Submitted:
26 March 2024
Posted:
27 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
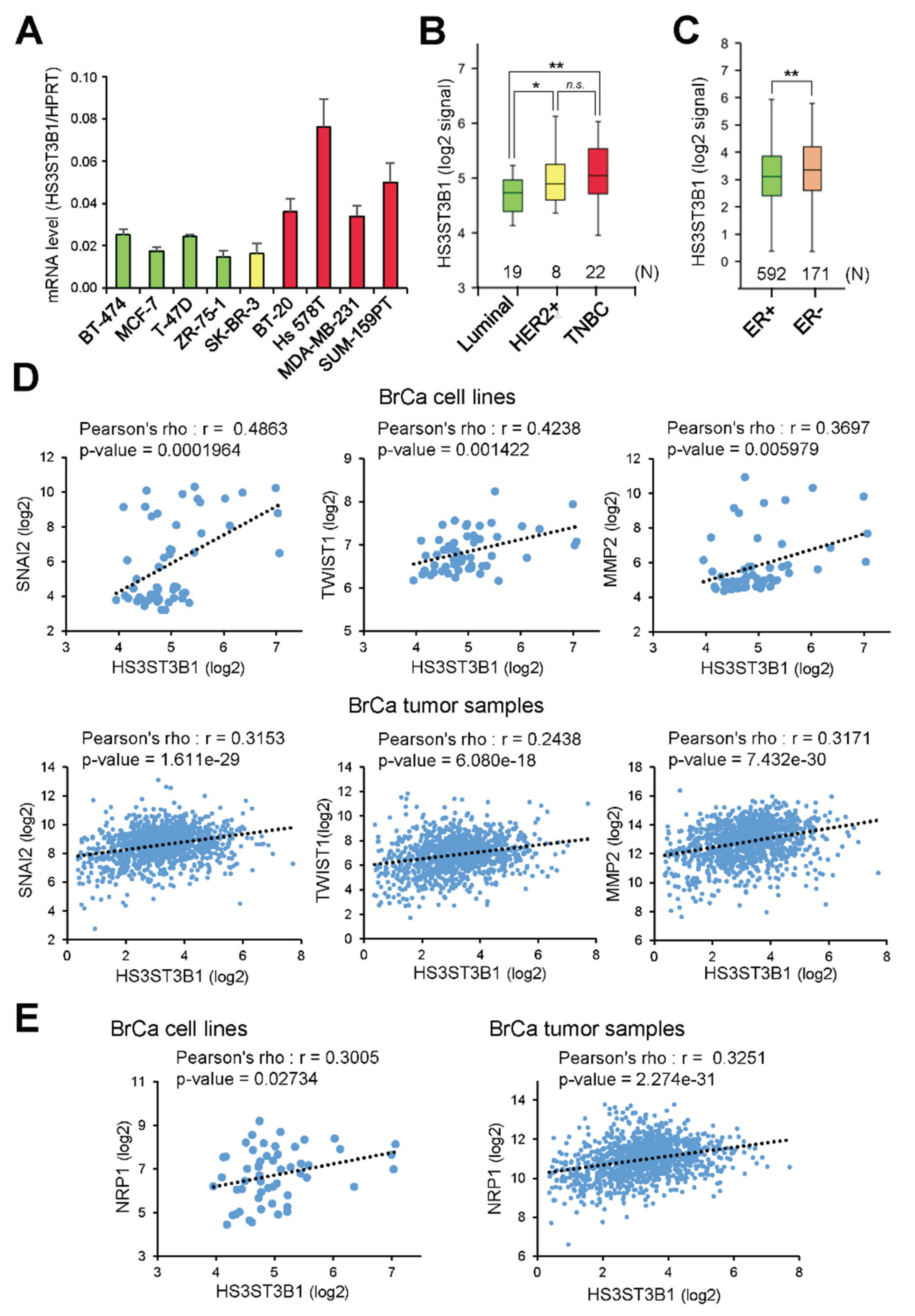
2.1. HS3ST3B1 Is Overexpressed in TNBC Subtype and Associates with Mesenchymal-Like and Invasive Phenotype in BrCa
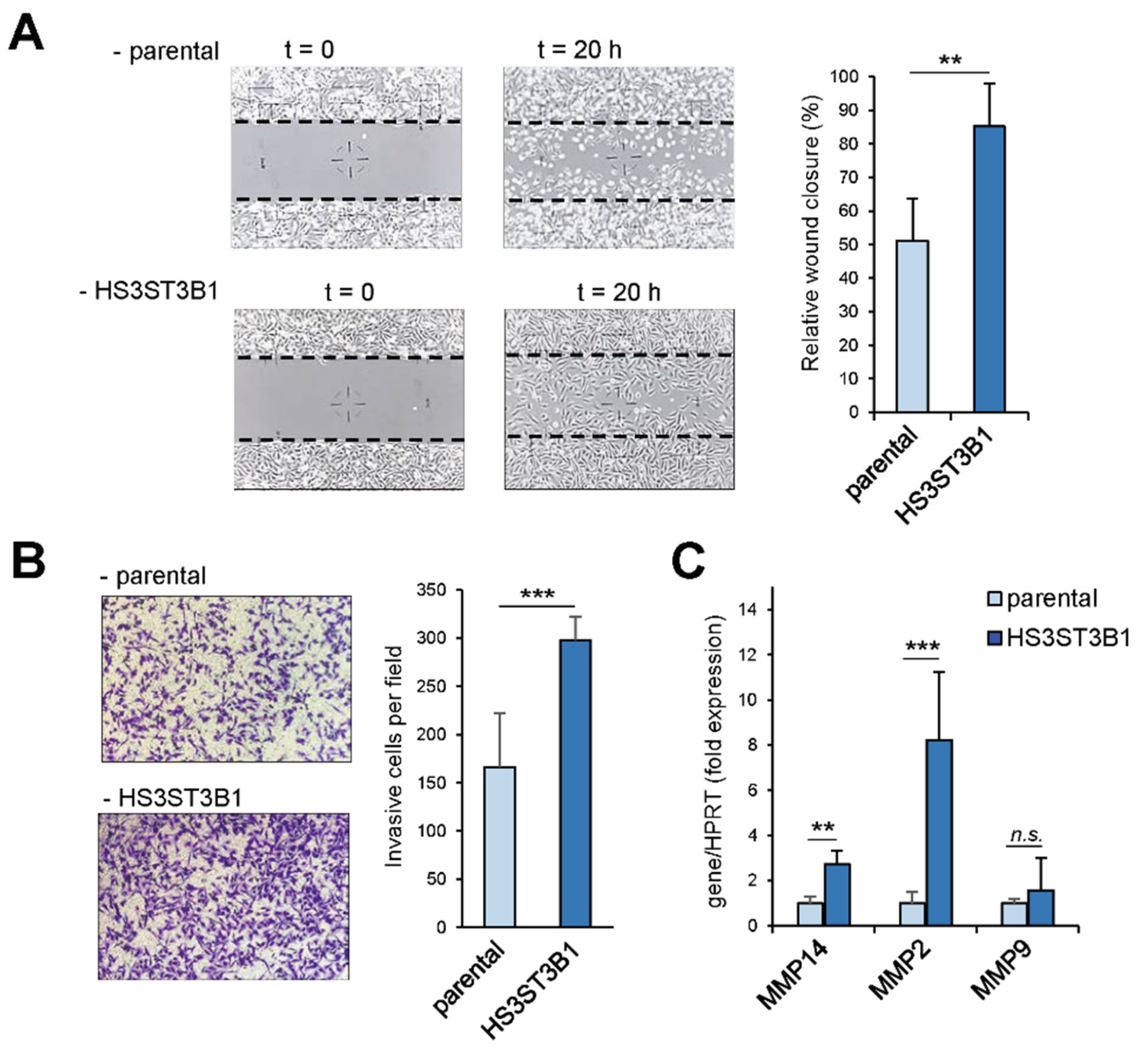
2.2. HS3ST3B1 Overexpression Enhances the Mesenchymal-Like and Invasive Phenotype of MDA-MB-231 Cells
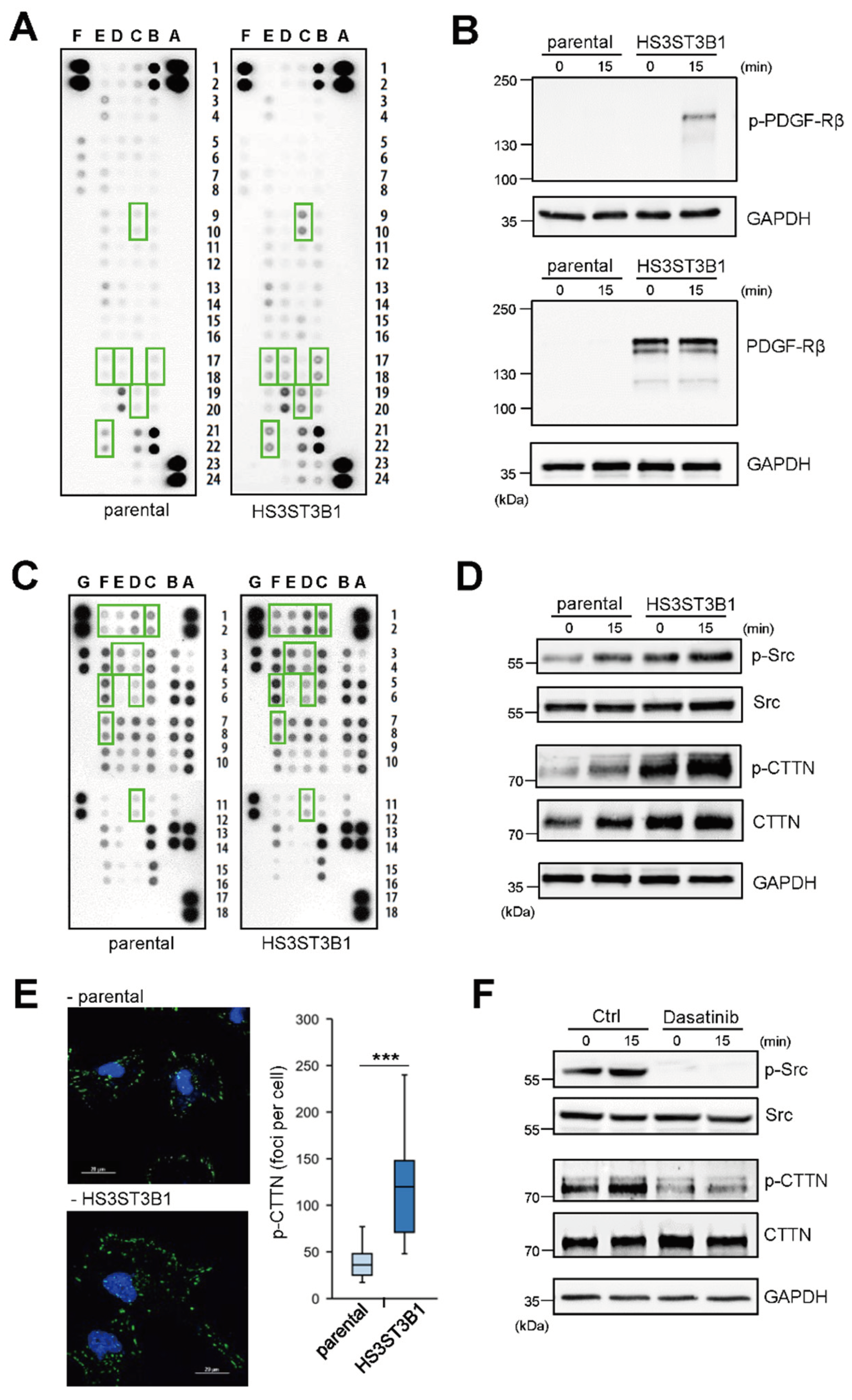
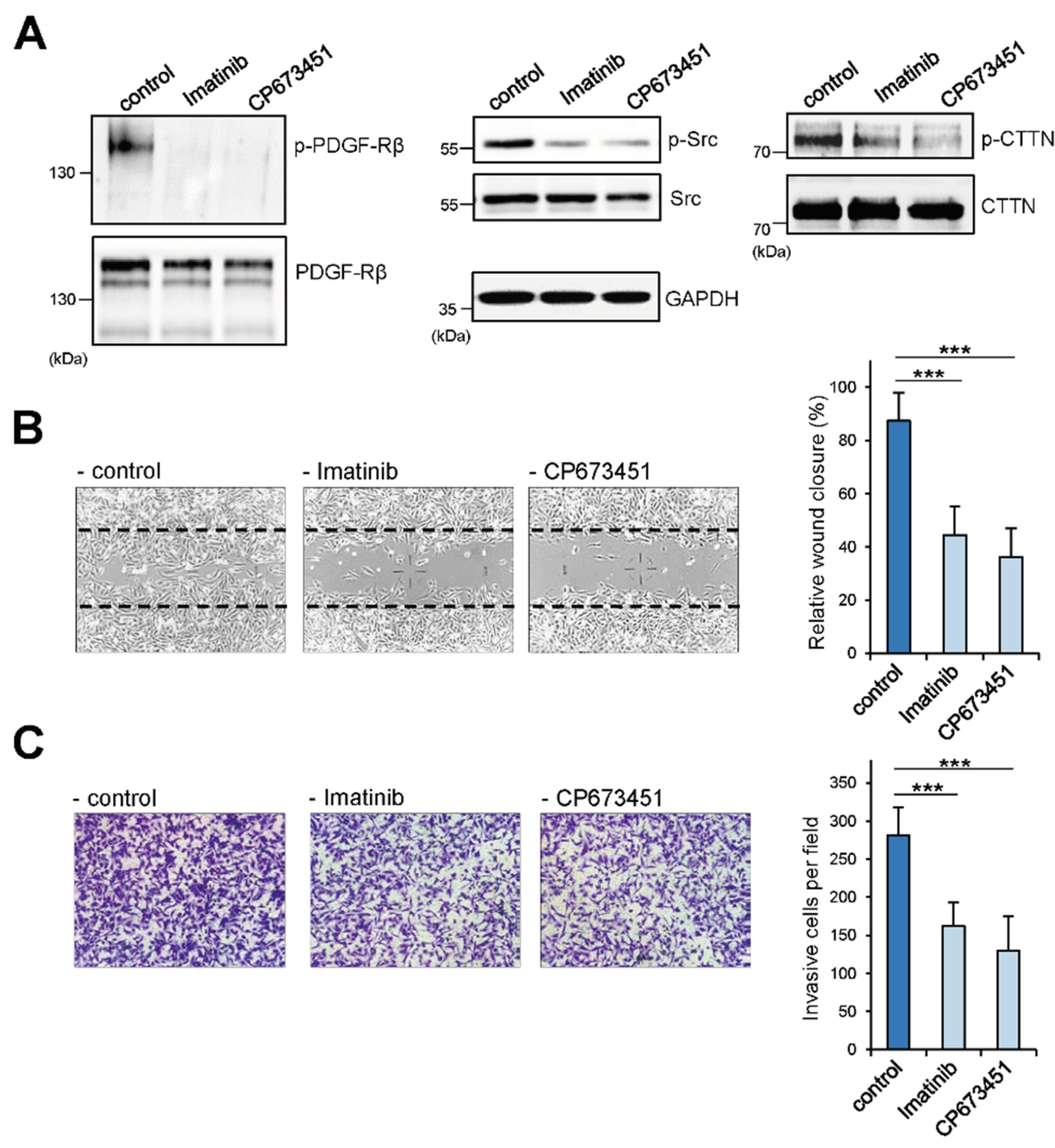
2.3. HS3ST3B1 Overexpression Enhances the Activation of PDGF-Rβ and Src/Cortactin Pathway
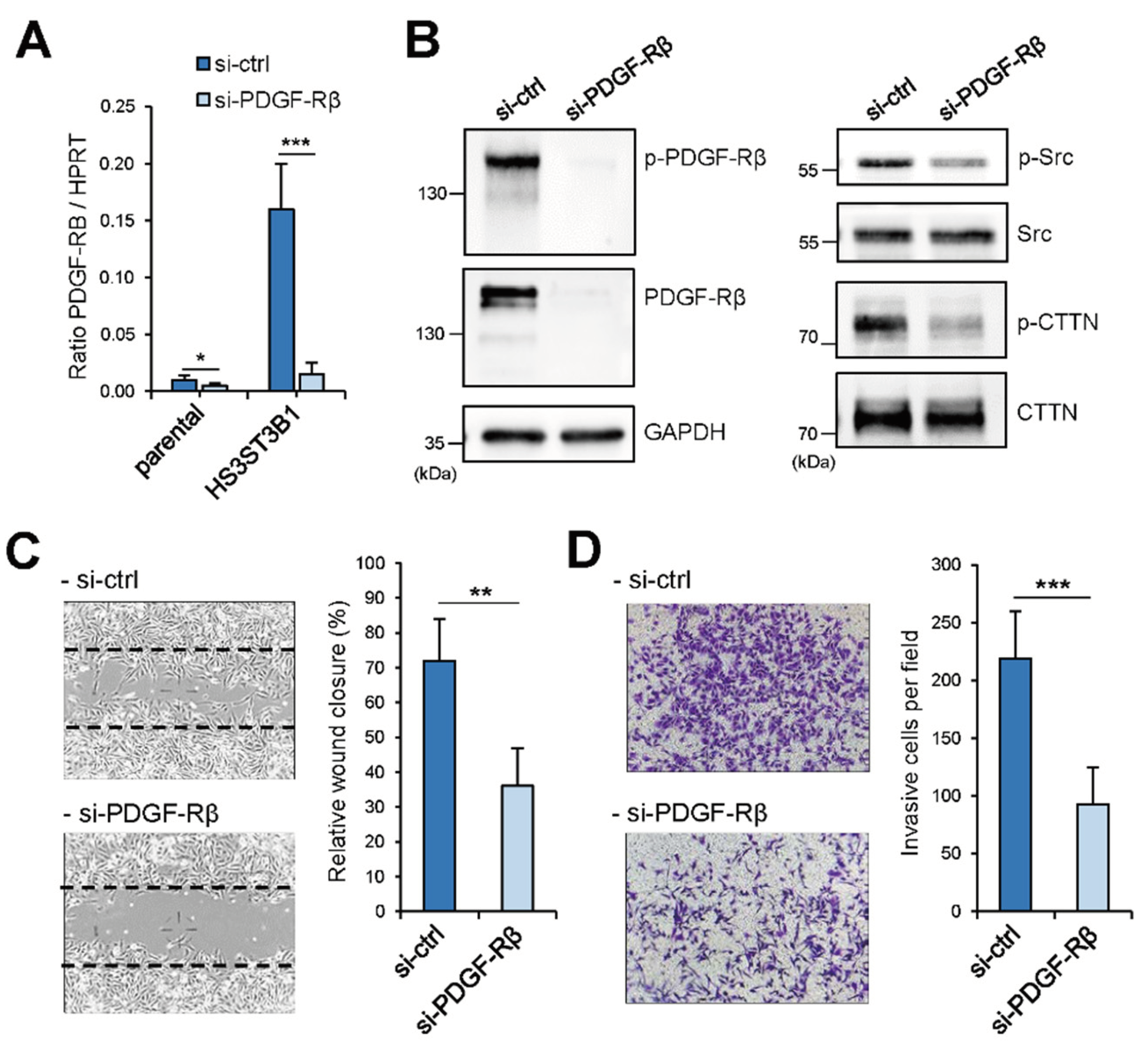
2.4. Enhanced Migration/Invasion of HS3STB1-Overexpressing MDA-MB-231 Cells is Dependent on the Expression of PDGF-Rβ
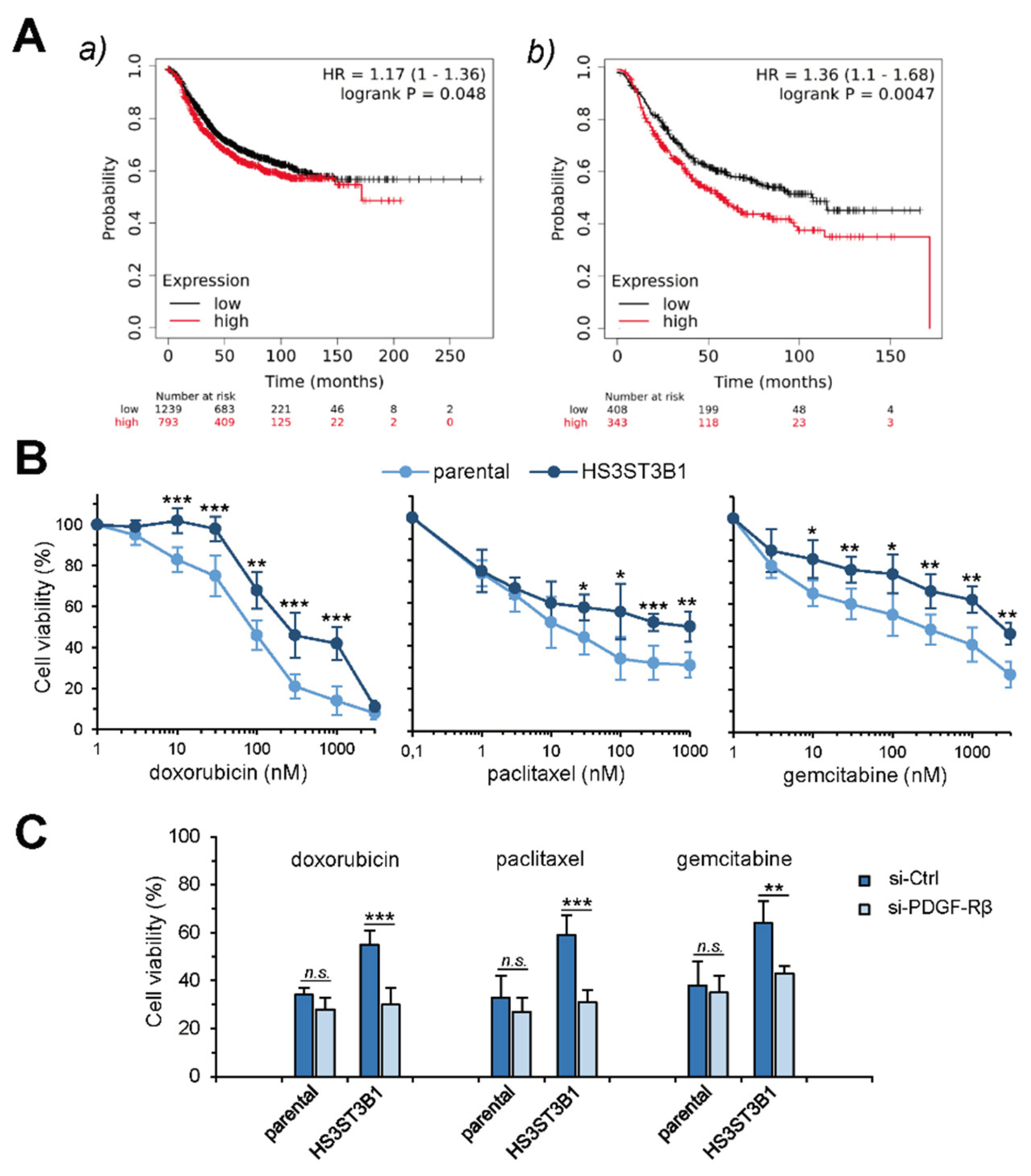
2.5. HS3ST3B1 Overexpression Is Associated with Poor Survival Outcome and Chemo-Resistance
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Microscopy Experiments
4.4. RNA Interference
4.5. RNA Isolation and Real-Time RT-PCR
4.6. SDS-PAGE and Western Blot
4.7. Microarray Experiments
4.8. Cell Migration and Invasion Assays
4.9. Measurement of Cell Viability
4.10. Data acquisition and Analysis from Public Open Databases
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Esko, J.D.; Selleck, S.B. Order out of chaos: assembly of ligand binding sites in heparan sulphate. Annu. Rev. Biochem. 2002, 71, 435–471. [Google Scholar] [CrossRef] [PubMed]
- Hayes, A.J.; Melrose, J. HS, an ancient molecular recognition and information storage glycosaminoglycan, equips HS-proteoglycans with diverse matrix and cell-interactive properties operative in tissue development and tissue function in health and disease. Int. J. Mol. Sci. 2023, 6, 1148. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.; Reis, C.A.; Vivès, R.R.; Magalhães, A. Heparan sulfate biosynthesis and sulfation profiles as modulators of cancer signalling and progression. Front. Oncol. 2021, 11, 778752. [Google Scholar] [CrossRef]
