Submitted:
20 March 2024
Posted:
21 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
2.1. Isolation of Endophytic Bacteria by Halotolerance
2.2. Molecular Identification
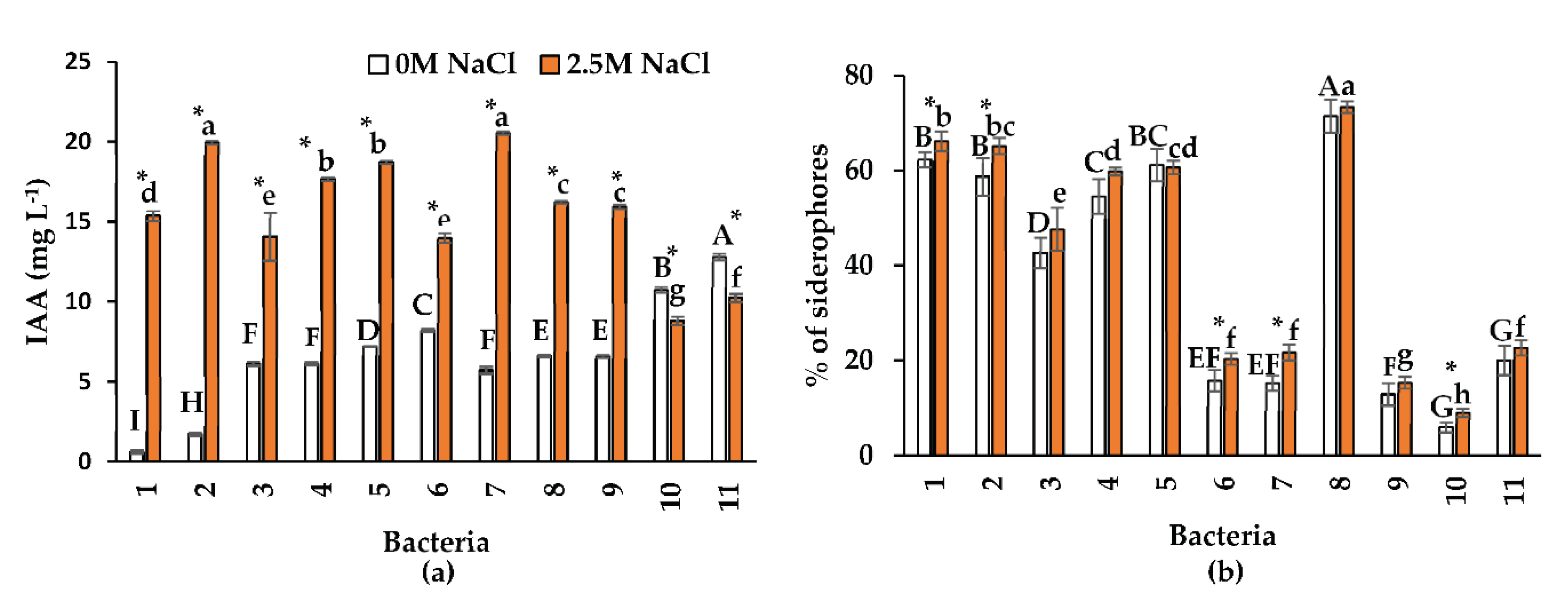
2.3. Indolacetic Acid and Siderophore Production
2.4. Nitrogenase Enzyme Activity and Exopolysaccharide Production
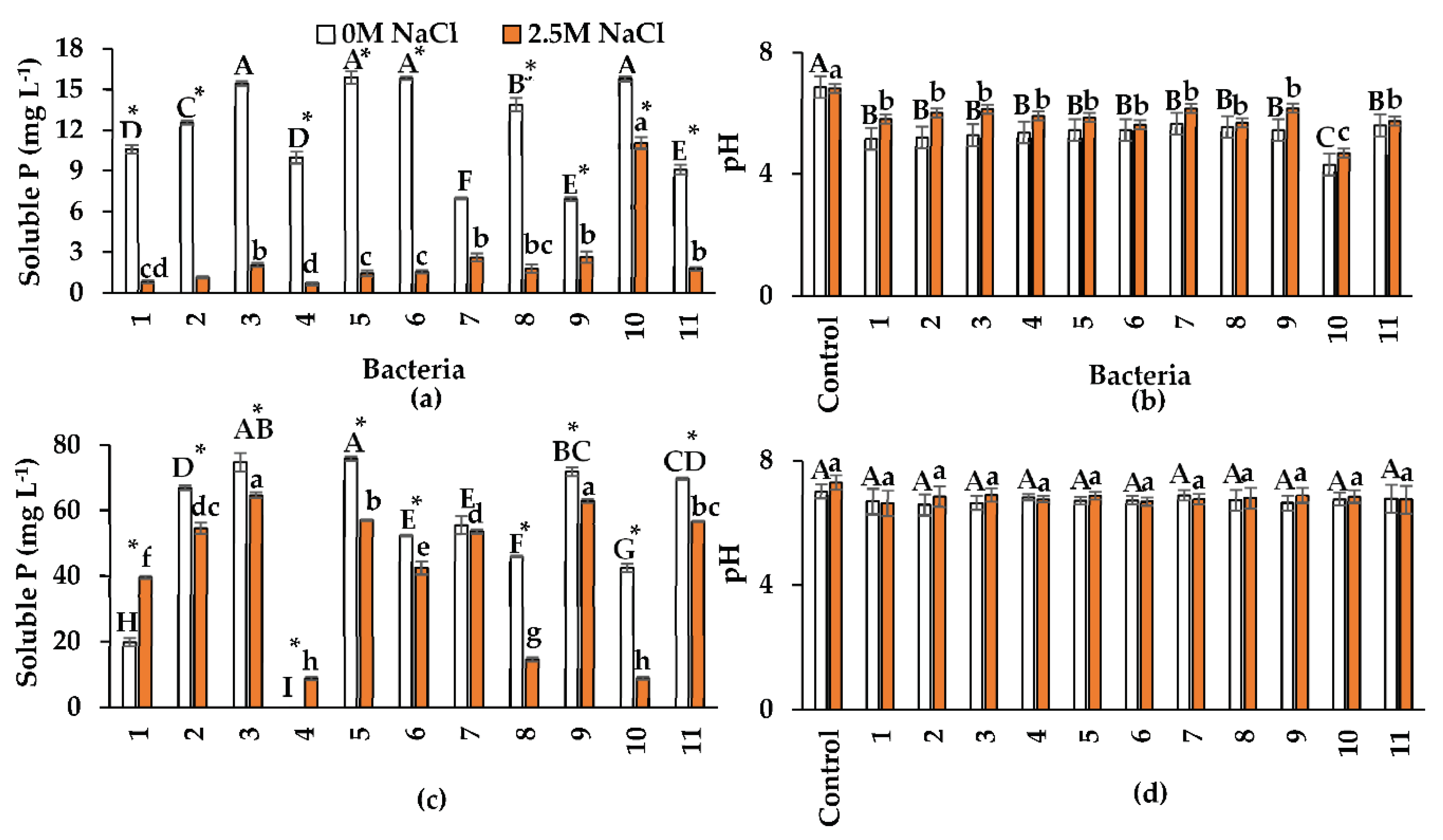
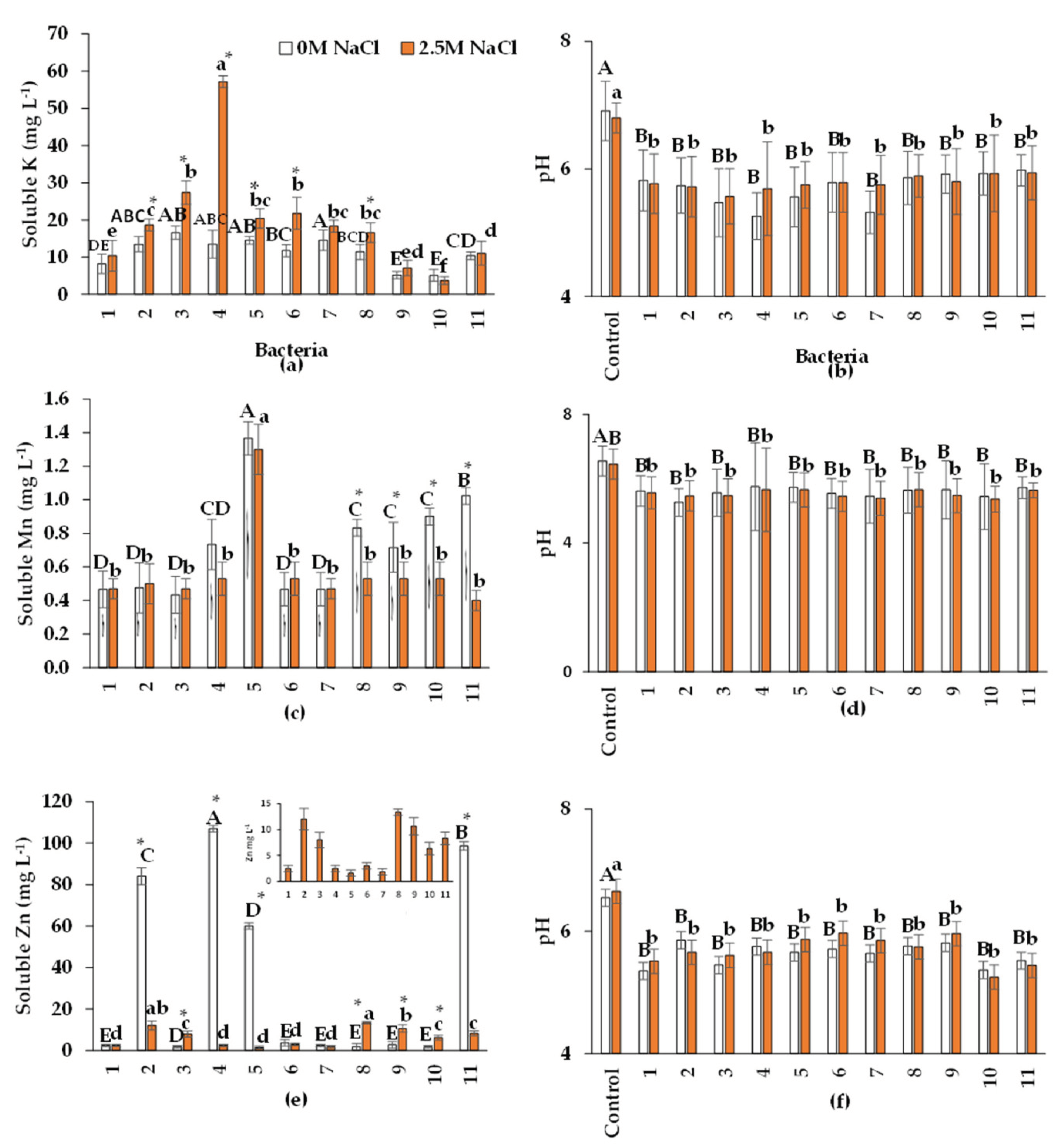
2.5. Nutrient Solubilization, Bacterial Growth, and pH Modification
2.5.1. Inorganic and Organic P
2.5.2. Solubilization of K, Mn, and Zn
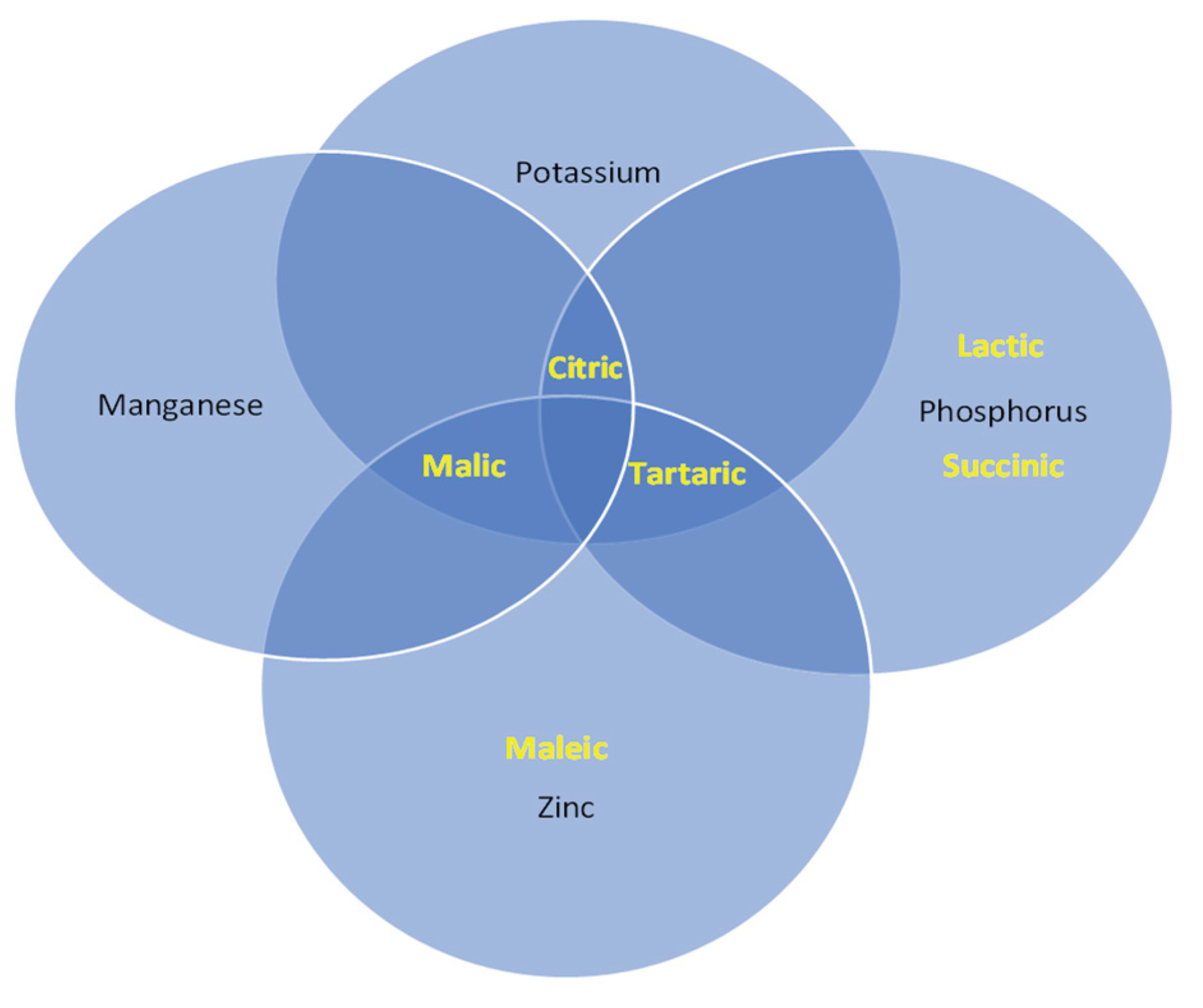
2.5.3. Interaction between Organic Acids and the Solubilization Ion
2.5.4. Production of Sugars in Nutrient Solubilization Tests
2.6. Enzyme Activity in Different Salinity Conditions
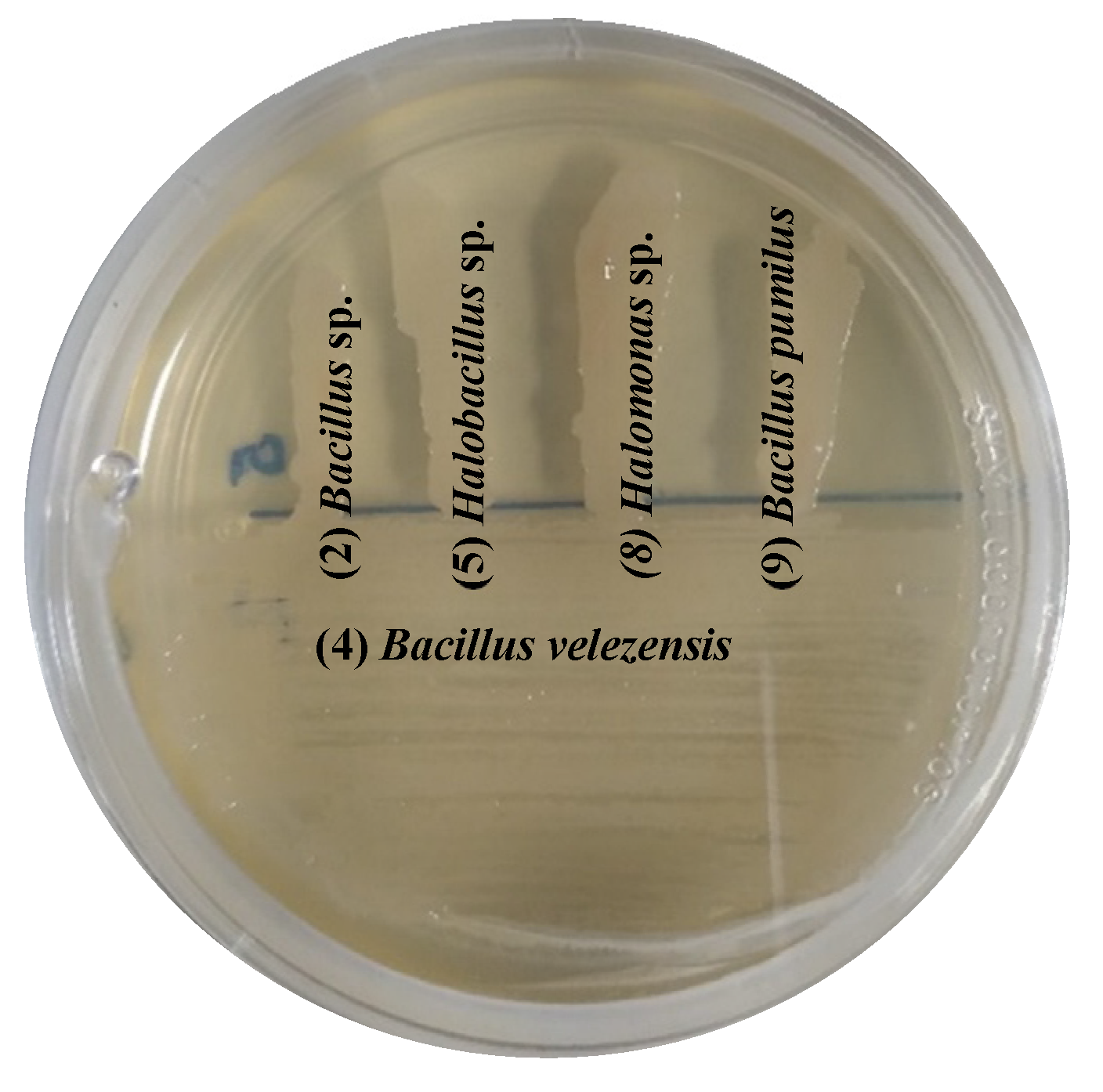
2.7. In vitro Compatibility between Bacterial Isolates
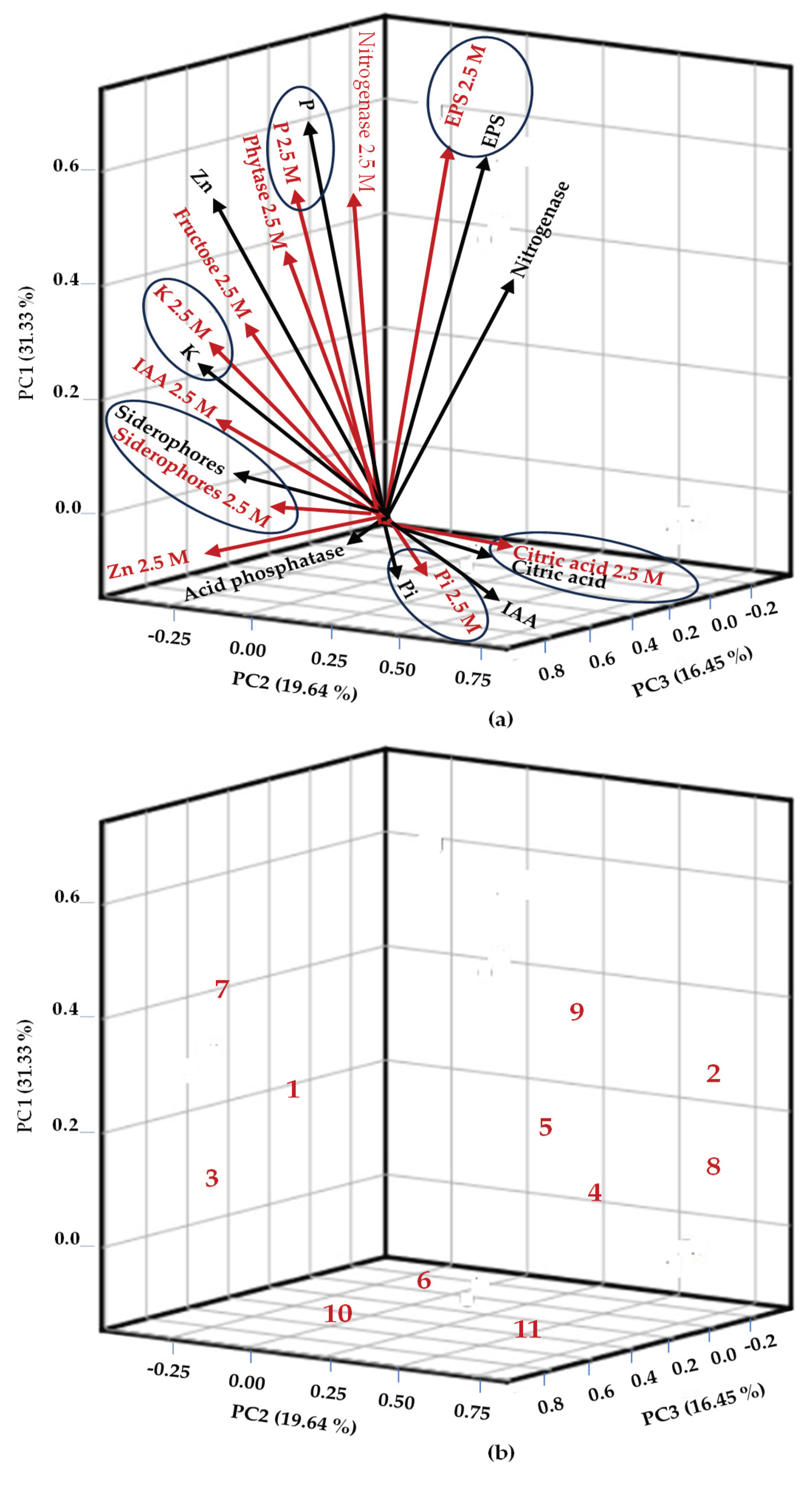
2.8. Principal Component Analysis
3. Materials and Methods
3.1. Selection of Halophytes, Sampling, and Root Disinfection
3.2. Isolation of Endophytic Bacteria by Halotolerance
3.3. Molecular Identification of Halophilic Endophytic Bacteria
3.4. Biochemical Characterization of Halophilic Endophytic Bacteria
3.4.1. Indole Acetic Acid and Siderophore Production
3.4.2. Exopolysaccharide and Nitrogenase Enzyme Production
3.4.3. Quantitative Determination of Nutrients Solubilization and Mechanisms Involved
3.4.3.1. Production of Organic Acids and Sugars
3.4.4. Extracellular Enzyme Activity at Different Salinity Conditions
3.5. Compatibility Between Bacterial Isolates
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siddikee, A.; Chauhan, P.S.; Anandham, R.; Han, G.-H.; Sa, T. Isolation, Characterization, and Use for Plant Growth Promotion Under Salt Stress, of ACC Deaminase-Producing Halotolerant Bacteria Derived from Coastal Soil. J. Microbiol. Biotechnol. 2010, 20, 1577–1584. [Google Scholar] [CrossRef]
