic Spectrum of Congenital Sensory and Autonomic Neuropathies Using Whole-Exome Sequencing. Neurology Genetics 2021, 7, e568. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.C.; Fu, W.M.; Huang, H.I.; Lai, Y.H.; Tsai, Y.F.; Guo, S.L.; Wu, T.J.; Ling, Q.D. Expression of Neurotrophic Factors in Neonatal Rats After Peripheral Inflammation. The Journal of Pain 2007, 8, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Sonkodi, B. Delayed Onset Muscle Soreness (DOMS): The Repeated Bout Effect and Chemotherapy-Induced Axonopathy May Help Explain the Dying-Back Mechanism in Amyotrophic Lateral Sclerosis and Other Neurodegenerative Diseases. Brain Sciences 2021, 11, 108. [Google Scholar] [CrossRef] [PubMed]
- Cherief, M.; Negri, S.; Qin, Q.; Pagani, C.A.; Lee, S.; Yang, Y.P.; Clemens, T.L.; Levi, B.; James, A.W. TrkA+ Neurons Induce Pathologic Regeneration After Soft Tissue Trauma. Stem Cells Translational Medicine 2022, 11, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Yigitturk, G.; Erbas, O.; Karabay Yavasoglu, N.U.; Acikgoz, E.; Buhur, A.; Gokhan, A.; Gurel, C.; Gunduz, C.; Yavasoglu, A. The neuro-restorative effect of adipose-derived mesenchymal stem cell transplantation on a mouse model of diabetic neuropathy. Neurological Research 2021, 44, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Allas, L.; Brochard, S.; Rochoux, Q.; Ribet, J.; Dujarrier, C.; Veyssiere, A.; Aury-Landas, J.; Grard, O.; Leclercq, S.; Vivien, D.; et al. EZH2 inhibition reduces cartilage loss and functional impairment related to osteoarthritis. Scientific Reports 2020, 10. [Google Scholar] [CrossRef]
- Scandolera, A.; Hubert, J.; Humeau, A.; Lambert, C.; De Bizemont, A.; Winkel, C.; Kaouas, A.; Renault, J.H.; Nuzillard, J.M.; Reynaud, R. GABA and GABA-Alanine from the Red Microalgae Rhodosorus marinus Exhibit a Significant Neuro-Soothing Activity through Inhibition of Neuro-Inflammation Mediators and Positive Regulation of TRPV1-Related Skin Sensitization. Marine Drugs 2018, 16, 96. [Google Scholar] [CrossRef]
- Romero, M.I.; Rangappa, N.; Li, L.; Lightfoot, E.; Garry, M.G.; Smith, G.M. Extensive Sprouting of Sensory Afferents and Hyperalgesia Induced by Conditional Expression of Nerve Growth Factor in the Adult Spinal Cord. The Journal of Neuroscience 2000, 20, 4435–4445. [Google Scholar] [CrossRef]
- Shaikh, S.S.; Chen, Y.; Halsall, S.; Nahorski, M.S.; Omoto, K.; Young, G.T.; Phelan, A.; Woods, C.G. A Comprehensive Functional Analysis ofNTRK1Missense Mutations Causing Hereditary Sensory and Autonomic Neuropathy Type IV (HSAN IV). Human Mutation 2016, 38, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, A.; Rahmati, M.; Eslami, R.; Sheibani, V. Activation of neurotrophins in lumbar dorsal root probably contributes to neuropathic pain after spinal nerve ligation. Iranian Journal of Basic Medical Sciences 2017, 20. [Google Scholar]
- Javed, S.; Petropoulos, I.N.; Alam, U.; Malik, R.A. Treatment of painful diabetic neuropathy. Therapeutic Advances in Chronic Disease 2014, 6, 15–28. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Jhang, J.F.; Kuo, H.C. Urinary Oxidative Stress Biomarker Levels Might Be Useful in Identifying Functional Bladder Disorders in Women with Frequency and Urgency Syndrome. Journal of Clinical Medicine 2023, 12, 2336. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Xie, W.; Kang, Z.; Jiang, C.; Liu, N. Administration of Curcumin Alleviates Neuropathic Pain in a Rat Model of Brachial Plexus Avulsion. Pharmacology 2019, 103, 324–332. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nature Structural & Molecular Biology 2003, 10, 980. [Google Scholar]