- Groux-Degroote, S.; Cavdarli, S.; Uchimura, K.; Allain, F.; Delannoy, P. Glycosylation changes in inflammatory diseases. Adv. Protein Chem. Struct. Biol. 2020, 119, 111–156. [Google Scholar] [CrossRef] [PubMed]
- Maïza, A.; Chantepie, S.; Vera, C.; Fifre, A.; Huynh, M.B.; Stettler, O.; Ouidja, M.O.; Papy-Garcia, D. The role of heparan sulfates in protein aggregation and their potential impact on neurodegeneration. FEBS Lett. 2018, 592, 3806–3818. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shworak, N.W.; Fritze, L.M.; Edelberg, J.M.; Rosenberg, R.D. Purification of heparan sulfate D-glucosaminyl 3-O-sulfotransferase. J. Biol. Chem. 1996, 271, 27072–27082. [Google Scholar] [CrossRef] [PubMed]
- Shukla, D.; Liu, J.; Blaiklock, P.; Shworak, N.W.; Bai, X.; Esko, J.D.; Cohen, G.H.; Eisenberg, R.J.; Rosenberg, R.D.; Spear, P.G. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 1999, 99, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Shworak, N.W.; Liu, J.; Petros, L.M.; Zhang, L.; Kobayashi, M.; Copeland, N.G.; Jenkins, N.A.; Rosenberg, R.D. Multiple isoforms of heparan sulfate D-glucosaminyl 3-O-sulfotransferase - isolation, characterization, and expression of human cDNAs and identification of distinct genomic loci. J. Biol. Chem. 1999, 274, 5170–5184. [Google Scholar] [CrossRef] [PubMed]
- Thacker, B.E.; Xu, D.; Lawrence, R.; Esko, J.D. Heparan sulfate 3-O-sulfation: a rare modification in search of a function. Matrix Biol. 2014, 35, 60–72. [Google Scholar] [CrossRef]
- Thacker, B.E.; Seamen, E.; Lawrence, R.; Parker, M.W.; Xu, Y.; Liu, J.; Vander Kooi, C.W.; Esko, J.D. Expanding the 3-O-sulfate proteome - Enhanced binding of neuropilin-1 to 3-O-sulfated heparan sulfate modulates its activity. ACS Chem. Biol. 2016, 11, 971–980. [Google Scholar] [CrossRef]
- Denys, A.; Allain, F. The emerging roles of heparan sulfate 3-O-sulfotransferases in cancer. Front. Oncol. 2019, 9, 507. [Google Scholar] [CrossRef]
