Submitted:
26 March 2024
Posted:
28 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Data Collections
2.2. Network Construction
2.3. Functional Enrichment of Drug Targets
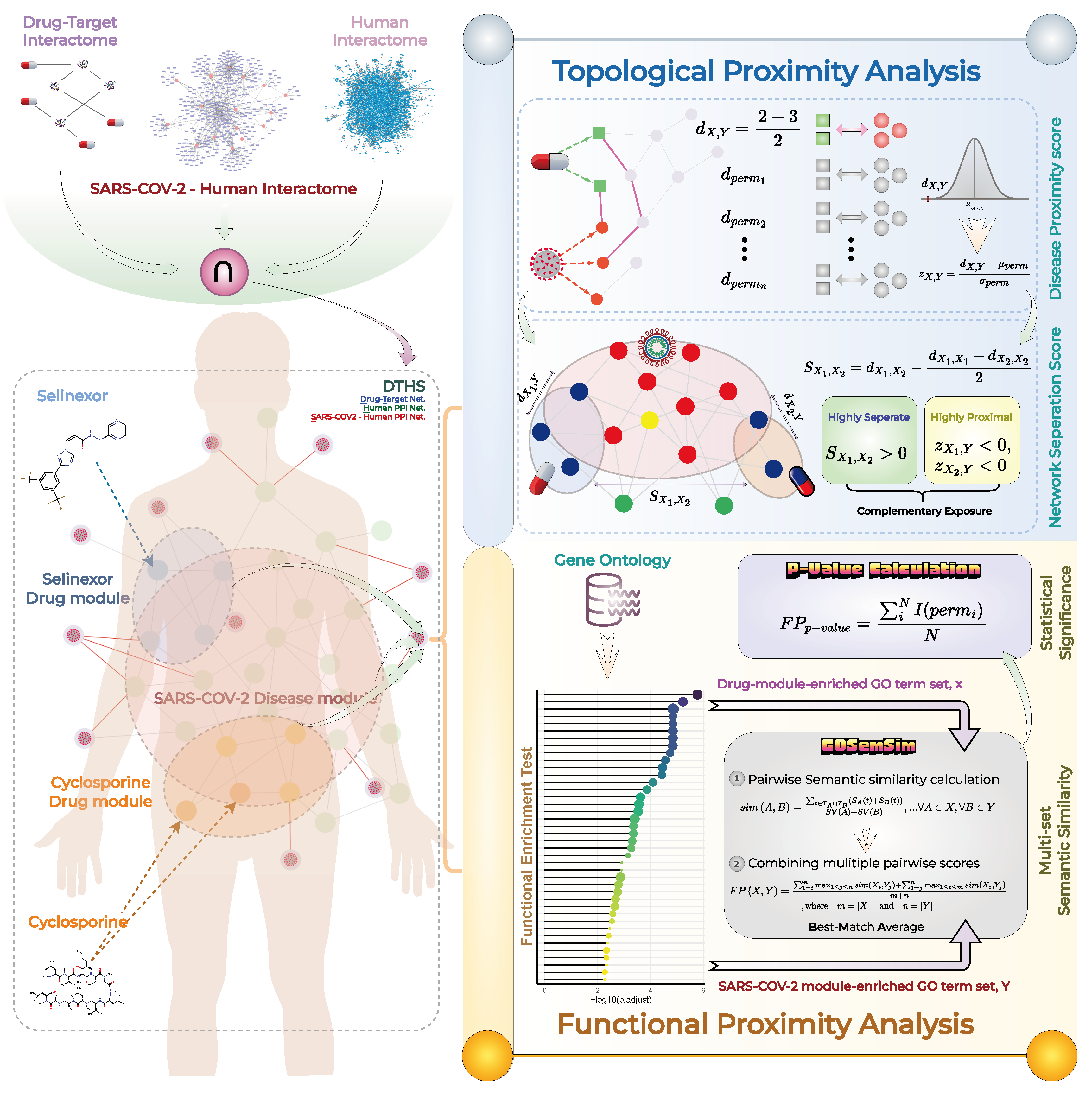
Measuring Topological and Functional Proximity Scores for Both Cyclosporine-SARS-COV-2 and Selinexor-SARS-COV-2
2.3.1. Topological Proximity Calculation
2.3.2. Functional Proximity Calculation
2.4. Measuring Network Separation Score for Cyclosporine and Selinexor Drugs
2.5. Statistical Significance Analysis