- Ramadoss, D.; Lakkineni, V.K.; Bose, P.; Ali, S.; Annapurna, K. Mitigation of salt stress in wheat seedlings by halotolerant bacteria isolated from saline habitats. SpringerPlus 2013, 2, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zandi, E. , Jafari, A. A., & Mirakhorli, R. Studying the effects of growth stages on forage quality of two halophytes in Garmsar. Iranian Journal of Range and Desert Research, 2017, 24, 467-473. [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef] [PubMed]
- Naz, I.; Bano, A. Biochemical, molecular characterization and growth promoting effects of phosphate solubilizing Pseudomonas sp. isolated from weeds grown in salt range of Pakistan. Plant Soil 2010, 334, 199–207. [Google Scholar] [CrossRef]
- Shin, W.; Siddikee, A.; Joe, M.M.; Benson, A.; Kim, K.; Selvakumar, G.; Kang, Y.; Jeon, S.; Samaddar, S.; Chatterjee, P.; et al. Halotolerant Plant Growth Promoting Bacteria Mediated Salinity Stress Amelioration in Plants. Korean J. Soil Sci. Fertil. 2016, 49, 355–367. [Google Scholar] [CrossRef]
- Coombs, J.T.; Franco, C.M.M. Isolation and Identification of Actinobacteria from Surface-Sterilized Wheat Roots. Appl. Environ. Microbiol. 2003, 69, 5603–5608. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, S.; Malik, K.; Mehnaz, S. Microbiome of Halophytes: Diversity and Importance for Plant Health and Productivity. Microbiol. Biotechnol. Lett. 2019, 47, 1–10. [Google Scholar] [CrossRef]
- Kushwaha, P.; Kashyap, P.L.; Bhardwaj, A.K.; Kuppusamy, P.; Srivastava, A.K.; Tiwari, R.K. Bacterial endophyte mediated plant tolerance to salinity: growth responses and mechanisms of action. World J. Microbiol. Biotechnol. 2020, 36, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Dodd, I.C.; Zinovkina, N.Y.; Safronova, V.I.; Belimov, A.A. Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 2010, 157, 361–379. [Google Scholar] [CrossRef]
- Abbas, G.; Saqib, M.; Akhtar, J.; Haq, M.A.U. Interactive effects of salinity and iron deficiency on different rice genotypes. J. Plant Nutr. Soil Sci. 2015, 178, 306–311. [Google Scholar] [CrossRef]