- Zhang, Z.; Jiang, H.; Wang, Y.; Shi, M. Heparan sulfate D-glucosamine 3-O-sulfotransferase 3B1 is a novel regulator of transforming growth factor-beta-mediated epithelial-to-mesenchymal transition and regulated by miR-218 in non-small cell lung cancer. J. Cancer Res. Ther. 2018, 14, 24–29. [Google Scholar] [CrossRef]
- Li, Y.; Shi, B.; Dong, F.; Zhu, X.; Liu, B.; Liu, Y. Long non-coding RNA DLEU1 promotes cell proliferation, invasion, and confers cisplatin resistance in bladder cancer by regulating the miR-99b/HS3ST3B1 axis. Front. Genet. 2019, 10, 280. [Google Scholar] [CrossRef]
- Qi, J., Yan; Idrees, M.; Almutairi, S.M.; Rasheed, R.A.; Hussein, U.A.; Abdel-Maksoud, M.A.; Wang, R.; Huang, J.; Huang, C.; et al. Five EMT-related gene signatures predict acute myeloid leukemia patient outcome. Dis. Markers 2022, 2022, 7826393. [Google Scholar] [CrossRef]
- Liu, G.; Lu, Y.; Gao, D.; Huang, Z.; Ma, L. Identification of an energy metabolism-related six-gene signature for distinguishing and forecasting the prognosis of low-grade gliomas. Ann. Transl. Med. 2023, 11, 146. [Google Scholar] [CrossRef]
- Cherfils-Vicini, J.; Iltis, C.; Cervera, L.; Pisano, S.; Croce, O.; Sadouni, N.; Győrffy, B.; Collet, R.; Renault, V.M.; Rey-Millet, M.; et al. Cancer cells induce immune escape via glycocalyx changes controlled by the telomeric protein TRF2. EMBO J. 2019, 38, e100012. [Google Scholar] [CrossRef]
- Li, Y.; Shi, B.; Dong, F.; Zhu, X.; Liu, B.; Liu, Y. LncRNA KCNQ1OT1 facilitates the progression of bladder cancer by targeting MiR-218-5p/HS3ST3B1. Cancer Gene Ther. 2021, 28, 212–220. [Google Scholar] [CrossRef]
- Song, K.; Li, Q.; Jiang, Z.Z.; Guo, C.W.; Li, P. Heparan sulfate D-glucosaminyl 3-O sulfotransferase-3B1, a novel epithelial-mesenchymal transition inducer in pancreatic cancer. Cancer Biol. Ther. 2011, 12, 388–398. [Google Scholar] [CrossRef]