3. Results
3.1. Monotherapeutic Targets of Cyclosporine and Selinexor Drugs for Treating SARS-COV-2 Patients
3.2. DTHS Network Construction for Evaluating Complementary Exposure of Cyclosporine and Selinexor Combination against SARS-COV-2
| Cyclosporine | Selinexor | |
|---|---|---|
| Drug Targets (Human) | PPP3R2, PPIA, CAMLG, PPIF | XPO1 |
| No. of PPI neighbours of Drug targets within the DTHS network | 356 | 1758 |
| No. of Disease genes as the PPI neighbours of Drug targets | 16 | 87 |
| Unique Disease genes targeted by drug targets’ PPI neighbourhood | Total = 7 (UBC, CWC27, PPIL3, TRAF6, PPIA, NDUFAF2, GRPEL1) | Total = 78 (ARF6, G3BP1, CDK1, CSNK2A2, DCAF7, MCM7, PRKACA, PRKDC, GNB1, RAB1A, FKBP15, SUZ12, RAB14, AP3B1, PABPC4, RIPK1, HNRNPK, NUP62, STOM, GORASP1, SLU7, GTF2F2, RHOA, DDX1, VPS39, EIF4E2, RAB18, CSNK2B, DDX10, RAB7A, SRP19, RBM28, NOL10, UBAP2L, MPHOSPH10, AP2M1, RAB2A, NUP214, RALA, HDAC2, GOLGA2, WASHC4, TUBGCP3, POR, NUP98, PRKAR2A, NUTF2, RAE1, PRIM2, DDX21, UPF1, POLA2, CEP350, EXOSC5, GIGYF2, NUP58, PLEKHA5, CHMP2A, EXOSC8, SRP72, MEPCE, SRP54, TBK1, TUBGCP2, CRTC3, LARP4B, TCF12, NUP88, NEK9, SEPSECS, AATF, FYCO1, AKAP8L, DNAJC11, MDN1, TYSND1, MAT2B, ZC3H7A) |
| Pathogen genes targeted by drug targets or its PPI neighbourhood | Total = 10 (SARS-COV2 R1A, SARS-COV2 R1AB, SARS-COV2 N, SARS-COV2 ORF9B, SARS-COV2 NSP7, SARS-COV2 SPIKE, SARS-COV2 E, SARS-COV2 NSP12, SARS-COV2 S, SARS-COV2 NSP10) | Total = 22 (SARS-COV2 N, SARS-COV2 ORF9B, SARS-COV2 NSP15, SARS-COV2 R1AB, SARS-COV2 NSP9, SARS-COV2 NSP13, SARS-COV2 R1A, SARS-COV2 NSP7, SARS-COV2 NSP2, SARS-COV2 E, SARS-COV2 NSP12, SARS-COV2 PLPRO, SARS-COV2 M, SARS-COV2 ORF3A, SARS-COV2 NSP8, SARS-COV2 SPIKE, SARS-COV2 NSP10, SARS-COV2 NSP5, SARS-COV2 ORF6, SARS-COV2 NSP1, SARS-COV2 NSP4, SARS-COV2 ORF7A) |
3.3. Both Cyclosporine and Selinexor Are Significantly Proximal to SARS-COV-2 Disease Modules
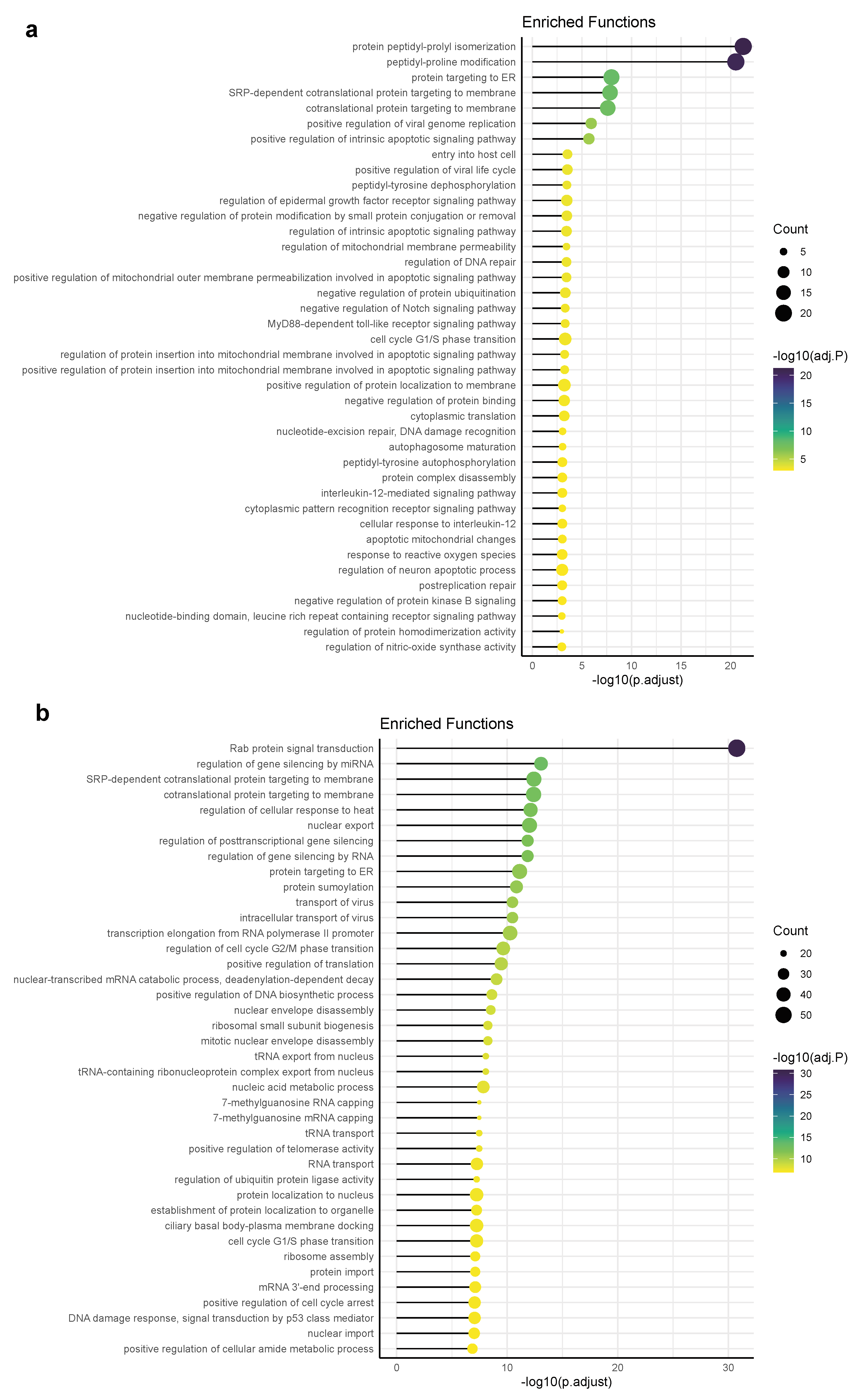
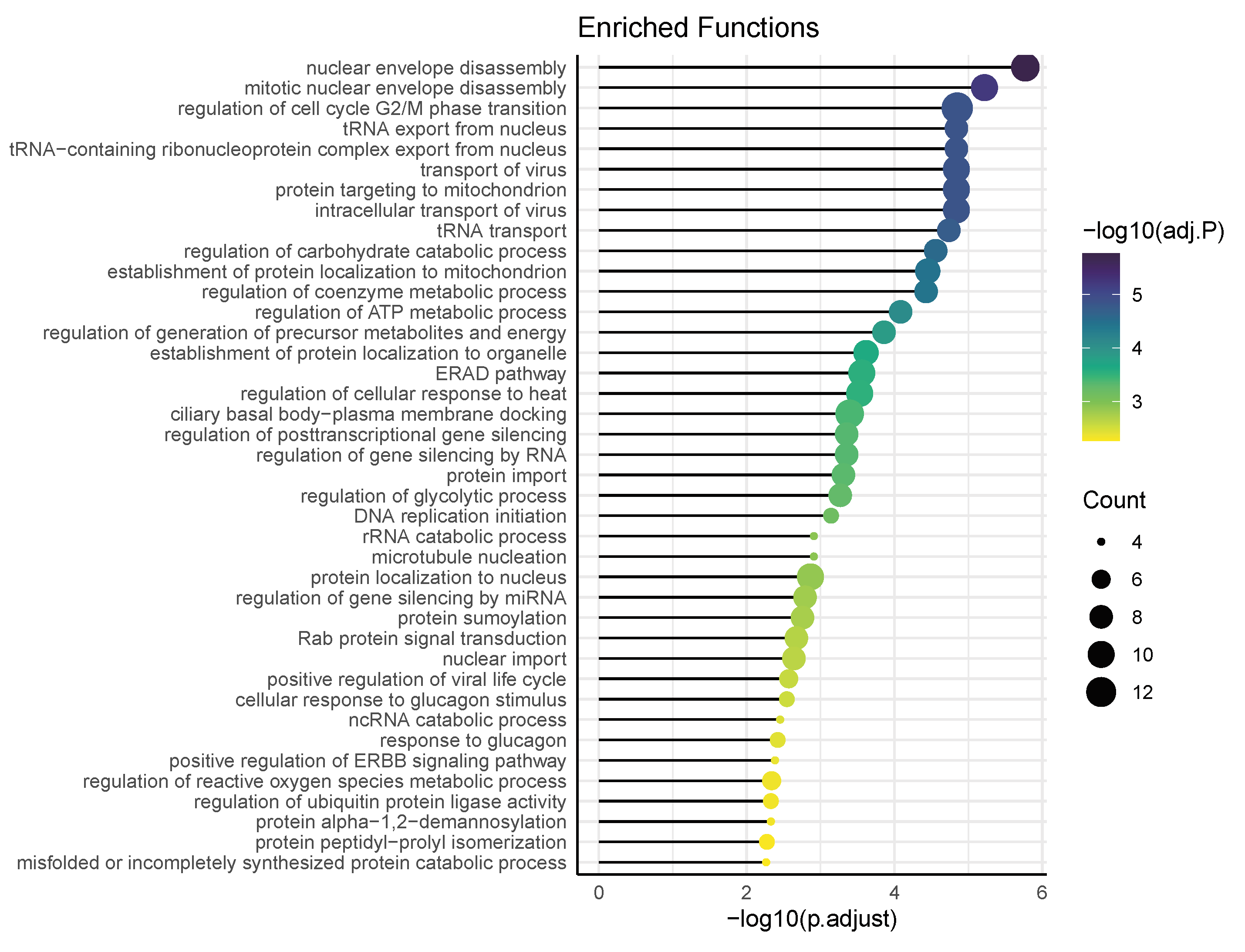
3.4. Functional Pathways/Terms Enriched by Cyclosporine and Selinexor Drug Modules and SARS-COV-2 Disease Module