- Wehrman, T.; He, X.; Raab, B.; Dukipatti, A.; Blau, H.; Garcia, K.C. Structural and Mechanistic Insights into Nerve Growth Factor Interactions with the TrkA and p75 Receptors. Neuron 2007, 53, 25–38. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsletter On Protein Crystallography 2002, 40, 82–92. [Google Scholar]
- Li, W. How do SMA-linked mutations of SMN1 lead to structural/functional deficiency of the SMA protein? PLOS ONE 2017, 12, e0178519. [Google Scholar] [CrossRef]
- Li, W. Towards a General Intermolecular Binding Affinity Calculator 2022. [CrossRef]
- Li, W.; Vottevor, G. Towards a Truly General Intermolecular Binding Affinity Calculator for Drug Discovery and Design 2023. [CrossRef]
- Webb, B.; Sali, A. Protein Structure Modeling with MODELLER. In Methods in Molecular Biology; Springer US, 2020; pp. 239–255.
- Vangone, A.; Bonvin, A.M. Contacts-based prediction of binding affinity in protein-protein complexes. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 2016, btw514. [Google Scholar]
- Fuji, H.; Qi, F.; Qu, L.; Takaesu, Y.; Hoshino, T. Prediction of Ligand Binding Affinity to Target Proteins by Molecular Mechanics Theoretical Calculation. Chemical and Pharmaceutical Bulletin 2017, 65, 461–468. [Google Scholar] [CrossRef]
- Sikandar, S.; Aasvang, E.K.; Dickenson, A.H. Scratching the surface: The processing of pain from deep tissues. Pain Management 2016, 6, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, S.; Bouhana, K.S.; Pheneger, J.; Andrews, S.W.; Walsh, D.A. Selective inhibition of tropomyosin-receptor-kinase A (TrkA) reduces pain and joint damage in two rat models of inflammatory arthritis. Arthritis Research: Therapy 2016, 18. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.F.; Cheng, J.K.; Chen, C.Y.; Lien, C.C.; Chu, D.; Wang, S.Y.; Tsaur, M.L. Mirror-image pain is mediated by nerve growth factor produced from tumor necrosis factor alpha-activated satellite glia after peripheral nerve injury. Pain 2014, 155, 906–920. [Google Scholar] [CrossRef] [PubMed]
- Schnakenberg, M.; Thomas, C.; Schmelz, M.; Rukwied, R. Nerve growth factor sensitizes nociceptors to C-fibre selective supra-threshold electrical stimuli in human skin. European Journal of Pain 2020, 25, 385–397. [Google Scholar] [CrossRef]
- Nees, T.A.; Rosshirt, N.; Zhang, J.A.; Reiner, T.; Sorbi, R.; Tripel, E.; Walker, T.; Schiltenwolf, M.; Hagmann, S.; Moradi, B. Synovial Cytokines Significantly Correlate with Osteoarthritis-Related Knee Pain and Disability: Inflammatory Mediators of Potential Clinical Relevance. Journal of Clinical Medicine 2019, 8, 1343. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Baylink, D.J.; Chen, C.S.; Reeves, M.E.; Xiao, J.; Lacy, C.; Lau, E.; Cao, H. The importance of vitamin d metabolism as a potential prophylactic, immunoregulatory and neuroprotective treatment for COVID-19. Journal of Translational Medicine 2020, 18. [Google Scholar] [CrossRef]