- Zhang, L.; Song, K.; Zhou, L.; Xie, Z.; Zhou, P.; Zhao, Y.; Han, Y.; Xu, X.; Li, P. Heparan sulfate D-glucosaminyl 3-O-sulfotransferase-3B1 (HS3ST3B1) promotes angiogenesis and proliferation by induction of VEGF in acute myeloid leukemia cells. J. Cell. Biochem. 2015, 116, 1101–1112. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Asada, K.; Fukutomi, T.; Okochi, E.; Yagi, Y.; Hasegawa, T.; Asahara, T.; Sugimura, T.; Ushijima, T. Methylation-associated silencing of heparan sulfate D-glucosaminyl 3-O-sulfotransferase-2 (3-OST-2) in human breast, colon, lung and pancreatic cancers. Oncogene 2003, 22, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Vijaya-Kumar, A.; Salem-Gassar, E.; Spillmann, D.; Stock, C.; Sen, Y.P.; Zhang, T.; Van Kuppevelt, T.H.; Hülsewig, C.; Koszlowski, E.O.; Pavao, M.S.G.; et al. HS3ST2 modulates breast cancer cell invasiveness via MAP kinase- and Tcf4 (Tcf7l2)-dependent regulation of protease and cadherin expression. Int. J. Cancer 2014, 135, 2579–2592. [Google Scholar] [CrossRef] [PubMed]
- Hellec, C.; Delos, M.; Carpentier, M.; Denys, A.; Allain, F. The heparan sulfate 3-O-sulfotransferases (HS3ST) 2, 3B and 4 enhance proliferation and survival in breast cancer MDA-MB-231 cells. PLoS One 2018, 13, e0194676. [Google Scholar] [CrossRef] [PubMed]
- Hellec, C.; Diawara, M.; Carpentier, M.; Denys, A.; Allain, F. The pro-tumoral activity of heparan sulfate 3-O-sulfotransferase 3B (HS3ST3B) in breast cancer MDA-MB-231 cells is dependent on the expression of neuropilin-1. Molecules 2018, 23, 2718. [Google Scholar] [CrossRef] [PubMed]
- Prud’homme, G.J.; Glinka, Y. Neuropilins are multifunctional coreceptors involved in tumor initiation, growth, metastasis and immunity. Oncotarget 2012, 3, 921–939. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J. Cancer. 2017, 8, 3131–3141. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.; Pineros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer, 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Heiser, L.M.; Sadanandam, A.; Kuo, W.L.; Benz, S.C.; Goldstein, T.C.; Ng, S.; Gibb, W.J.; Wang, N.J.; Ziyad, S.; Tong, F.; et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 2724–2729. [Google Scholar] [CrossRef]
- Barzaman, K.; Karami, J.; Zarei, Z.; Hosseinzadeh, A.; Kazemi, M.H.; Moradi-Kalbolandi, S.; Safari, E.; Farahmand, L. Breast cancer: biology, biomarkers, and treatments. Int. Immunopharmacol. 2020, 84, 106535. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Jézéquel, P.; Campone, M.; Gouraud, W.; Guérin-Charbonnel, C.; Leux, C.; Ricolleau, G.; Campion, L. bc-GenExMiner: an easy to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res. Treat. 2012, 131, 765–775. [Google Scholar] [CrossRef]
- Park, S.J.; Yoon, B.H.; Kim, S.K.; Kim, S.Y. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med. Genomics 2019, 12, 101. [Google Scholar] [CrossRef]
- Franke, T.F.; Yang, S.I.; Chan, T.O.; Datta, K.; Kazlauskas, A.; Morrison, D.K.; Kaplan, D.R.; Tsichlis, P.N. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell 1995, 81, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Schaller, M.D.; Hildebrand, J.D.; Shannon, J.D.; Fox, J.W.; Vines, R.R.; Parsons, J.T. Autophosphorylation of the focal adhesion kinase, pp125FAK, directs SH2-dependent binding of pp60src. Mol. Cell. Biol. 1994, 14, 1680–1688. [Google Scholar] [CrossRef]
- Yin, M.; Ma, W.; An, L. Cortactin in cancer cell migration and invasion. Oncotarget 2017, 8, 88232–88243. [Google Scholar] [CrossRef]
- Lanczky, A.; Gyorffy, B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Meng, F.; Speyer, C.L.; Zhang, B.; Zhao, Y.; Chen, W.; Gorski, D.H.; Miller, F.R.; Wu, G. PDGFRα and β play critical roles in mediating Foxq1-driven breast cancer stemness and chemoresistance. Cancer Res. 2015, 75, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Kalimutho, M.; Sinha, D.; Mittal, D.; Srihari, S.; Nanayakkara, D.; Shafique, S.; Raninga, P.; Nag, P.; Parsons, K.; Khanna, K.K. Blockade of PDGFRβ circumvents resistance to MEK-JAK inhibition via intratumoral CD8+ T-cells infiltration in triple-negative breast cancer. J. Exp. Clin. Cancer Res. 2019, 38, 85. [Google Scholar] [CrossRef]
- Xu, J.; Lamouille, S.; Derynck, R. TGF-β-induced epithelial to mesenchymal transition. Cell Res. 2009, 19, 156–172. [Google Scholar] [CrossRef]
- Krenn, E.C.; Wille, I.; Gesslbauer, B.; Poteser, M.; van Kuppevelt, T.H.; Kungl, A.J. Glycanogenomics: a qPCR-approach to investigate biological glycan function. Biochem. Biophys. Res. Commun. 2008, 375, 297–302. [Google Scholar] [CrossRef]
- Martinez, P.; Denys, A.; Delos, M.; Sikora, A.S.; Carpentier, M.; Julien, S.; Pestel, J.; Allain, F. Macrophage polarization alters the expression and sulfation pattern of glycosaminoglycans. Glycobiology 2015, 25, 502–513. [Google Scholar] [CrossRef]
- Sikora, A.S.; Delos, M.; Martinez, P.; Carpentier, M.; Allain, F.; Denys, A. Regulation of the expression of heparan sulfate 3-O-sulfotransferase 3B (HS3ST3B) by inflammatory stimuli in human monocytes. J. Cell. Biochem. 2016, 117, 1529–1542. [Google Scholar] [CrossRef]
- Sikora, A.S.; Hellec, C.; Carpentier, M.; Martinez, P.; Delos, M.; Denys, A.; Allain, F. Tumour-necrosis factor-α induces heparan sulfate 6-O-endosulfatase 1 (Sulf-1) expression in fibroblasts. Int. J. Biochem. Cell. Biol. 2016, 80, 57–65. [Google Scholar] [CrossRef]
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar] [CrossRef]
- Liao, S.J.; Luo, J.; Li, D.; Zhou, Y.H.; Yan, B.; Wei, J.J.; Tu, J.C.; Li, Y.R.; Zhang, G.M.; Feng, Z.H. TGF-β1 and TNF-α synergistically induce epithelial to mesenchymal transition of breast cancer cells by enhancing TAK1 activation J. Cell Commun. Signal 2019, 13, 369–380. [Google Scholar] [CrossRef]