- Etesami, H.; Alikhani, H.A. Bacillus species as the most promising bacterial biocontrol agents in rhizosphere and endorhiza of plants grown in rotation with each other. Eur. J. Plant Pathol. 2017, 150, 497–506. [Google Scholar] [CrossRef]
- Puente, M.E.; Bashan, Y.; Li, C.Y.; Lebsky, V.K. Microbial Populations and Activities in the Rhizoplane of Rock-Weathering Desert Plants. I. Root Colonization and Weathering of Igneous Rocks. Plant Biol. 2004, 6, 629–642. [Google Scholar] [CrossRef]
- Wei, Y.; Zhao, Y.; Shi, M.; Cao, Z.; Lu, Q.; Yang, T.; Fan, Y.; Wei, Z. Effect of organic acids production and bacterial community on the possible mechanism of phosphorus solubilization during composting with enriched phosphate-solubilizing bacteria inoculation. Bioresour. Technol. 2017, 247, 190–199. [Google Scholar] [CrossRef]
- Seshachala, U.; Tallapragada, P. Phosphate Solubilizers from the Rhizospher of Piper nigrum L. in Karnataka, India. Chil. J. Agric. Res. 2012, 72, 397–403. [Google Scholar] [CrossRef]
- Gontia-Mishra, I.; Sapre, S.; Kachare, S.; Tiwari, S. Molecular diversity of 1-aminocyclopropane-1-carboxylate (ACC) deaminase producing PGPR from wheat (Triticum aestivum L.) rhizosphere. Plant Soil 2016, 414, 213–227. [Google Scholar] [CrossRef]
- Alloway, B.J. Soil factors associated with zinc deficiency in crops and humans. Environ. Geochem. Health 2009, 31, 537–548. [Google Scholar] [CrossRef]
- Bradáčová, K.; Florea, A.S.; Bar-Tal, A.; Minz, D.; Yermiyahu, U.; Shawahna, R.; Kraut-Cohen, J.; Zolti, A.; Erel, R.; Dietel, K.; et al. Microbial Consortia versus Single-Strain Inoculants: An Advantage in PGPM-Assisted Tomato Production? Agronomy 2019, 9, 105. [Google Scholar] [CrossRef]
- Liu, X.; Mei, S.; Salles, J.F. Inoculated microbial consortia perform better than single strains in living soil: A meta-analysis. Appl. Soil Ecol. 2023, 190. [Google Scholar] [CrossRef]
- Shenoy, V. V. , & Kalagudi, G. M. Meta-bug and near-isogenic strain consortia concepts for plant growth promoting rhizobacteria. In 6th International PGPR workshop, India, section VII—mechanism of biocontrol. 2003, p (Vol. 108).