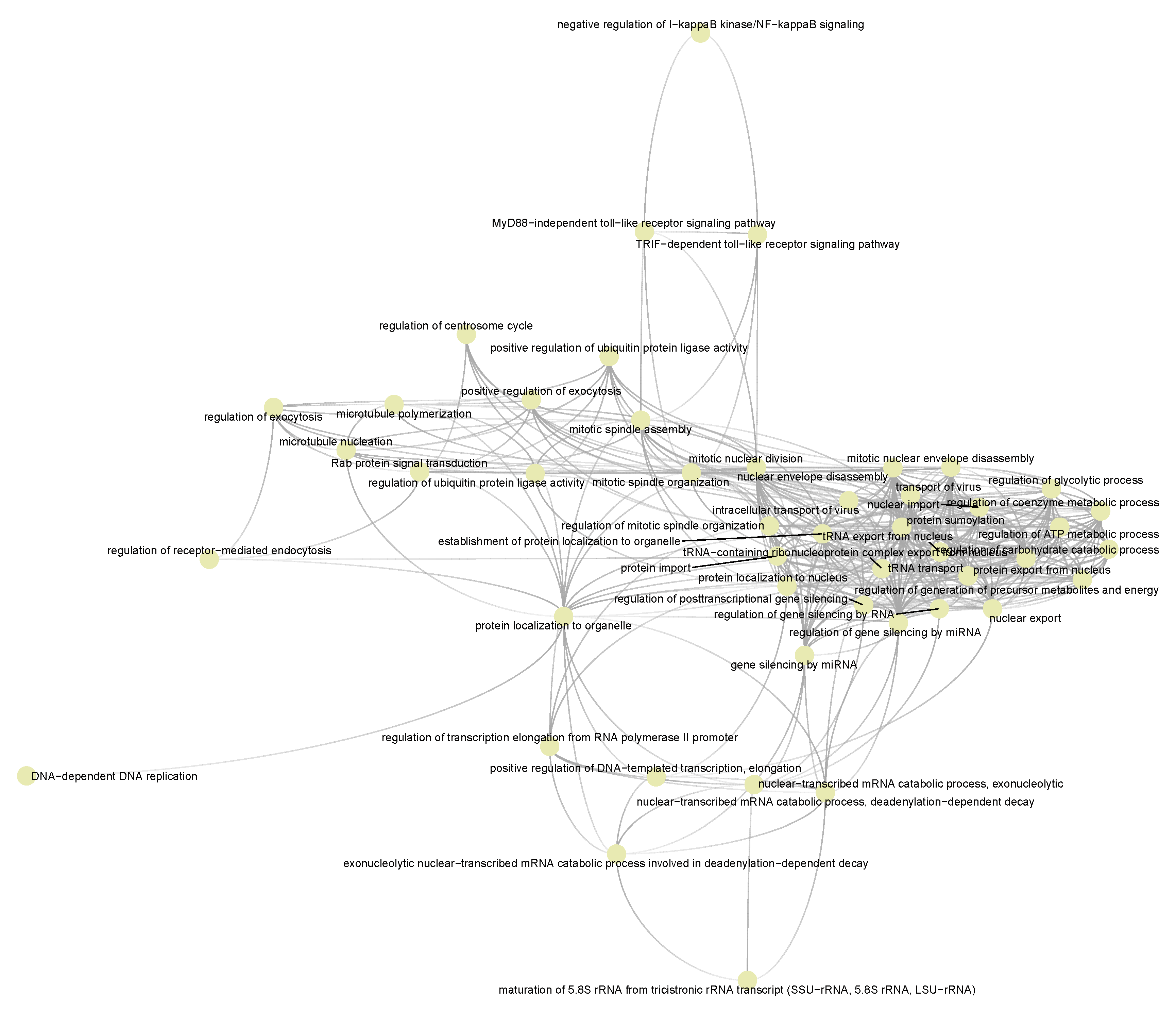
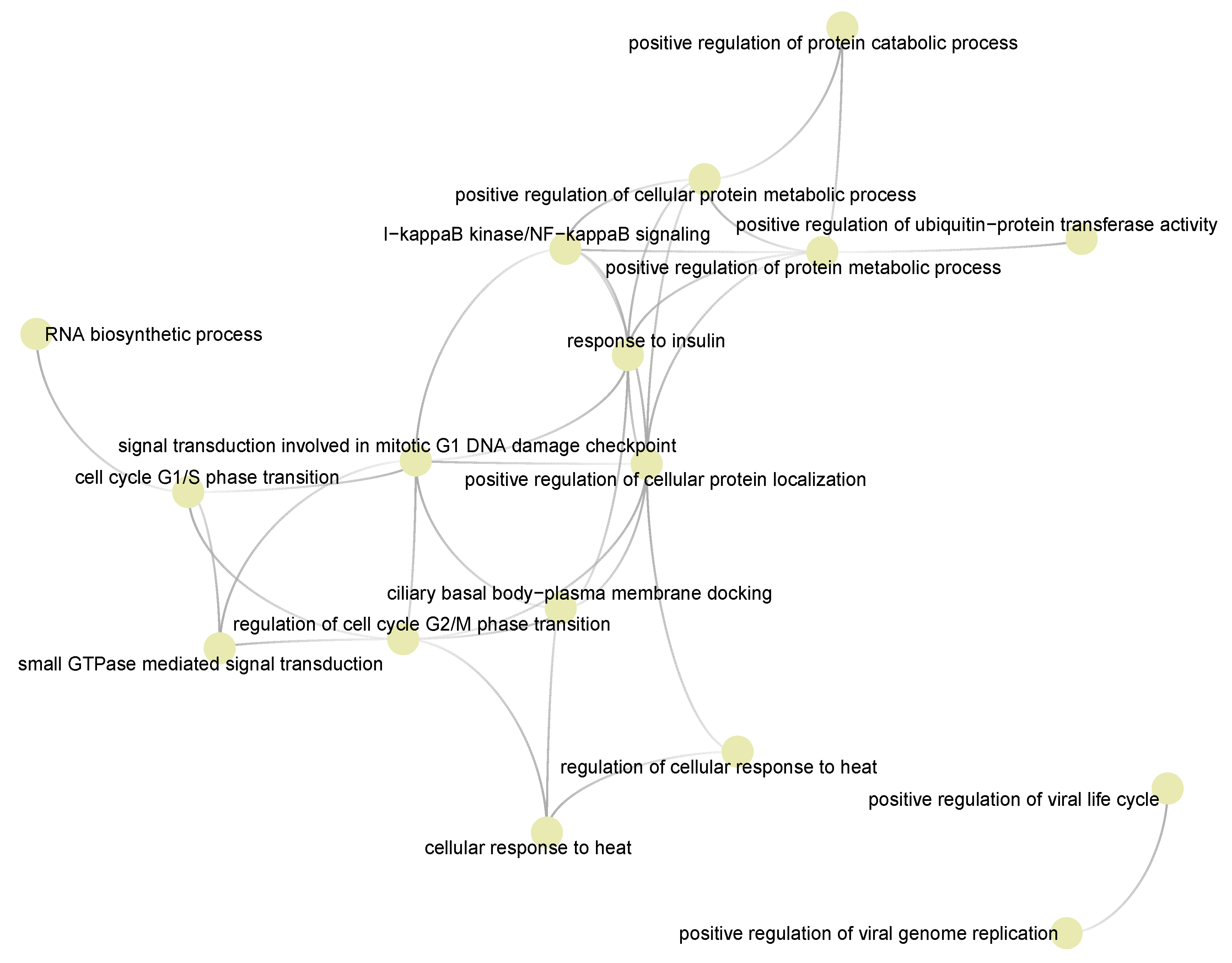
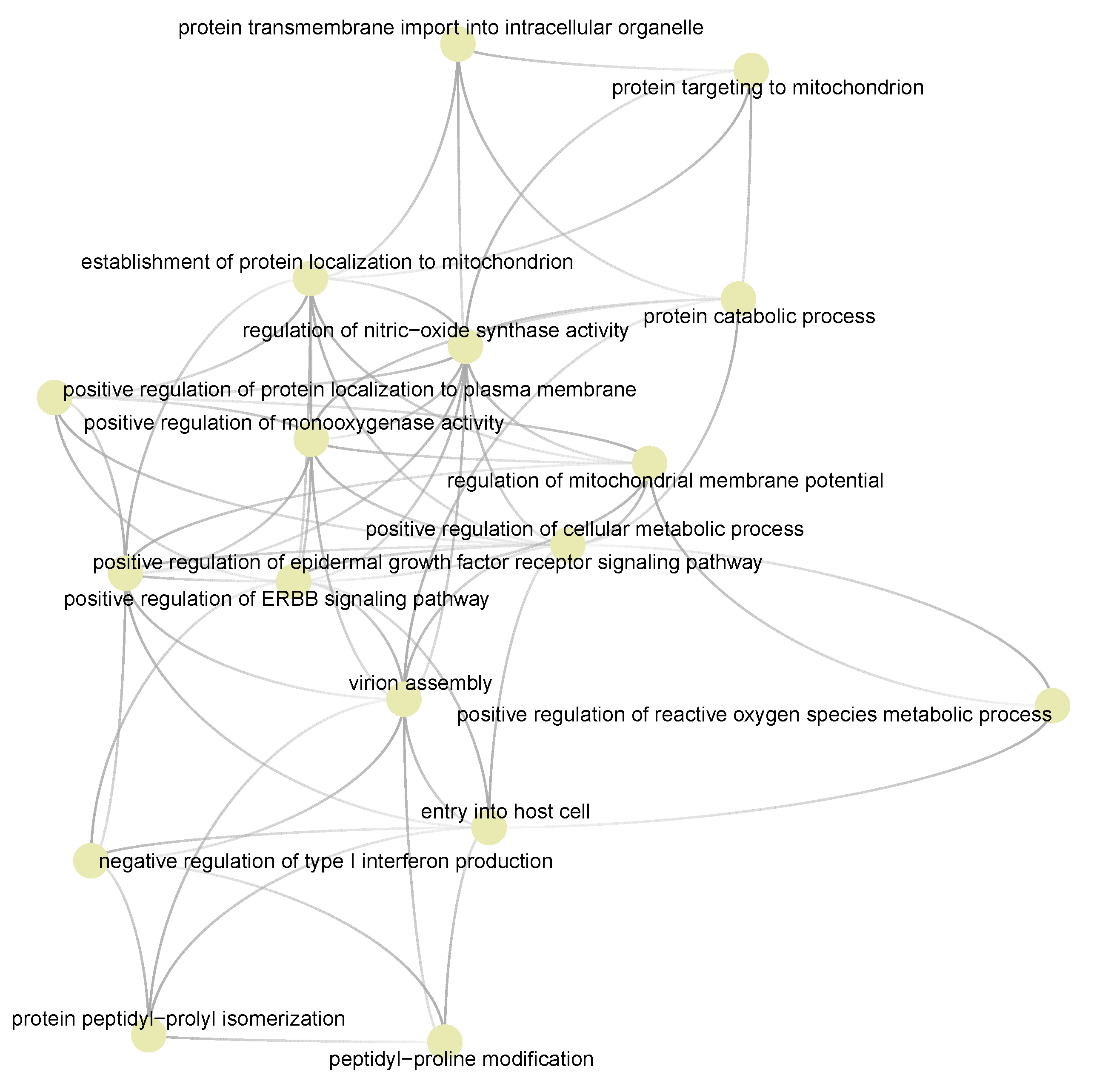
3.5. Semantic Similarities Shown a Profound Functional Overlap between Drug-Target Enriched and SARS-COV-2 Enriched GO Term Sets
3.6. Cyclosporine and Selinexor Drug Modules Reveal Their Complementary Exposure Pattern in Targeting SARS-COV-2 Disease Module
4. Discussions
5. Conclusions
References
- COVID-19 deaths | WHO COVID-19 dashboard.