- Biroccio, A.; Cherfils-Vicini, J.; Augereau, A.; Pinte, S.; Bauwens, S.; Ye, J.; Simonet, T.; Horard, B.; Jamet, K.; Cervera, L.; et al. TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat. Cell Biol. 2013, 15, 818–828. [Google Scholar] [CrossRef]
- Mao, X.; Gauche, C.; Coughtrie, M.W.; Bui, C.; Gulberti, S.; Merhi-Soussi, F.; Ramalanjaona, N.; Bertin-Jung, I.; Diot, A.; Dumas, A.D.; et al. The heparan sulfate sulfotransferase 3-OST3A (HS3ST3A) is a novel tumor regulator and a prognostic marker in breast cancer. Oncogene 2016, 35, 5043–5055. [Google Scholar] [CrossRef]
- Paulsson, J.; Sjöblom, T.; Micke, P.; Pontén, F.; Landberg, G.; Heldin, C.H.; Bergh, J.; Brennan, D.J.; Jirström, K.; Ostman, A. Prognostic significance of stromal platelet-derived growth factor beta-receptor expression in human breast cancer. Am. J. Pathol. 2009, 175, 334–341. [Google Scholar] [CrossRef]
- Hollier, B.G.; Tinnirello, A.A.; Werden, S.J.; Evans, K.W.; Taube, J.H.; Sarkar, T.R.; Sphyris, N.; Shariati, M.; Kumar, S.V.; Battula, V.L.; et al. FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer. Cancer Res. 2013, 73, 1981–1992. [Google Scholar] [CrossRef]
- Jechlinger, M.; Sommer, A.; Moriggl, R.; Seither, P.; Kraut, N.; Capodiecci, P.; Donovan, M.; Cordon-Cardo, C.; Beug, H.; Grünert, S. Autocrine PDGFR signaling promotes mammary cancer metastasis J. Clin. Invest. 2006, 116, 1561–1570. [Google Scholar] [CrossRef]
- Chopra, P.; Joshi, A.; Wu, J.; Lu, W.; Yadavalli, T.; Wolfert, M.A.; Shukla, D.; Zaia, J.; Boons, G.J. The 3-O-sulfation of heparan sulfate modulates protein binding and lyase degradation. Proc. Natl. Acad. Sci. U.S.A. 2021, 118, e2012935118. [Google Scholar] [CrossRef]
- West, D.; Rees, C.G.; Duchesne, L.; Patey, S.J.; Terry, C.J.; Turnbull, J.E.; Delehedde, M.; Heegaard, C.W.; Allain, F.; Vanpouille, C.; et al. Interactions of multiple heparin binding growth factors with neuropilin-1 and potentiation of the activity of fibroblast growth factor-2. J. Biol. Chem. 2005, 280, 13457–13464. [Google Scholar] [CrossRef] [PubMed]