- Vaishnav, A. , Varma, A., Tuteja, N., Choudhary, D.K. PGPR-mediated amelioration of crops under salt stress. In: Choudhary, D., Varma, A., Tuteja, N. (eds) Plant-Microbe Interaction: An Approach to Sustainable Agriculture. Springer, Singapore, 2016. [CrossRef]
- Marghoob, M.U.; Nawaz, A.; Ahmad, M.; Waheed, M.Q.; Khan, M.H.; Imtiaz, M.; Islam, E.U.; Imran, A.; Mubeen, F. Assessment of halotolerant bacterial and fungal consortia for augmentation of wheat in saline soils. Front. Microbiol. 2023, 14, 1207784. [Google Scholar] [CrossRef]
- Rajput, L., Imran, A., Mubeen, F., & Hafeez, F. Y. Wheat (Triticum aestivum L.) growth promotion by halo-tolerant PGPR-consortium. Soil Environment 2018, 37, 178–189. [CrossRef]
- Damodaran, T.; Jha, S.K.; Kumari, S.; Gupta, G.; Mishra, V.K.; Sharma, P.C.; Gopal, R.; Singh, A.; Jat, H.S. Development of Halotolerant Microbial Consortia for Salt Stress Mitigation and Sustainable Tomato Production in Sodic Soils: An Enzyme Mechanism Approach. Sustainability 2023, 15, 5186. [Google Scholar] [CrossRef]
- Merino, N.; Aronson, H.S.; Bojanova, D.P.; Feyhl-Buska, J.; Wong, M.L.; Zhang, S.; Giovannelli, D. Living at the Extremes: Extremophiles and the Limits of Life in a Planetary Context. Front. Microbiol. 2019, 10, 780. [Google Scholar] [CrossRef] [PubMed]
- Albdaiwi, R.N.; Khyami-Horani, H.; Ayad, J.Y.; Alananbeh, K.M.; Al-Sayaydeh, R. Isolation and Characterization of Halotolerant Plant Growth Promoting Rhizobacteria From Durum Wheat (Triticum turgidum subsp. durum) Cultivated in Saline Areas of the Dead Sea Region. Front. Microbiol. 2019, 10, 1639. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, S.; Mehnaz, S.; Mirza, M.S.; Malik, K.A. Isolation and characterization of bacteria associated with the rhizosphere of halophytes (Salsola stocksii and Atriplex amnicola) for production of hydrolytic enzymes. Braz. J. Microbiol. 2019, 50, 85–97. [Google Scholar] [CrossRef]
- Mora-Ruiz, M.d.R.; Font-Verdera, F.; Orfila, A.; Rita, J.; Rosselló-Móra, R. Endophytic microbial diversity of the halophyteArthrocnemum macrostachyumacross plant compartments. FEMS Microbiol. Ecol. 2016, 92, fiw145. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Gao, J.; Chen, X.; Zhang, M.; Yang, F.; Du, Y.; Moe, T.S.; Munir, I.; Xue, J.; Zhang, X. The Endophytic Bacteria Bacillus velezensis Lle-9, Isolated from Lilium leucanthum, Harbors Antifungal Activity and Plant Growth-Promoting Effects. J. Microbiol. Biotechnol. 2020, 30, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Brahim, A.H.; Ben Ali, M.; Daoud, L.; Jlidi, M.; Akremi, I.; Hmani, H.; Feto, N.A.; Ben Ali, M. Biopriming of Durum Wheat Seeds with Endophytic Diazotrophic Bacteria Enhances Tolerance to Fusarium Head Blight and Salinity. Microorganisms 2022, 10, 970. [Google Scholar] [CrossRef] [PubMed]
- Szymańska, S.; Dąbrowska, G.B.; Tyburski, J.; Niedojadło, K.; Piernik, A.; Hrynkiewicz, K. Boosting the Brassica napus L. tolerance to salinity by the halotolerant strain Pseudomonas stutzeri ISE12. Environ. Exp. Bot. 2019, 163, 55–68. [Google Scholar] [CrossRef]
- Mahgoub, H.A.M.; Fouda, A.; Eid, A.M.; Ewais, E.E.-D.; Hassan, S.E.-D. Biotechnological application of plant growth-promoting endophytic bacteria isolated from halophytic plants to ameliorate salinity tolerance of Vicia faba L. Plant Biotechnol. Rep. 2021, 15, 819–843. [Google Scholar] [CrossRef]
- Shurigin, V.; Egamberdieva, D.; Li, L.; Davranov, K.; Panosyan, H.; Birkeland, N.-K.; Wirth, S.; Bellingrath-Kimura, S.D. Endophytic bacteria associated with halophyte Seidlitzia rosmarinus Ehrenb. ex Boiss. from saline soil of Uzbekistan and their plant beneficial traits. J. Arid. Land 2020, 12, 730–740. [Google Scholar] [CrossRef]
- Huang, Q.-S.; Yan, Z.-F.; Chen, X.-Q.; Du, Y.-Y.; Li, J.; Liu, Z.-Z.; Xia, W.; Chen, S.; Wu, J. Accelerated biodegradation of polyethylene terephthalate by Thermobifida fusca cutinase mediated by Stenotrophomonas pavanii. Sci. Total. Environ. 2022, 808, 152107. [Google Scholar] [CrossRef]
- Bhutani, N.; Maheshwari, R.; Sharma, N.; Kumar, P.; Dang, A.S.; Suneja, P. Characterization of halo-tolerant plant growth promoting endophytic Bacillus licheniformis MHN 12. J. Genet. Eng. Biotechnol. 2022, 20, 113–12. [Google Scholar] [CrossRef]
- Dhungana, S.A.; Itoh, K. Effects of Co-Inoculation of Indole-3-Acetic Acid-Producing and -Degrading Bacterial Endophytes on Plant Growth. Horticulturae 2019, 5, 17. [Google Scholar] [CrossRef]
- Mahdi, I.; Hafidi, M.; Allaoui, A.; Biskri, L. Halotolerant Endophytic Bacterium Serratia rubidaea ED1 Enhances Phosphate Solubilization and Promotes Seed Germination. Agriculture 2021, 11, 224. [Google Scholar] [CrossRef]
- Patel, R.; Parekh, V.B. Culturable endophytic bacteria from halotolerant Salicornia brachata L. : Isolation and plant growth promoting traits. Indian J. Appl. Microbiol. 2018, 21, 10–21. [Google Scholar] [CrossRef]
- Panwar, M.; Tewari, R.; Nayyar, H. Native halo-tolerant plant growth promoting rhizobacteria Enterococcus and Pantoea sp. improve seed yield of Mungbean (Vigna radiata L.) under soil salinity by reducing sodium uptake and stress injury. Physiol. Mol. Biol. Plants 2016, 22, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Kearl, J.; McNary, C.; Lowman, J.S.; Mei, C.; Aanderud, Z.T.; Smith, S.T.; West, J.; Colton, E.; Hamson, M.; Nielsen, B.L. Salt-Tolerant Halophyte Rhizosphere Bacteria Stimulate Growth of Alfalfa in Salty Soil. Front. Microbiol. 2019, 10, 1849. [Google Scholar] [CrossRef] [PubMed]
- Abbas, R.; Rasul, S.; Aslam, K.; Baber, M.; Shahid, M.; Mubeen, F.; Naqqash, T. Halotolerant PGPR: A hope for cultivation of saline soils. J. King Saud Univ. - Sci. 2019, 31, 1195–1201. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Leong, J.; Teintze, M.; Schroth, M.N. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 1980, 286, 885–886. [Google Scholar] [CrossRef]
- Vyacheslav, S.; Begali, A.; Kakhramon, D.; Zafar, I. Bacterial endophytes from halophyte black saxaul (Haloxylon aphyllum Minkw.) and their plant growth-promoting properties. J. Appl. Biol. Biotechnol. 2022, 10, 45–53. [Google Scholar] [CrossRef]
- Enquahone, S.; van Marle, G.; Simachew, A. Plant growth-promoting characteristics of halotolerant endophytic bacteria isolated from Sporobolus specatus (Vahr) Kunth and Cyperus laevigatus L. of Ethiopian rift valley lakes. Arch. Microbiol. 2022, 204, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ruppel, S.; Franken, P.; Witzel, K. Properties of the halophyte microbiome and their implications for plant salt tolerance. Funct. Plant Biol. 2013, 40, 940–951. [Google Scholar] [CrossRef]
- Shahid, M.; Zeyad, M.T.; Syed, A.; Singh, U.B.; Mohamed, A.; Bahkali, A.H.; Elgorban, A.M.; Pichtel, J. Stress-Tolerant Endophytic Isolate Priestia aryabhattai BPR-9 Modulates Physio-Biochemical Mechanisms in Wheat (Triticum aestivum L.) for Enhanced Salt Tolerance. Int. J. Environ. Res. Public Heal. 2022, 19, 10883. [Google Scholar] [CrossRef]
- Paredes-Mendoza, M. , & Espinosa-Victoria, D. Ácidos orgánicos producidos por rizobacterias que solubilizan fosfato: una revisión crítica. Terra Latinoamericana, 2010, 28, 61–70. [Google Scholar]
- Rodrı́guez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef] [PubMed]
- Dey, G.; Banerjee, P.; Sharma, R.K.; Maity, J.P.; Etesami, H.; Shaw, A.K.; Huang, Y.-H.; Huang, H.-B.; Chen, C.-Y. Management of Phosphorus in Salinity-Stressed Agriculture for Sustainable Crop Production by Salt-Tolerant Phosphate-Solubilizing Bacteria—A Review. Agronomy 2021, 11, 1552. [Google Scholar] [CrossRef]
- Li, W.C.; Ye, Z.H.; Wong, M.H. Metal mobilization and production of short-chain organic acids by rhizosphere bacteria associated with a Cd/Zn hyperaccumulating plant, Sedum alfredii. Plant Soil 2009, 326, 453–467. [Google Scholar] [CrossRef]
- Rasul, M.; Yasmin, S.; Yahya, M.; Breitkreuz, C.; Tarkka, M.; Reitz, T. The wheat growth-promoting traits of Ochrobactrum and Pantoea species, responsible for solubilization of different P sources, are ensured by genes encoding enzymes of multiple P-releasing pathways. Microbiol. Res. 2021, 246, 126703. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial Phosphorus Solubilization and Its Potential for Use in Sustainable Agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef]
- Rosales, E., Pasoz, M., Sanroman, M. A. Solid-state fermentation for food applications. In Current Developments in Biotechnology and Bioengineering; Pandey, A., Larroche, C., Soccol, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2018, 319–355. [CrossRef]
- Etesami, H.; Beattie, G.A. Plant-Microbe Interactions in Adaptation of Agricultural Crops to Abiotic Stress Conditions. In Probiotics and Plant Health; Springer Singapore: Singapore, Singapore, 2017; pp. 163–200. [Google Scholar] [CrossRef]
- Patel, P. K. , Jadav, N. J., & Gohil, V. K. N. Direct effects of potassium, sulphur and KSB on growth, yield and quality of direct seeded rice under direct seeded rice-chickpea cropping system. The Pharma Innovation Journal 2023, SP-12, 846-851.
