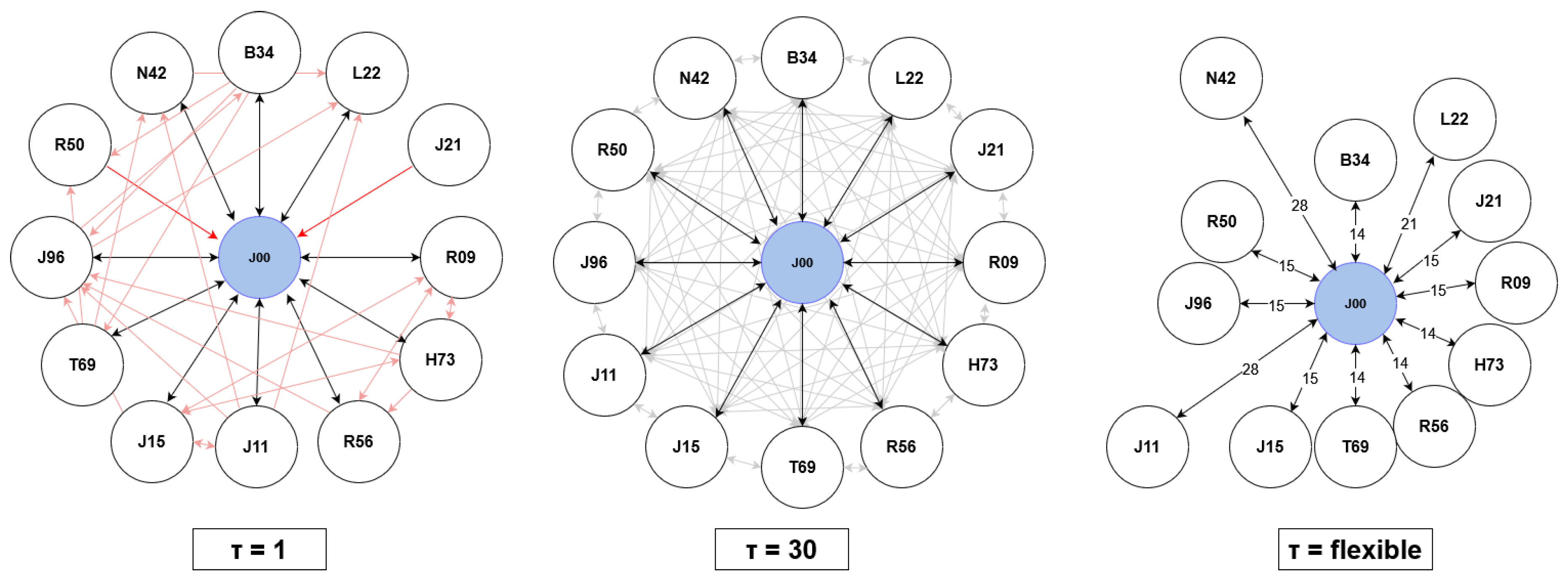
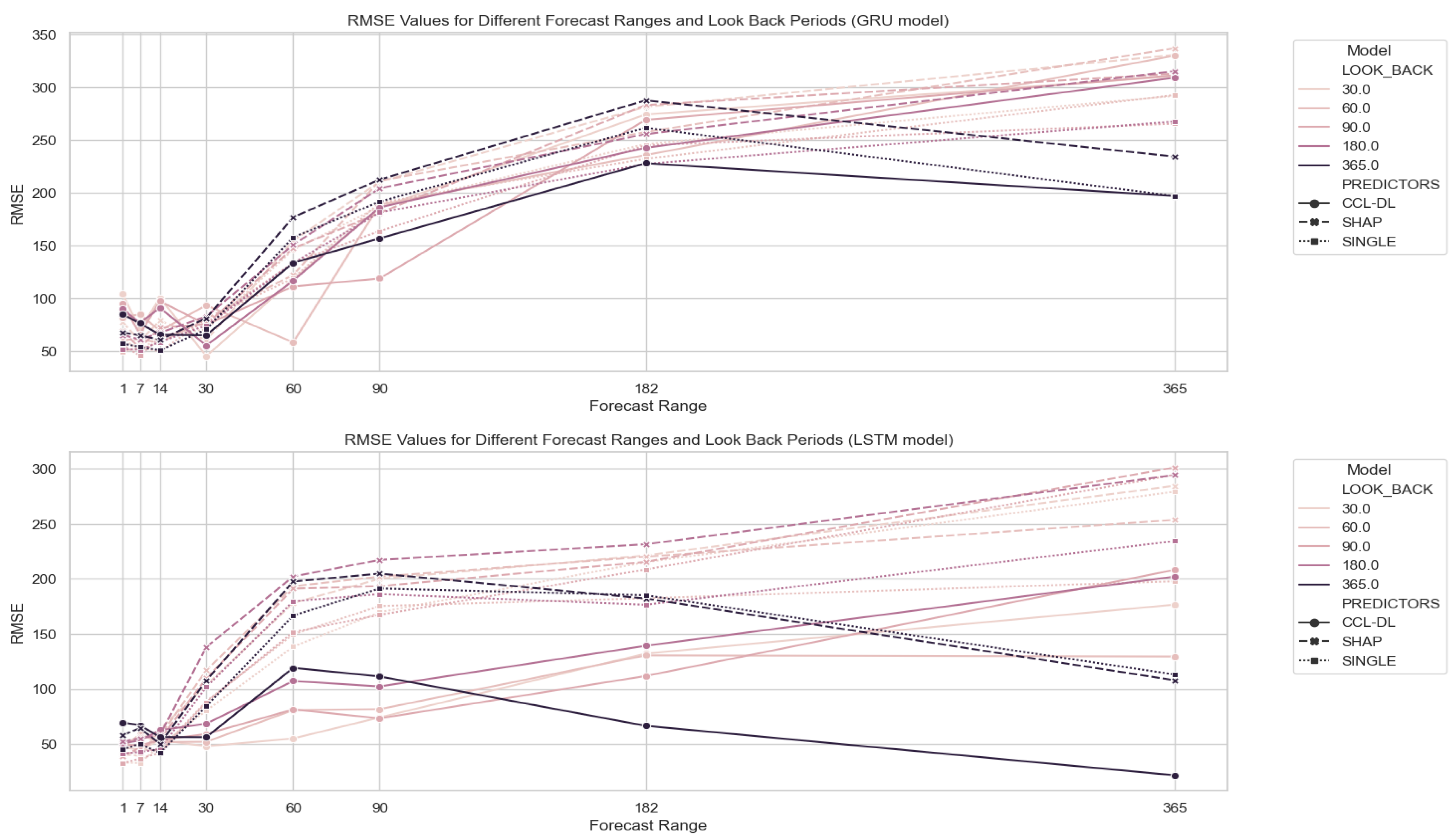
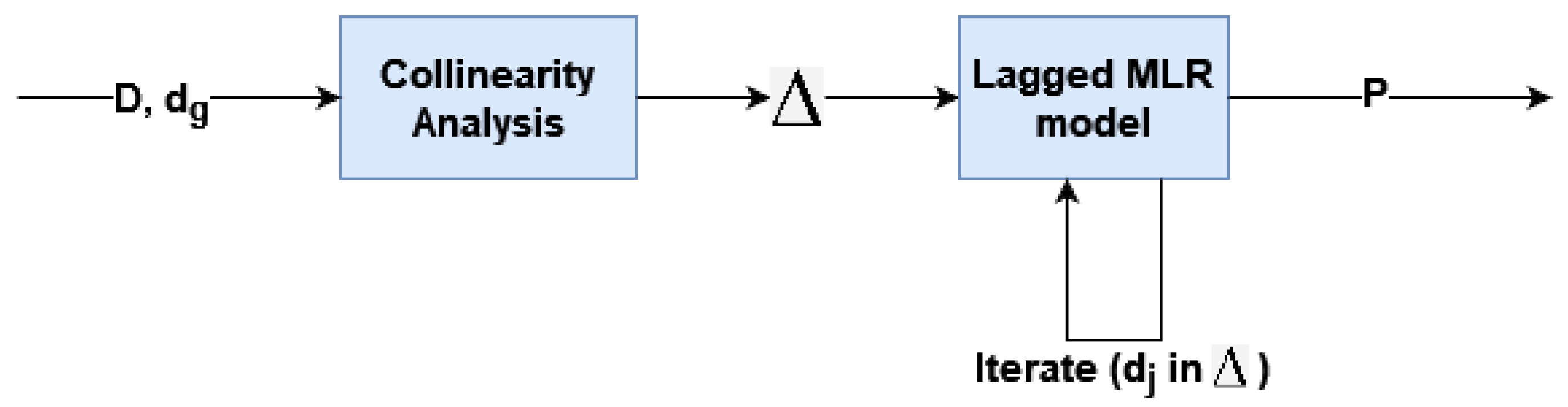
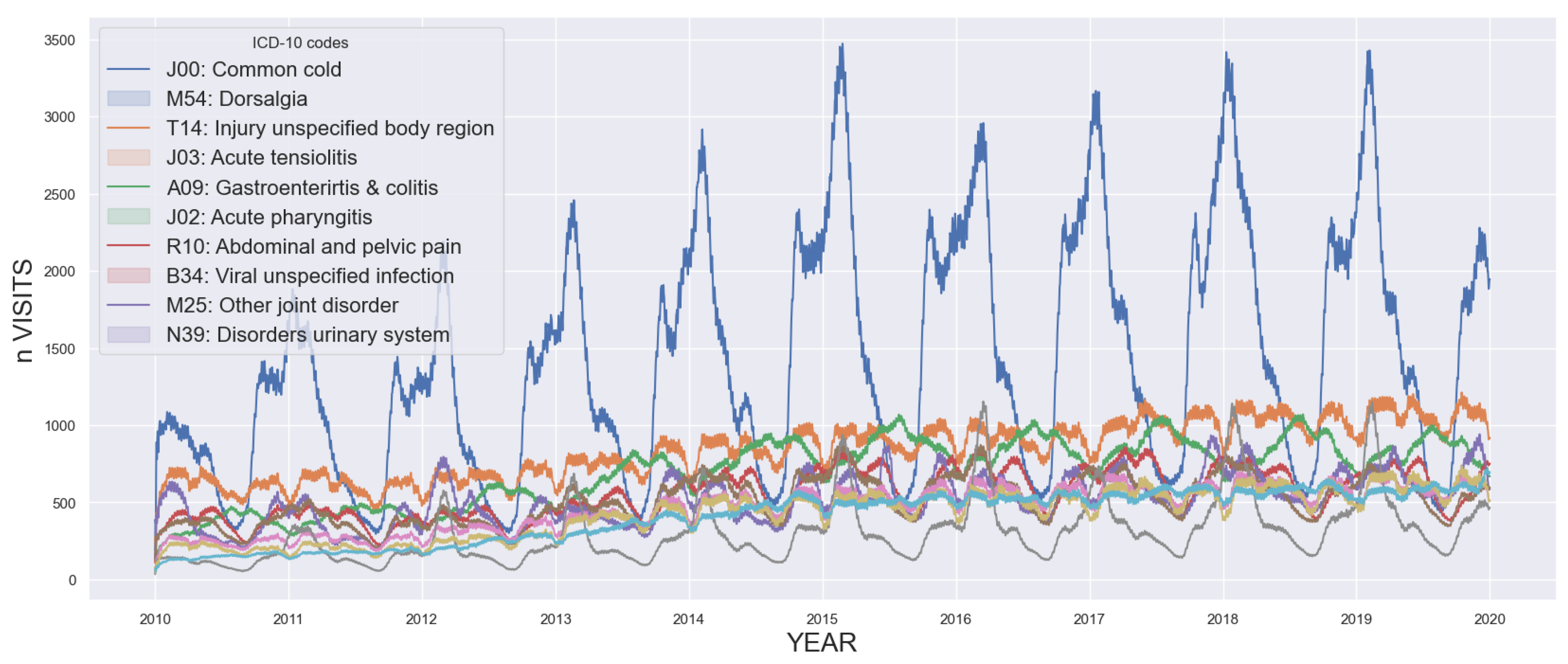
Mitigating future uncertainty to refine decision-making is a pivotal aspect in diverse fields, processes, and businesses. The temporal dimension is intrinsic to many prediction challenges, requiring the extrapolation of time series data. Precise forecasting not only enables effective planning, optimization, and risk identification but also opens avenues for seizing opportunities. Consequently, accurate time series forecasting has evolved into a cornerstone of data science, prompting substantial efforts to advance and refine forecasting methods. In this context, initial proposals were based on statistical models. This encompasses exponential smoothing, seasonal, ARIMA, and state space models, among others [
6]. These methods are based on their prescription of the underlying data-generating process, assuming the structural components of the series and the correlation of historical observations. These models require a small number of parameters and can handle cases with limited historical observations. In recent years, methods based on DL