- Vander Kooi, C.W.; Jusino, M.A.; Perman, B.; Neau, D.B.; Bellamy, H.D.; Leahy, D.J. Structural basis for ligand and heparin binding to neuropilin B domains. Proc. Nat.l Acad. Sci. U.S.A. 2007, 104, 6152–6157. [Google Scholar] [CrossRef] [PubMed]
- Sarabipour, S.; Mac Gabhann, F. VEGF-A121a binding to neuropilins - A concept revisited. Cell. Adh. Migr. 2018, 12, 204–214. [Google Scholar] [CrossRef]
- Muhl, L.; Folestad, E.B.; Gladh, H.; Wang, Y.; Moessinger, C.; Jakobsson, L.; Eriksson, U. Neuropilin 1 binds PDGF-D and is a co-receptor in PDGF-D-PDGFRβ signaling. J. Cell Sci. 2017, 130, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Lanzi, C.; Cassinelli, G. Heparan sulfate mimetics in cancer therapy: the challenge to define structural determinants and the relevance of targets for optimal activity. Molecules 2018, 23, 36. [Google Scholar] [CrossRef] [PubMed]
- Morla, S. Glycosaminoglycans and glycosaminoglycan mimetics in cancer and inflammation. Int. J. Mol. Sci. 2019, 20, 1963. [Google Scholar] [CrossRef] [PubMed]
- Cassinelli, G.; Favini, E.; Dal Bo, L.; Tortoreto, M.; De Maglie, M.; Dagrada, G.; Pilotti, S.; Zunino, F.; Zaffaroni, N.; Lanzi, C. Antitumor efficacy of the heparan sulfate mimic Roneparstat (SST0001) against sarcoma models involves multi-target inhibition of receptor tyrosine kinases. Oncotarget 2016, 7, 47848–47863. [Google Scholar] [CrossRef]
- Byrne, D.P.; Li, Y.; Ramakrishnan, K.; Barsukov, I.L.; Yates, E.A.; Eyers, C.E.; Papy-Garcia, D.; Chantepie, S.; Pagadala, V.; Liu, J.; et al. New tools for carbohydrate sulfation analysis: heparan sulfate 2-O sulfotransferase (HS2ST) is a target for small-molecule protein kinase Inhibitors. Biochem. J. 2018, 475, 2417–2433. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