- Bakelli, A., Amrani, S., Nacer, A., Bouri, M., & Sahin, F. Klebsiella sp. S6. A halotolerant rhizosphere bacterium of Phragmites communis L. with potential plant-growth promotion of pepper. Analele Universitatii din Oradea, Fascicula Biologie, 2022, 29, 92-100.
- Ranawat, B.; Bachani, P.; Singh, A.; Mishra, S. Enterobacter hormaechei as Plant Growth-Promoting Bacteria for Improvement in Lycopersicum esculentum. Curr. Microbiol. 2021, 78, 1208–1217. [Google Scholar] [CrossRef] [PubMed]
- Ashfaq, M.; Hassan, H.M.; Ghazali, A.H.A.; Ahmad, M. Halotolerant potassium solubilizing plant growth promoting rhizobacteria may improve potassium availability under saline conditions. Environ. Monit. Assess. 2020, 192, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Zarjani, J.K.; Aliasgharzad, N.; Oustan, S.; Emadi, M.; Ahmadi, A. Isolation and characterization of potassium solubilizing bacteria in some Iranian soils. Arch. Agron. Soil Sci. 2013, 59, 1713–1723. [Google Scholar] [CrossRef]
- Tiwari, S.; Singh, P.; Tiwari, R.; Meena, K.K.; Yandigeri, M.; Singh, D.P.; Arora, D.K. Salt-tolerant rhizobacteria-mediated induced tolerance in wheat (Triticum aestivum) and chemical diversity in rhizosphere enhance plant growth. Biol. Fertil. Soils 2011, 47, 907–916. [Google Scholar] [CrossRef]
- Yamaji, K.; Nagata, S.; Haruma, T.; Ohnuki, T.; Kozaki, T.; Watanabe, N.; Nanba, K. Root endophytic bacteria of a 137Cs and Mn accumulator plant, Eleutherococcus sciadophylloides, increase 137Cs and Mn desorption in the soil. J. Environ. Radioact. 2016, 153, 112–119. [Google Scholar] [CrossRef]
- Dixit, S. J. , Kuttan, A. K., & Shrivastava, R. M. Genetic characterization of sulphur and iron oxidizing bacteria in manganese mining area of Balaghat and Chhindwara, Madhya Pradesh, 2018, India.
- Uren, N. Chemical reduction of an insoluble higher oxide of manganese by plant roots. J. Plant Nutr. 1981, 4, 65–71. [Google Scholar] [CrossRef]
- Ijaz, A.; Mumtaz, M.Z.; Wang, X.; Ahmad, M.; Saqib, M.; Maqbool, H.; Zaheer, A.; Wang, W.; Mustafa, A. Insights Into Manganese Solubilizing Bacillus spp. for Improving Plant Growth and Manganese Uptake in Maize. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.B.; Jensen, P.E.; Husted, S. Manganese Deficiency in Plants: The Impact on Photosystem II. Trends Plant Sci. 2016, 21, 622–632. [Google Scholar] [CrossRef]
- Schmidt, S.B.; Husted, S. The Biochemical Properties of Manganese in Plants. Plants 2019, 8, 381. [Google Scholar] [CrossRef]
- Sanket, A.S.; Ghosh, S.; Sahoo, R.; Nayak, S.; Das, A.P. Molecular Identification of Acidophilic Manganese (Mn)-Solubilizing Bacteria from Mining Effluents and Their Application in Mineral Beneficiation. Geomicrobiol. J. 2016, 34, 71–80. [Google Scholar] [CrossRef]
- Fatima, T.; Mishra, I.; Verma, R.; Arora, N.K. Mechanisms of halotolerant plant growth promoting Alcaligenes sp. involved in salt tolerance and enhancement of the growth of rice under salinity stress. 3 Biotech 2020, 10, 361. [Google Scholar] [CrossRef]
- Tewari, S.; Arora, N.K. Fluorescent Pseudomonas sp. PF17 as an efficient plant growth regulator and biocontrol agent for sunflower crop under saline conditions. Symbiosis 2016, 68, 99–108. [Google Scholar] [CrossRef]
- Ramesh, A.; Sharma, S.K.; Sharma, M.P.; Yadav, N.; Joshi, O.P. Inoculation of zinc solubilizing Bacillus aryabhattai strains for improved growth, mobilization and biofortification of zinc in soybean and wheat cultivated in Vertisols of central India. Appl. Soil Ecol. 2014, 73, 87–96. [Google Scholar] [CrossRef]
- Upadhyay, H.; Gangola, S.; Sharma, A.; Singh, A.; Maithani, D.; Joshi, S. Contribution of zinc solubilizing bacterial isolates on enhanced zinc uptake and growth promotion of maize (Zea mays L.). Folia Microbiol. 2021, 66, 1–11. [Google Scholar] [CrossRef]
- Li, W.C.; Ye, Z.H.; Wong, M.H. Metal mobilization and production of short-chain organic acids by rhizosphere bacteria associated with a Cd/Zn hyperaccumulating plant, Sedum alfredii. Plant Soil 2009, 326, 453–467. [Google Scholar] [CrossRef]
- Zhang, L.; Shi, N.; Fan, J.; Wang, F.; George, T.S.; Feng, G. Arbuscular mycorrhizal fungi stimulate organic phosphate mobilization associated with changing bacterial community structure under field conditions. Environ. Microbiol. 2018, 20, 2639–2651. [Google Scholar] [CrossRef]
- Sharma, V.; Kumar, V.; Archana, G.; Kumar, G.N. Substrate specificity of glucose dehydrogenase (GDH) ofEnterobacter asburiaePSI3 and rock phosphate solubilization with GDH substrates as C sources. Can. J. Microbiol. 2005, 51, 477–482. [Google Scholar] [CrossRef]
- Etesami, H.; Glick, B.R. Halotolerant plant growth–promoting bacteria: Prospects for alleviating salinity stress in plants. Environ. Exp. Bot. 2020, 178, 104124. [Google Scholar] [CrossRef]
- Shabaan, M. Preliminary evaluation of halotolerant PGPRs for promoting maize growth under salt stressed gnotobiotic conditions. Pak. J. Agric. Sci. 2021, 58, 1491–1500. [Google Scholar] [CrossRef]
- Cummings, S.P.; Gilmour, D.J. The effect of NaCl on the growth of a Halomonas species: accumulation and utilization of compatible solutes. Microbiology 1995, 141, 1413–1418. [Google Scholar] [CrossRef]
- Barrera, M.C.; Jakobs-Schoenwandt, D.; Gómez, M.I.; Becker, M.; Patel, A.V.; Ruppel, S. Salt stress and hydroxyectoine enhance phosphate solubilisation and plant colonisation capacity of Kosakonia radicincitans. J. Adv. Res. 2019, 19, 91–97. [Google Scholar] [CrossRef]
- Sharma, S.B.; Sayyed, R.Z.; Trivedi, M.H.; Gobi, T.A. Phosphate solubilizing microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. Springer Plus 2013, 2, 587. [Google Scholar] [CrossRef]
- Belkebla, N.; Bessai, S.A.; Melo, J.; Caeiro, M.F.; Cruz, C.; Nabti, E.-H. Restoration of Triticum aestivum Growth under Salt Stress by Phosphate-Solubilizing Bacterium Isolated from Southern Algeria. Agronomy 2022, 12, 2050. [Google Scholar] [CrossRef]
- Wejse, P.L.; Ingvorsen, K.; Mortensen, K.K. Purification and characterisation of two extremely halotolerant xylanases from a novel halophilic bacterium. Extremophiles 2003, 7, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Rohban, R.; Amoozegar, M.A.; Ventosa, A. Screening and isolation of halophilic bacteria producing extracellular hydrolyses from Howz Soltan Lake, Iran. J. Ind. Microbiol. Biotechnol. 2008, 36, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Sohail, M.; Ali, S.T. Arthrocnemum macrostachyum biomass: a novel substrate for multienzyme preparation by endophytic Bacillus species. Biomass- Convers. Biorefinery, 2023. [Google Scholar] [CrossRef]
- Giridhar, P.V.; Chandra, T. Production of novel halo-alkali-thermo-stable xylanase by a newly isolated moderately halophilic and alkali-tolerant Gracilibacillus sp. TSCPVG. Process. Biochem. 2010, 45, 1730–1737. [Google Scholar] [CrossRef]
- Siah, A.; Deweer, C.; Duyme, F.; Sanssené, J.; Durand, R.; Halama, P.; Reignault, P. Correlation of in planta endo-beta-1,4-xylanase activity with the necrotrophic phase of the hemibiotrophic fungus Mycosphaerella graminicola. Plant Pathol. 2010, 59, 661–670. [Google Scholar] [CrossRef]
- Moreno, M.D.L.; Pérez, D.; García, M.T.; Mellado, E. Halophilic Bacteria as a Source of Novel Hydrolytic Enzymes. Life 2013, 3, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Badiaa, E.; Abdeljabbar, H.; Mohamed, R.H.; Abdellatif, B.; Najla, S.-Z. In vivo and in vitro evaluation of antifungal activities from a halotolerant Bacillus subtilis strain J9. Afr. J. Microbiol. Res. 2012, 6, 4073–4083. [Google Scholar] [CrossRef]
- Santiago, C.D.; Yagi, S.; Ijima, M.; Nashimoto, T.; Sawada, M.; Ikeda, S.; Asano, K.; Orikasa, Y.; Ohwada, T. Bacterial Compatibility in Combined Inoculations Enhances the Growth of Potato Seedlings. Microbes Environ. 2017, 32, 14–23. [Google Scholar] [CrossRef]
- Gaiero, J.R.; McCall, C.A.; Thompson, K.A.; Day, N.J.; Best, A.S.; Dunfield, K.E. Inside the root microbiome: Bacterial root endophytes and plant growth promotion. Am. J. Bot. 2013, 100, 1738–1750. [Google Scholar] [CrossRef]
- Valenzuela-Encinas, C.; Neria-González, I.; Alcántara-Hernández, R.J.; Enríquez-Aragón, J.A.; Estrada-Alvarado, I.; Hernández-Rodríguez, C.; Dendooven, L.; Marsch, R. Phylogenetic analysis of the archaeal community in an alkaline-saline soil of the former lake Texcoco (Mexico). Extremophiles 2007, 12, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Töpper, B.; Larsen, A.; Thingstad, T.F.; Thyrhaug, R.; Sandaa, R.-A. Bacterial community composition in an Arctic phytoplankton mesocosm bloom: the impact of silicate and glucose. Polar Biol. 2010, 33, 1557–1565. [Google Scholar] [CrossRef]
- Bano, N.; Musarrat, J. Characterization of a New Pseudomonas aeruginosa Strain NJ-15 as a Potential Biocontrol Agent. Curr. Microbiol. 2003, 46, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Hussein, K.A.; Joo, J.H. Isolation and characterization of rhizomicrobial isolates for phosphate solubilization and indole acetic acid production. J. Korean Soc. Appl. Biol. Chem. 2015, 58, 847–855. [Google Scholar] [CrossRef]
- Arora, N.K.; Verma, M. Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Payne, S.M. Iron acquisition in microbial pathogenesis. Trends Microbiol. 1993, 1, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, R., Hajoori, M., Suthar, M., Desai, S., & Bharthana-Vesu, S. Isolation, screening and characterization of marine bacteria for exopolysaccharide production. Bioscience Discovery 2017, 8, 643–9.