- Sharma, P.; Rani, N.; Gangwar, A.; Singh, R.; Kaur, R.; Upadhyaya, K. Diabetic Neuropathy: A Repercussion of Vitamin D Deficiency. Current Diabetes Reviews 2023, 19. [Google Scholar] [CrossRef]
- Tanaka, Y.; Niwa, S.; Dong, M.; Farkhondeh, A.; Wang, L.; Zhou, R.; Hirokawa, N. The Molecular Motor KIF1A Transports the TrkA Neurotrophin Receptor and Is Essential for Sensory Neuron Survival and Function. Neuron 2016, 90, 1215–1229. [Google Scholar] [CrossRef]
- Subedi, L.; Gaire, B.P.; Do, M.H.; Lee, T.H.; Kim, S.Y. Anti-neuroinflammatory and neuroprotective effects of the Lindera neesiana fruit in vitro. Phytomedicine 2016, 23, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Anand, U.; Otto, W.; Facer, P.; Zebda, N.; Selmer, I.; Gunthorpe, M.; Chessell, I.; Sinisi, M.; Birch, R.; Anand, P. TRPA1 receptor localisation in the human peripheral nervous system and functional studies in cultured human and rat sensory neurons. Neuroscience Letters 2008, 438, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Araújo-filho, H.G.; Pereira, E.W.; Heimfarth, L.; Souza Monteiro, B.; Santos Passos, F.R.; Siqueira-Lima, P.; Gandhi, S.R.; Viana dos Santos, M.R.; Guedes da Silva Almeida, J.R.; Picot, L.; et al. Limonene, a food additive, and its active metabolite perillyl alcohol improve regeneration and attenuate neuropathic pain after peripheral nerve injury: Evidence for IL-1β, TNF-α, GAP, NGF and ERK involvement. International Immunopharmacology 2020, 86, 106766. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.J.; Loescher, A.R.; Boissonade, f.M.; Whawell, S.A.; Robinson, P.P.; Andrew, D. Temporal mismatch between pain behaviour, skin Nerve Growth Factor and intra-epidermal nerve fibre density in trigeminal neuropathic pain. BMC Neuroscience 2014, 15. [Google Scholar] [CrossRef]
- Nauta, H.J.W.; Wehman, J.C.; Koliatsos, V.E.; Terrell, M.A.; Chung, K. Intraventricular infusion of nerve growth factor as the cause of sympathetic fiber sprouting in sensory ganglia. Journal of Neurosurgery 1999, 91, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Spezia Adachi, L.N.; Vercelino, R.; de Oliveira, C.; Scarabelot, V.L.; de Souza, A.; Medeiros, L.F.; Cioato, S.G.; Caumo, W.; Torres, I.L. Isoflurane and the Analgesic Effect of Acupuncture and Electroacupuncture in an Animal Model of Neuropathic Pain. Journal of Acupuncture and Meridian Studies 2018, 11, 97–106. [Google Scholar] [CrossRef]
- Obata, K.; Yamanaka, H.; Dai, Y.; Mizushima, T.; Fukuoka, T.; Tokunaga, A.; Noguchi, K. Activation of extracellular signal-regulated protein kinase in the dorsal root ganglion following inflammation near the nerve cell body. Neuroscience 2004, 126, 1011–1021. [Google Scholar] [CrossRef] [PubMed]
- Malerba, F.; Paoletti, F.; Bruni Ercole, B.; Materazzi, S.; Nassini, R.; Coppi, E.; Patacchini, R.; Capsoni, S.; Lamba, D.; Cattaneo, A. Functional Characterization of Human ProNGF and NGF Mutants: Identification of NGF P61SR100E as a ’Painless’ Lead Investigational Candidate for Therapeutic Applications. PLOS ONE 2015, 10, e0136425. [Google Scholar] [CrossRef]
- Jönhagen, M.E. Nerve Growth Factor Treatment in Dementia. Alzheimer Disease and Associated Disorders 2000, 14, S31–S38. [Google Scholar] [CrossRef] [PubMed]