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; Cheng, Z.; Yu, T.; Xia, J.; Wei, Y.; Wu, W.; Xie, X.; Yin, W.; Li, H.; Liu, M.; Xiao, Y.; Gao, H.; Guo, L.; Xie, J.; Wang, G.; Jiang, R.; Gao, Z.; Jin, Q.; Wang, J.; Cao, B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. 395, 497–506. [CrossRef]
- Gysi, D.M.; Valle, Í.D.; Zitnik, M.; Ameli, A.; Gan, X.; Varol, O.; Ghiassian, S.D.; Patten, J.J.; Davey, R.; Loscalzo, J.; Barabási, A.L. Network Medicine Framework for Identifying Drug Repurposing Opportunities for COVID-19. [2004.07229].
- Barabási, A.L.; Gulbahce, N.; Loscalzo, J. Network medicine: a network-based approach to human disease. 12, 56–68. [CrossRef]
- Cheng, F.; Desai, R.J.; Handy, D.E.; Wang, R.; Schneeweiss, S.; Barabasi, A.L.; Loscalzo, J. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nature Communications 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Rudnicka, L.; Glowacka, P.; Goldust, M.; Sikora, M.; Sar-Pomian, M.; Rakowska, A.; Samochocki, Z.; Olszewska, M. Cyclosporine therapy during the COVID-19 pandemic. 83, e151–e152.
- Kashyap, T.; Murray, J.; Walker, C.J.; Chang, H.; Tamir, S.; Hou, B.; Shacham, S.; Kauffman, M.G.; Tripp, R.A.; Landesman, Y. Selinexor, a novel selective inhibitor of nuclear export, reduces SARS-CoV-2 infection and protects the respiratory system in vivo. 192, 105115. [CrossRef]
- Mostafa-Hedeab, G.; Al-kuraishy, H.M.; Al-Gareeb, A.I.; Welson, N.N.; El-Saber Batiha, G.; Conte-Junior, C.A. Selinexor and COVID-19: The Neglected Warden. 13, 884228. [CrossRef]
- Sharma, T.; Mondal, T.; Khan, S.; Churqui, M.P.; Nyström, K.; Thombare, K.; Baig, M.H.; Dong, J.J. Identifying novel inhibitors targeting Exportin-1 for the potential treatment of COVID-19. Archives of Microbiology 2024, 206. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; Assempour, N.; Iynkkaran, I.; Liu, Y.; Maciejewski, A.; Gale, N.; Wilson, A.; Chin, L.; Cummings, R.; Le, D.; Pon, A.; Knox, C.; Wilson, M. DrugBank 5.0: a major update to the DrugBank database for 2018. 46, D1074–D1082. [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al.A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. 583, 459–468. [CrossRef]
- Brown, K.R.; Jurisica, I. Unequal evolutionary conservation of human protein interactions in interologous networks. 8, R95. [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; McDermott, M.G.; Monteiro, C.D.; Gundersen, G.W.; Ma’ayan, A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. 44, W90–W97. [CrossRef]
- The UniProt Consortium. UniProt: a worldwide hub of protein knowledge. 47, D506–D515.
- Menche, J.; Sharma, A.; Kitsak, M.; Ghiassian, S.D.; Vidal, M.; Loscalzo, J.; Barabási, A.L. Uncovering disease-disease relationships through the incomplete interactome. 347, 1257601. [CrossRef]
- Yu, G.; Li, F.; Qin, Y.; Bo, X.; Wu, Y.; Wang, S. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. 26, 976–978. [CrossRef]
- Wang, J.Z.; Du, Z.; Payattakool, R.; Yu, P.S.; Chen, C.F. A new method to measure the semantic similarity of GO terms. 23, 1274–1281. [CrossRef]
- Cheng, F.; Kovács, I.A.; Barabási, A.L. Network-based prediction of drug combinations. 10, 1197. [CrossRef]
- Pizzorno, A.; Padey, B.; Dubois, J.; Julien, T.; Traversier, A.; Dulière, V.; Brun, P.; Lina, B.; Rosa-Calatrava, M.; Terrier, O. In vitro evaluation of antiviral activity of single and combined repurposable drugs against SARS-CoV-2. 181, 104878. [CrossRef]
- Uddin, M.H.; Zonder, J.A.; Azmi, A.S. Exportin 1 inhibition as antiviral therapy. 25, 1775–1781.