- Arciola, C.R.; Campoccia, D.; Montanaro, L. Detection of biofilm-forming strains of Staphylococcus epidermidis and S. aureus. Expert Rev. Mol. Diagn. 2002, 2, 478–484. [Google Scholar] [CrossRef]
- Xie, G.H.; Cui, Z.; Yu, J.; Yan, J.; Hai, W.; Steinberger, Y. Identification of nif genes in N2-fixing bacterial strains isolated from rice fields along the Yangtze River Plain. J. Basic Microbiol. 2006, 46, 56–63. [Google Scholar] [CrossRef]
- Battini, F.; Cristani, C.; Giovannetti, M.; Agnolucci, M. Multifunctionality and diversity of culturable bacterial communities strictly associated with spores of the plant beneficial symbiont Rhizophagus intraradices. Microbiol. Res. 2016, 183, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Parmar, P.; Sindhu, S.S. The novel and efficient method for isolating potassium solubilizing bacteria from rhizosphere soil. Geomicrobiol. J. 2018, 36, 130–136. [Google Scholar] [CrossRef]
- de Freitas, J.R.; Banerjee, M.R.; Germida, J.J. Phosphate-solubilizing rhizobacteria enhance the growth and yield but not phosphorus uptake of canola (Brassica napus L.). Biol. Fertil. Soils 1997, 24, 358–364. [Google Scholar] [CrossRef]
- Murkovic, M.; Derler, K. Analysis of amino acids and carbohydrates in green coffee. J. Biochem. Biophys. Methods 2006, 69, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Singh, P.; Jorquera, M.A.; Sangwan, P.; Kumar, P.; Verma, A.K.; Agrawal, S. Isolation of phytase-producing bacteria from Himalayan soils and their effect on growth and phosphorus uptake of Indian mustard (Brassica juncea). World J. Microbiol. Biotechnol. 2013, 29, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Ramı́rez, M.G.; Avelizapa, L.R.; Avelizapa, N.R.; Camarillo, R.C. Colloidal chitin stained with Remazol Brilliant Blue R®, a useful substrate to select chitinolytic microorganisms and to evaluate chitinases. J. Microbiol. Methods 2004, 56, 213–219. [Google Scholar] [CrossRef]
- Rojas-Avelizapa, L.; Cruz-Camarillo, R.; Guerrero, M.; Rodríguez-Vázquez, R.; Ibarra, J. Selection and characterization of a proteo-chitinolytic strain of Bacillus thuringiensis, able to grow in shrimp waste media. World J. Microbiol. Biotechnol. 1999, 15, 299–308. [Google Scholar] [CrossRef]
- Sundaramoorthy, S.; Raguchander, T.; Ragupathi, N.; Samiyappan, R. Combinatorial effect of endophytic and plant growth promoting rhizobacteria against wilt disease of Capsicum annum L. caused by Fusarium solani. Biol. Control. 2012. [Google Scholar] [CrossRef]
- Kassambara, A., Mundt, F., Package ‘factoextra’. Extract and visualize the results of multivariate data analyses. 2017, Available online: http://www.sthda.com/english/rpkgs/factoextra.
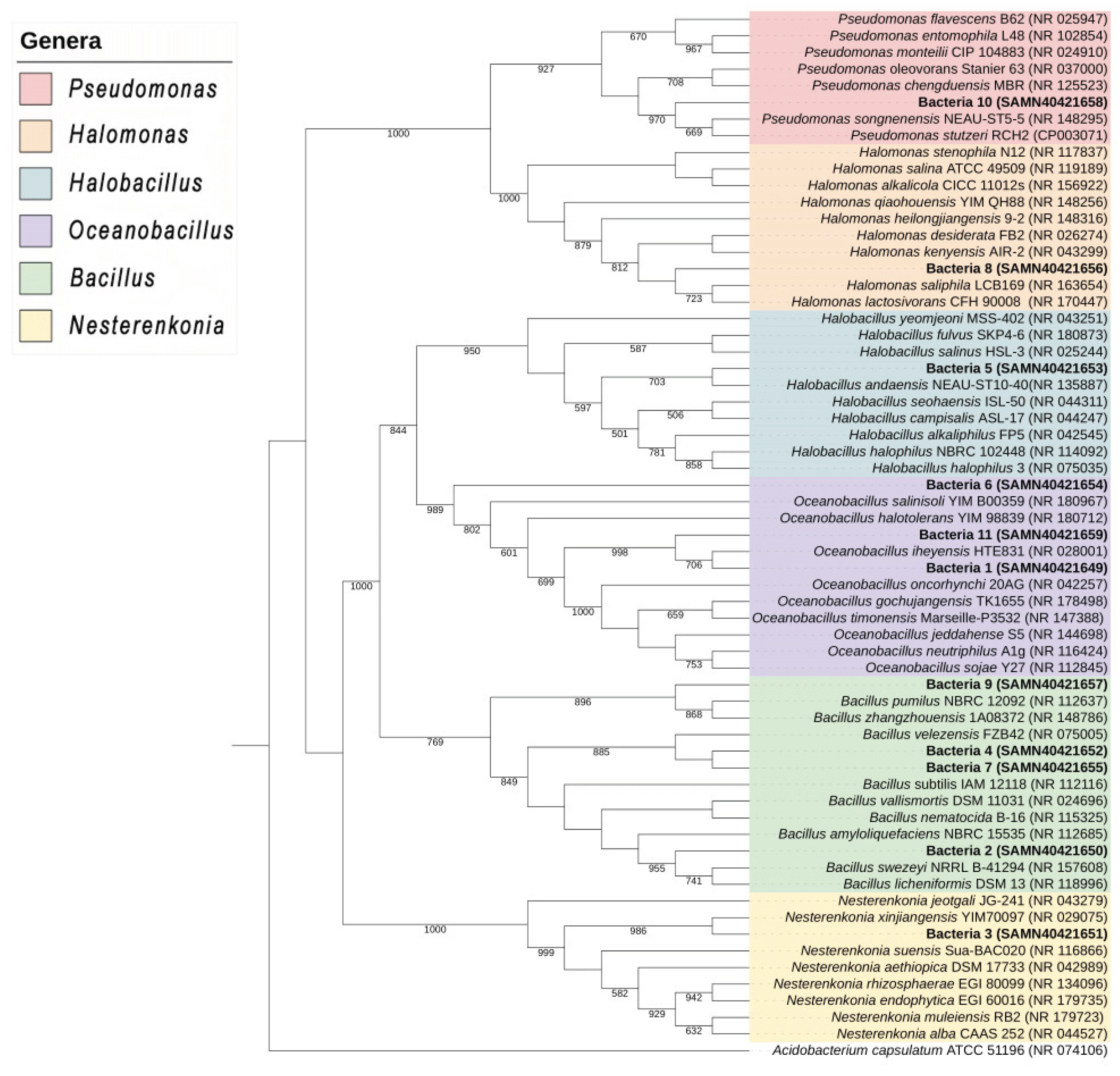
| Halophyte | EC (dS m-1) | Bacterial code | Molecular identification | |
| Sequence homology (% identity) | NCBI accession number | |||
| D. spicata | 16 | 1 | 99 | SAMN40421649 |
| D. spicata | 12 | 2 | 99 | SAMN40421650 |
| K. scoparia | 7 | 3 | 99 | SAMN40421651 |
| S. torreyana | 12 | 4 | 99 | SAMN40421652 |
| C. dactylon | 6 | 5 | 99 | SAMN40421653 |
| D. spicata | 8 | 6 | 99 | SAMN40421654 |
| K. scoparia | 5 | 7 | 99 | SAMN40421655 |
| K. scoparia | 8 | 8 | 99 | SAMN40421656 |
| E. obtusiflora | 9 | 9 | 99 | SAMN40421657 |
| S. torreyana | 7 | 10 | 99 | SAMN40421658 |
| B. salicifolia | 6 | 11 | 99 | SAMN40421659 |
| Bacteria | Nitrogenase enzyme | Exopolysaccharide | ||
| NaCl 0M | NaCl 2.5M |
NaCl 0M | NaCl 2.5M | |
| 1 | - | - | + | + |
| 2 | - | - | - | - |
| 3 | + | + | - | - |
| 4 | + | +++ | + | + |
| 5 | - | - | + | + |
| 6 | - | - | + | + |
| 7 | + | + | - | - |
| 8 | +++ | ++ | - | - |
| 9 | ++ | +++ | - | - |
| 10 | - | +++ | - | - |
| 11 | - | - | - | - |
| 0 M NaCl | 2.5 M NaCl | |||||||
| Bacteria | Citric | Succinic | Lactic | Tartaric | Citric | Succinic | Lactic | Tartaric |
| 1 | 6.3±0.4B* | 1.8±0.2A | 4.7±0.3A | nd | 7.2±0.1b* | 1.3±0.1a | 4.20±0.2a | nd |
| 2 | 6.2±0.4B* | 1.4±0.4A | 5.2±0.3A* | nd | 4.6±0.1b* | 1.0±0.2a | 4.0±0.2a* | nd |
| 3 | 1.4±0.2C | 1.2±0.1A | 0.4±0.2B* | 1.8±0.1A | 1.9±0.2b | 1.2±0.1a | nd* | 1.4±0.1a |
| 4 | 15.5±0.5A | 0.4±0.1A | 0.8±0.2B | nd | 15.2±0.3a | 0.8±0.3a | 1.2±0.2b | nd |
| 5 | 5.6±0.3B | 0.6±0.3A | 1.0±0.1B | nd | 5.2±0.1b | 0.4±0.3a | 1.0±0.2b | nd |
| 6 | 5.6±0.6B | 0.8±0.3A | 0.6±0.1B* | nd | 6.5±0.3b | 1.0±0.4a | 1.8±0.2b* | nd |
| 7 | 2.2±0.1C | 1.4±0.2A | 0.6±0.2B* | nd | 2.8±0.3b | 1.2±0.4a | 1.4±0.3b* | nd |