- Yasui, M.; Shiraishi, Y.; Ozaki, N.; Hayashi, K.; Hori, K.; Ichiyanagi, M.; Sugiura, Y. Nerve growth factor and associated nerve sprouting contribute to local mechanical hyperalgesia in a rat model of bone injury. European Journal of Pain 2011, 16, 953–965. [Google Scholar] [CrossRef]
- Andres, C.; Meyer, S.; Dina, O.A.; Levine, J.D.; Hucho, T. Quantitative Automated Microscopy (QuAM) Elucidates Growth Factor Specific Signalling in Pain Sensitization. Molecular Pain 2010, 6, 1744–8069–6–98. [Google Scholar] [CrossRef] [PubMed]
- Ramer, M.S.; Thompson, S.W.; McMahon, S.B. Causes and consequences of sympathetic basket formation in dorsal root ganglia. Pain 1999, 82, S111–S120. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.M.; Wang, Q.; Sun, W.B. Silencing of FKBP51 alleviates the mechanical pain threshold, inhibits DRG inflammatory factors and pain mediators through the NF-kappaB signaling pathway. Gene 2017, 627, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Severini, C.; Petrocchi Passeri, P.; Ciotti, M.; Florenzano, F.; Petrella, C.; Malerba, F.; Bruni, B.; D’Onofrio, M.; Arisi, I.; Brandi, R.; et al. Nerve growth factor derivative NGF61100 promotes outgrowth of primary sensory neurons with reduced signs of nociceptive sensitization. Neuropharmacology 2017, 117, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Alhilou, A.M.; Shimada, A.; Svensson, C.I.; Svensson, P.; Ernberg, M.; Cairns, B.E.; Christidis, N. Sex-related differences in response to masseteric injections of glutamate and nerve growth factor in healthy human participants. Scientific Reports 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Krock, E.; Rosenzweig, D.H.; Chabot-Doré, A.; Jarzem, P.; Weber, M.H.; Ouellet, J.A.; Stone, L.S.; Haglund, L. Painful, degenerating intervertebral discs up-regulate neurite sprouting and CGRP- through nociceptive factors. Journal of Cellular and Molecular Medicine 2014, 18, 1213–1225. [Google Scholar] [CrossRef] [PubMed]
- Handa, J.; Sekiguchi, M.; Krupkova, O.; Konno, S.i. The effect of serotonin-noradrenaline reuptake inhibitor duloxetine on the intervertebral disk-related radiculopathy in rats. European Spine Journal 2015, 25, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Moon, S.; Suh, H.; Hochman, S.; Lee, M.G.; Kim, Y.; Jang, I.; Han, H. Disc degeneration induces a mechano-sensitization of disc afferent nerve fibers that associates with low back pain. Osteoarthritis and Cartilage 2019, 27, 1608–1617. [Google Scholar] [CrossRef] [PubMed]
- Hepner, D.L.; Arriaga, A.F.; Cooper, J.B.; Goldhaber-fiebert, S.N.; Gaba, D.M.; Berry, W.R.; Boorman, D.J.; Bader, A.M. Operating Room Crisis Checklists and Emergency Manuals. Anesthesiology 2017, 127, 384–392. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Jhang, J.F.; Hsu, Y.H.; Ho, H.C.; Wu, Y.H.; Kuo, H.C. Urine biomarkers in ESSIC type 2 interstitial cystitisbladder pain syndrome and overactive bladder with developing a novel diagnostic algorithm. Scientific Reports 2021, 11. [Google Scholar]
- Hasenauer, J.; Hasenauer, C.; Hucho, T.; Theis, F.J. ODE Constrained Mixture Modelling: A Method for Unraveling Subpopulation Structures and Dynamics. PLoS Computational Biology 2014, 10, e1003686. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Guo, X.; Yan, X.; Tian, Z.; Jiang, W.; He, X. TSG-6 inhibits IL-1β-induced inflammatory responses and extracellular matrix degradation in nucleus pulposus cells by activating the PI3KAkt signaling pathway. Journal of Orthopaedic Surgery and Research 2022, 17. [Google Scholar] [CrossRef]