- Wang, T.; Cao, Y.; Zhang, H.; Wang, Z.; Man, C.H.; Yang, Y.; Chen, L.; Xu, S.; Yan, X.; Zheng, Q.; Wang, Y. COVID-19 metabolism: Mechanisms and therapeutic targets. 3, e157. [CrossRef]
- Caillet, C.; Stofberg, M.L.; Muleya, V.; Shonhai, A.; Zininga, T. Host cell stress response as a predictor of COVID-19 infectivity and disease progression. 9, 938099. [CrossRef]
- Gupta, R.K.; Mlcochova, P. Cyclin D3 restricts SARS-CoV-2 envelope incorporation into virions and interferes with viral spread. 41, e111653. [CrossRef]
- Nilsson-Payant, B.E.; Uhl, S.; Grimont, A.; Doane, A.S.; Cohen, P.; Patel, R.S.; Higgins, C.A.; Acklin, J.A.; Bram, Y.; Chandar, V.; Blanco-Melo, D.; Panis, M.; Lim, J.K.; Elemento, O.; Schwartz, R.E.; Rosenberg, B.R.; Chandwani, R.; tenOever, B.R. The NF-kB Transcriptional Footprint Is Essential for SARS-CoV-2 Replication. 95, e01257–21. [CrossRef]
- Shi, F.S.; Yu, Y.; Li, Y.L.; Cui, L.; Zhao, Z.; Wang, M.; Wang, B.; Zhang, R.; Huang, Y.W. Expression Profile and Localization of SARS-CoV-2 Nonstructural Replicase Proteins in Infected Cells. 10, e00744–22. [CrossRef]
- Pizzato, M.; Baraldi, C.; Boscato Sopetto, G.; Finozzi, D.; Gentile, C.; Gentile, M.D.; Marconi, R.; Paladino, D.; Raoss, A.; Riedmiller, I.; Ur Rehman, H.; Santini, A.; Succetti, V.; Volpini, L. SARS-CoV-2 and the Host Cell: A Tale of Interactions. 1, 815388. [CrossRef]
- Yamamotoya, T.; Nakatsu, Y.; Kanna, M.; Hasei, S.; Ohata, Y.; Encinas, J.; Ito, H.; Okabe, T.; Asano, T.; Sakaguchi, T. Prolyl isomerase Pin1 plays an essential role in SARS-CoV-2 proliferation, indicating its possibility as a novel therapeutic target. 11, 18581. [CrossRef]
- Kanna, M.; Nakatsu, Y.; Yamamotoya, T.; Encinas, J.; Ito, H.; Okabe, T.; Asano, T.; Sakaguchi, T. Roles of peptidyl prolyl isomerase Pin1 in viral propagation. 10, 1005325. [CrossRef]
- Atik, N.; Wirawan, F.; Amalia, R.; Khairani, A.F.; Pradini, G.W. Differences in endosomal Rab gene expression between positive and negative COVID-19 patients. 15, 252. [CrossRef]
- Hayn, M.; Hirschenberger, M.; Koepke, L.; Nchioua, R.; Straub, J.H.; Klute, S.; Hunszinger, V.; Zech, F.; Prelli Bozzo, C.; Aftab, W.; Christensen, M.H.; Conzelmann, C.; Müller, J.A.; Srinivasachar Badarinarayan, S.; Stürzel, C.M.; Forne, I.; Stenger, S.; Conzelmann, K.K.; Münch, J.; Schmidt, F.I.; Sauter, D.; Imhof, A.; Kirchhoff, F.; Sparrer, K.M.J. Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. 35, 109126. [CrossRef]
- Sajidah, E.S.; Lim, K.; Wong, R.W. How SARS-CoV-2 and Other Viruses Build an Invasion Route to Hijack the Host Nucleocytoplasmic Trafficking System. 10, 1424. [CrossRef]
- Krishnan, S.; Nordqvist, H.; Ambikan, A.T.; Gupta, S.; Sperk, M.; Svensson-Akusjärvi, S.; Mikaeloff, F.; Benfeitas, R.; Saccon, E.; Ponnan, S.M.; et al. Metabolic Perturbation Associated With COVID-19 Disease Severity and SARS-CoV-2 Replication. 20, 100159. [CrossRef]
- Wang, X.; Chang, Z.; Zhao, T.; Zhong, W.; Shi, J.; Wang, G.; Xu, X. The role of post-transcriptional regulation in SARS-CoV-2 infection and pathogenicity. 14, 1256574. [CrossRef]
- Katanski, C.D.; Alshammary, H.; Watkins, C.P.; Huang, S.; Gonzales-Reiche, A.; Sordillo, E.M.; Van Bakel, H.; Mount Sinai PSP study group.; Lolans, K.; Simon, V.; Pan, T. tRNA abundance, modification and fragmentation in nasopharyngeal swabs as biomarkers for COVID-19 severity. 10, 999351. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