| 8 | 1.4±0.1C* | 0.4±0.2A* | 1.4±0.2B* | nd | 2.1±0.5b* | 1.0±0.1a* | nd* | nd |
| 9 | 2.6±0.1C | 1.0±0.1A | 1.0±0.3B* | 1.4±0.1A | 2.4±0.2b | 1.4±0.1a | nd* | 1.0±0.2a |
| 10 | 3.2±0.3C | 1.2±0.1A | 3.2±0.5A* | nd | 3.8±0.2c | 1.0±0.2a | 4.0±0.1a* | nd |
| 11 | 3.1±0.1C | 1.0±0.2A | 1.8±0.2B | nd | 3.6±0.1c | 1.0±0.2a | 2.0±0.1b | nd |
| 0 M NaCl | 2.5 M NaCl | |||||||||
| Bacteria | Citric | Malic | Tartaric | Maleic | Vanillic | Citric | Malic | Tartaric | Maleic | Vanillic |
| K | ||||||||||
| 1 | 5.2±0.1B* | 0.4±0.1C | 1.7±0.2B* | Nd | Nd | 4.0±0.2b* | 0.8±0.1b | 2.8±0.1b* | Nd | Nd |
| 2 | 4.8±0.1B | 0.8±0.1C* | 1.3±0.2B | Nd | Nd | 4.7±0.3b | 1.6±0.2b* | 2.0±0.1b | Nd | Nd |
| 3 | 1.9±0.3C | 1.0±0.1C* | 8.9±0.1A* | Nd | Nd | 2.2±0.1b | 1.8±0.2b* | 2.2±0.1b* | Nd | Nd |
| 4 | 0.5±0.5C* | 3.2±0.2B | 2.0±0.1B* | Nd | Nd | 1.2±0.1c* | 2.8±0.1b | 10.8±0.2a* | Nd | Nd |
| 5 | 4.2±0.2B | 2.0±0.3B | 2.0±0.3B* | Nd | Nd | 3.9±0.2b | 1.6±0.3b | 3.7±0.2b* | Nd | Nd |
| 6 | 1.2±0.2C | 2.0±0.2B | 1.8±0.5B* | Nd | Nd | 0.7±0.2c | 1.8±0.3b | 4.2±0.3b* | Nd | Nd |
| 7 | 4.3±0.1C* | 1.2±0.2C | 8.8±0.5A* | Nd | Nd | 1.0±0.3c* | 1.7±0.4b | 4.7±0.4b* | Nd | Nd |
| 8 | 4.7±0.1B | 1.5±0.1C | 1.0±0.2B* | Nd | 6.0±0.1 | 4.6±0.5b | 1.6±0.4b | 7.5±0.6a* | Nd | 6.2±0.2 |
| 9 | 11.1±0.2A | 7.6±0.1A* | 1.7±0.1B | Nd | Nd | 10.5±0.5a | 8.7±0.1a* | 2.4±0.4b | Nd | Nd |
| 10 | 14.0±0.2* | 0.5±0.3C* | 1.3±0.2B* | Nd | Nd | 4.7±0.2b* | 1.2±0.2b* | 2.8±0.3b* | Nd | Nd |
| 11 | 4.9±0.1B | 1.0±0.2C | 1.0±0.3B* | Nd | Nd | 5.0±0.1b | 1.5±0.1b | 2.2±0.1b* | Nd | Nd |
| Mn | ||||||||||
| 1 | 8.9±0.3B* | 4.8±0.2B* | Nd | Nd | Nd | 10.5±0.3b* | 7.4±0.1a* | Nd | Nd | Nd |
| 2 | 9.0±0.4B* | 4.9±0.2B* | Nd | Nd | Nd | 10.2±0.4b* | 7.2±0.2a* | Nd | Nd | Nd |
| 3 | 3.8±0.1C | 0.4±0.1C | Nd | Nd | Nd | 3.4±0.2c | 0.6±0.2b | Nd | Nd | Nd |
| 4 | 5.0±0.1C | Nd | Nd | Nd | Nd | 5.4±0.1c | Nd | Nd | Nd | Nd |
| 5 | 4.7±0.2C | Nd | Nd | Nd | Nd | 5.0±0.1c | Nd | Nd | Nd | Nd |
| 6 | 4.0±0.1C | Nd | Nd | Nd | Nd | 4.8±0.2c | Nd | Nd | Nd | Nd |
| 7 | 4.2±0.2C | Nd | Nd | Nd | Nd | 4.0±0.4c | Nd | Nd | Nd | Nd |
| 8 | 3.0±0.2C* | Nd | Nd | Nd | Nd | 4.6±0.2c* | Nd | Nd | Nd | Nd |
| 9 | 4.0±0.1C | 7.8±0.2A* | Nd | Nd | Nd | 4.6±0.3c | 6.8±0.2a* | Nd | Nd | Nd |
| 10 | 15.8±0.1A | Nd | Nd | Nd | Nd | 16.0±0.3a | Nd | Nd | Nd | Nd |
| 11 | 9.2±0.2B | Nd | Nd | Nd | Nd | 10.0±0.1b | Nd | Nd | Nd | Nd |
| Zn | ||||||||||
| 1 | Nd | 3.0±0.1A | 4.5±0.2B | 3.4±0.1B* | Nd | Nd | 3.2±0.1a | 4.0±0.1b | 2.8±0.1b* | Nd |
| 2 | Nd | 3.5±0.2A* | 3.6±0.2B | 3.0±0.2B | Nd | Nd | 2.0±0.2a* | 3.2±0.2b | 2.6±0.2b | Nd |
| 3 | Nd | 2.6±0.1A* | 2.8±0.1B | 1.2±0.4C | Nd | Nd | 4.7±0.1a* | 3.0±0.1b | 1.6±0.5c | Nd |
| 4 | Nd | 2.8±0.2A* | Nd | 11.8±0.1A | Nd | Nd | 3.4±0.2a* | Nd | 10.4±0.4a | Nd |
| 5 | 8.0±0.3A | 4.3±0.3A | Nd | 5.0±0.2B* | Nd | 7.8±0.3a | 4.7±0.3a | Nd | 3.6±0.3b* | Nd |
| 6 | Nd | 1.2±0.3B* | Nd | 4.8±0.2B | Nd | Nd | 4.0±0.3a* | Nd | 5.6±0.2b | Nd |
| 7 | Nd | 1.0±0.4B* | 3.4±0.1B | 1.0±0.3C* | Nd | Nd | 3.7±0.5a* | 4.0 | 2.8±0.3b* | Nd |
| 8 | 9.2±0.2A | 3.4±0.1A | 11.8±0.1A | 5.4±0.4B* | Nd | 9.3±0.1a | 3.2±0.4a | 11.6±0.3a | 6.4±0.1a* | Nd |
| 9 | Nd | 3.0±0.2A | Nd | 1.4±0.1C* | Nd | Nd | 3.8±0.5a | Nd | 10.5±0.2a* | Nd |
| 10 | 9.2±0.1A | 1.3±0.1B | Nd | 2.9±0.4C* | Nd | 8.6±0.2a | 2.0±0.3a | Nd | 4.7±0.1a* | Nd |
| 11 | Nd | 2.8±0.1A | 5.0±0.2B | 3.0±0.2B | Nd | Nd | 2.2±0.1a | 4.6±0.2b | 3.8±0.2b | Nd |
| 0 M NaCl | 2.5 M NaCl | |||||||||
| Bacteria | Ca3(PO4)2 | Phytate | K | Mn | Zn | Ca3(PO4)2 | Phytate | K | Mn | Zn |
| Fructose | ||||||||||
| 1 | 55.3±0.4D* | 20.4±1.8B | 129.7±0.1B* | 3.6±0.1C | 3.7±0.4G | 61.3±1.6d* | 21.6±0.8b | 160.5±1.9b* | 3.4±0.1d | 4.0±0.4h |
| 2 | 57.5±0.5D* | 2.0±0.6D | 122.8±0.7B* | 4.7±0.2C | 4.7±0.5G | 65.7±2.5d* | 1.8±0.5c | 151.0±2.5b* | 5.0±0.5d | 4.7±0.3h |
| 3 | 295.4±1.2B* | 1.7±0.1D | 16.6±0.5E | 16.5±0.4B | 111.1±0.2D | 333.4±1.2b* | 2.8±0.6c | 16.8±0.1e | 18.2±0.2c | 114.8±05c |
| 4 | 458.8±0.3A | 2.3±0.5D | 157.5±1.5A* | 32.0±0.2A | 414.0±0.3A* | 459.4±3.0a | 4.7±1.0c | 267.6±0.2a* | 33.4±0.4a | 349.4±0.5a* |
| 5 | Nd | 1.7±0.1D | 3.5±1.0F* | 24.7±0.2B | 343.5±0.2B* | Nd | 2.2±0.3c | 8.4±1.0g* | 23.9±0.3b | 83.3±1.0d* |
| 6 | 153.1±0.2C* | 93.1±1.0A | 113.3±1.4B* | 18.7±0.5B | 275.3±0.4C* | 308.6±2.0b* | 95.1±0.5a | 128.3±0.1c* | 20.6±0.3b | 282.3±0.5b* |
| 7 | 463.8±0.4A* | 2.2±0.3D | 153.1±0.7A* | 35.0±0.3A* | 417.9±0.4A* | 456.2±0.5a | 2.4±0.3c | 264.8±0.1a* | 32.0±0.5a* | 347.4±0.5a* |