- Kashima, S.; Roberto, P.G.; Soares, A.M.; Astolfi-filho, S.; Pereira, J.O.; Giuliati, S.; Faria, M.F., Jr.; Xavier, M.A.; Fontes, M.R.; Giglio, J.R.; et al. Analysis of Bothrops jararacussu venomous gland transcriptome focusing on structural and functional aspects11All sequence data reported in this paper will appear in the GenBank database under the following accession numbers: BOJU-I (AY 185200), BOJU-II (AY 185206), BOJU-III (AY 145836), BOJUMET-I (AY 55005), BOJUMET-II (AY 25584), BOJUMET-III (AY 258153), C-type lectin (AY 251283), serine-proteases (AY 251282).: I-gene expression profile of highly expressed phospholipases A2. Biochimie 2004, 86, 211–219. [Google Scholar] [PubMed]
- Xu, P.; Van Slambrouck, C.; Berti-Mattera, L.; Hall, A.K. Activin Induces Tactile Allodynia and Increases Calcitonin Gene-Related Peptide after Peripheral Inflammation. The Journal of Neuroscience 2005, 25, 9227–9235. [Google Scholar] [CrossRef] [PubMed]
- Kovačič, U.; Tesovnik, B.; Molnar, N.; Cör, A.; Skalerič, U.; Gašperšič, R. Dental pulp and gingivomucosa in rats are innervated by two morphologically and neurochemically different populations of nociceptors. Archives of Oral Biology 2013, 58, 788–795. [Google Scholar] [CrossRef]
- De Oliveira, C.C.; Gouveia, F.V.; de Castro, M.C.; Kuroki, M.A.; dos Santos, L.C.T.; Fonoff, E.T.; Teixeira, M.J.; Otoch, J.P.; Martinez, R.C.R. A Window on the Study of Aversive Instrumental Learning: Strains, Performance, Neuroendocrine, and Immunologic Systems. Frontiers in Behavioral Neuroscience 2016, 10. [Google Scholar] [CrossRef]
- Anand, U.; Oldfield, C.; Pacchetti, B.; Anand, P.; Sodergren, M.H. Dose-Related Inhibition of Capsaicin Responses by Cannabinoids CBG, CBD, THC and their Combination in Cultured Sensory Neurons. Journal of Pain Research 2021, Volume 14, 3603–3614. [Google Scholar] [CrossRef]
| PDB ID | Structure Title (release date from newest to oldest) |
|---|---|
| 8DWN | Crystal structure of bis-phosphorylated insulin receptor kinase domain |
| 6PL1 | TRK-A IN COMPLEX WITH LIGAND 1B |
| 6NPT | TRK-A IN COMPLEX WITH LIGAND 1 |
| 6NSP | TRK-A IN COMPLEX WITH LIGAND 9 |
| 6NSS | TRK-A IN COMPLEX WITH LIGAND 6 |
| 5WR7 | Crystal structure of Trk-A complexed with a selective inhibitor CH7057288 |
| 4XPJ | Crystal structure of Nerve growth factor in complex with lysophosphatidylinositol |
| 4NWT | Crystal structure of the anti-human NGF Fab APE1531 |
| 4NWU | Crystal structure of APE1551, an anti-human NGF Fab with a nine amino acid insertion in CDR H1 |
| 2LPN | Solution Structure of N-Terminal domain of human Conserved Dopamine Neurotrophic Factor (CDNF) |
| 4EFV | Crystal structure of OIF from Llama seminal plasma |
| 2IFG | Structure of the extracellular segment of human TRKA in complex with nerve growth factor |
| 1SG1 | Crystal Structure of the Receptor-Ligand Complex between Nerve Growth Factor and the Common Neurotrophin Receptor p75 |
| 1HE7 | Human Nerve growth factor receptor TrkA |
| 1WWW | NGF IN COMPLEX WITH DOMAIN 5 OF THE TRKA RECEPTOR |
| 1BTG | CRYSTAL STRUCTURE OF BETA NERVE GROWTH FACTOR AT 2.5 A RESOLUTION IN C2 SPACE GROUP WITH ZN IONS BOUND |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