| 8 | 2.3±0.5F* | 1.9±0.3D | 35.3±0.2D* | 16.4±0.2B | 16.0±0.5F | 6.7±0.6e | 1.5±0.4c | 43.8±0.1e* | 17.4±0.3c | 16.1±0.5g |
| 9 | 151.7±0.6C* | 16.2±1.4B | 70.8±0.1C* | 1.9±0.6D* | 3.5±0.6G | 322.8±1.0b* | 18.5±0.9b | 113.1±0.2d* | 3.6±0.2d* | 2.9±0.3h |
| 10 | 184.5±0.2C* | 7.0±0.3C | 78.1±0.4C* | 13.1±0.1B* | 38.1±0.2E | 226.7±0.4c* | 7.3±0.1c | 104.0±0.2d* | 16.3±0.5c* | 39.1±0.1e |
| 11 | 8.7±0.1E* | 1.8±0.4D | 16.7±0.4E | 9.5±0.3C | 18.8±0.1F* | 19.6±0.6e* | 2.5±0.2c | 18.7±0.1f* | 8.7±0.1d | 25.8±0.1f* |
| Xylose | ||||||||||
| 1 | Nd | Nd | 6.4±0.3B | Nd | Nd | Nd | Nd | 7.4±0.1b | Nd | Nd |
| 2 | Nd | Nd | 8.6±0.4B | Nd | Nd | Nd | Nd | 9.6±0.2b | Nd | Nd |
| 3 | Nd | 8.5±0.2A | 67.0±0.4A | Nd | 23.2±0.4A | Nd | 8.0±0.4a | 68.0±0.4a | Nd | 24.6±0.6a |
| 4 | Nd | 6.4±0.1AB | 65.7±0.1A | 3.6±0.1A | Nd | Nd | 7.8±0.4ab | 67.7±0.5a | 4.8±0.1a | Nd |
| 5 | 29.0±1.5B* | 5.70.1B | 5.4±0.2B | Nd | Nd | 228.1±2.8a* | 6.40.2b | 6.2±0.6b | Nd | Nd |
| 6 | Nd | Nd | 8.2±0.1B | Nd | Nd | Nd | Nd | 9.4±0.1b | Nd | Nd |
| 7 | Nd | 3.4±0.2C | 70.1±0.3A | 3.8±0.2A | Nd | Nd | 4.2±0.1b | 73.1±0.1a | 4.6±0.3a | Nd |
| 8 | Nd* | Nd | Nd | Nd | Nd | 4.4±0.4b* | Nd | Nd | Nd | Nd |
| 9 | Nd | Nd | 66.6±0.4A | Nd | 24.6±0.3A | Nd | Nd | 68.6±0.2a | Nd | 25.7±1.0a |
| 10 | 229.5±1.8A* | Nd | Nd | Nd | Nd | Nd* | Nd | Nd | Nd | Nd |
| 11 | Nd | Nd | 6.4±0.6B | Nd | Nd | Nd | Nd | 7.8±0.3b | Nd | Nd |
|
Alkaline phosphatase (Nitrophenylphosphate µg mL-1 h-1) |
Acid phosphatase (Nitrophenylphosphate µg mL-1 h-1) |
|||||
| NaCl (M) | NaCl (M) | |||||
| Bacteria | 0 | 1.5 | 2.5 | 0 | 1.5 | 2.5 |
| 1 | 27.62±2.3bA | 8.38±2.8aB | 3.1±0.9cC | Nd | 52.56±2.7bA | 42.36±1.8cB |
| 2 | 34.45±2.7aA | 7.75±2.8aB | 3.1±0.9cC | Nd | 59.67±3.2aA | 48.85±2.4bB |
| 3 | 18.00±2.3cA | 8.62±2.3aB | 7.75±1.2aB | Nd | 49.78±2.7bA | 49.27±3.8bA |
| 4 | 28.24±2.8bA | 6.20±2.4aB | 4.64±1.9bB | 47.00±1.4bB | 60.29±4.1aA | 57.82±2.9aA |
| 5 | 16.44±1.9cA | 9.86±1.9aB | 4.34±1.4bC | 44.94±1.4bB | 62.87±2.8aA | 62.43±2.3aA |
| 6 | 27.00±2.4bA | 6.44±1.8aB | 6.82±1.8bB | 27.11±2.3dC | 60.29±4.1aA | 50.71±2.4bB |
| 7 | 23.27±1.8bA | 3.58±1.4bB | 3.79±1.9cB | 23.81±3.2dC | 60.29±4.1aA | 42.67±1.9bB |
| 8 | Nd | 8.51±1.9aA | 9.24±1.9aA | Nd | 62.87±2.8aA | 42.05±1.4bB |
| 9 | 29.79±2.3bA | 9.31±1.9aB | 8.69±1.9aB | 33.19±1.7cC | 63.80±2.8aA | 51.94±2.3bB |
| 10 | 25.14±1.8bA | 5.51±1.9aB | 9.55±1.9aB | 51.32±1.2aC | 62.15±2.9aA | 51.33±1.4bB |
| 11 | 17.69±2.4cA | 9.55±1.9aB | 7.44±1.4abB | 39.27±5.2cC | 51.33±1.7bA | 44.21±1.8cB |
| Interval | 16.44-34.45 | 3.58-9.86 | 3.1-9.25 | 23.81-51.32 | 49.78-63.80 | 42.05-62.43 |
|
Phytase (Phosphate µg mL-1 h-1) |
||||||
| NaCl (M) | ||||||
| Bacteria | 0 | 1.5 | 2.5 | |||
| 1 | 80.21±1.8bC | 107.12±2.1bB | 124.16±2.8cA | |||
| 2 | 70.01±1.2cC | 100.15±2.8bB | 122.18±2.4cA | |||
| 3 | 90.12±2.8aA | 87.56±3.2cA | 94.75±2.5dA | |||
| 4 | 4.24±2.4fC | 67.24±3.6dB | 117.74±2.8cA | |||
| 5 | 90.38±2.2aC | 100.08±3.8bB | 144.53±94bA | |||
| 6 | 70.45±2.6cC | 87.34±3.3cB | 110.22±2.9cA | |||
| 7 | 70.56±2.1cC | 100.75±3.1bB | 164.72±3.1aA | |||
| 8 | Nd | 84.74±3.4cB | 144.94±3.5bA | |||
| 9 | 90.01±2.6aC | 120.85±3.6aB | 154.42±3.4aA | |||
| 10 | 50.83±2.8dC | 104.16±3.1bB | 140.74±3.1bA | |||
| 11 | 40.12±2.6eC | 97.12±3.5bB | 147.48±3.2bA | |||
| Interval | 4.24-90.38 | 67.24-107.12 | 94.75-164.72 | |||
|
Xylanase (µg mL-1 h-1) |
Chitinase (µg mL-1 h-1) |
|||||
| NaCl (M) | NaCl (M) | |||||
| Bacteria | 0 | 1.5 | 2.5 | 0 | 1.5 | 2.5 |
| 1 | Nd | Nd | Nd | 1.62±0.931aB | 6.85±1.021aA | 6.70±0.923aA |
| 2 | Nd | Nd | Nd | 1.90±0.161aB | 6.43±0.212aA | 5.48±0.546aA |
| 3 | 0.09±0.007aB | 0.76±0.004bB | 3.14±0.005bA | 1.67±0.112aB | 6.92±0.283aA | 6.54±0.283aA |
| 4 | 0.14±0.012aB | 0.68±0.005bB | 3.01±0.010bA | 1.24±0.932aB | 6.48±0.827aA | 5.45±1.093aA |
| 5 | 0.06±0.008aC | 0.62±0.004bB | 2.14±0.003bA | 1.45±0.836aB | 6.87±0.927aA | 6.30±0.864aA |
| 6 | 0.06±0.008aC | 0.67±0.006bB | 2.92±0.005bA | 1.86±0.732aB | 6.56±1.052aA | 6.28±0.834aA |
| 7 | 0.02±0.034aC | 0.76±0.005bB | 2.74±0.008bA | 1.90±0.623aB | 5.68±1.024aA | 6.34±0.941aA |
| 8 | Nd | 0.79±0.004bB | 2.80±0.005bA | Nd | 6.14±0.962aA | 5.85±0.841aA |
| 9 | 0.09±0.005aC | 0.83±0.004bB | 3.22±0.007bA | 1.78±0.924aB | 6.24±0.863aA | 6.35±0.974aA |
| 10 | Nd | Nd | Nd | Nd | Nd | Nd |
| 11 | 0.08±0.015aC | 1.89±0.010aB | 5.32±0.008aA | Nd | Nd | Nd |
| Interval | 0.02-0.14 | 0.62-1.89 | 2.14-5.32 | 1.24-1.90 | 5.68-6.92 | 5.45-6.70 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