have surpassed statistical approaches, particularly in multivariate datasets and for longer forecast horizons. These methods can capture both short-term dependencies among variables and discover long-term patterns [
10]. In the healthcare context, there is literature focused on forecasting demand. An area of significant research interest involves predicting emergency attendance for precise resource planning. Studies such as [
11], employ LR analysis to link emergency departments with specific resources, highlighting the significant enhancements simple tools can bring to healthcare systems in terms of resource management. The work by [
12] use RNN to predict patient visits using previous diagnosing information. Some predict visits using DL, while others forecast unplanned visits for diabetic patients [
13]. Numerous studies predict epidemiological visits, like flu outbreaks [
14]. Particularly within the context of the COVID-19 pandemic, there has been a notable increase in research efforts due to the availability of datasets. In the research conducted by [
15], LSTM models outperformed statistical models such as ARIMA and the Nonlinear Autoregression Neural Network (NARNN) when applied to modeling the spread of the pandemic in countries like Germany, France, Belgium, and Denmark [
16]. In [
17], various LSTM models (e.g., Bi-LSTM, ed-LSTM) were employed for short-term forecasting in India. The study suggests that including external factors such as population density, travel logistics, or sociodemographic data could enhance predictions. Other works also employ standard implementations of LSTM on Canadian databases [
18]. In [
19], various models were compared, including LSTM, Bi-LSTM, GRU, SVR, and ARIMA. Bi-LSTM performed best with lower Mean Absolute Error (MAE) and Root Mean Square Error (RMSE). In multivariate studies, climate variables are used for improving COVID-19 outbreak predictions [
20,
21,
22,
23]. These methods, despite obtaining promising results, focus on improving prediction and do not aim to discover the underlying relationship of variables, select predictors, and enhance explainability in the models as we are proposing in the present work.
Hybrid models that integrate statistical and deep learning methods, as the one proposed in this work, have been previously proposed achieving promising results. In [
24], a combination of Multiple Linear Regression (MLR) and Artificial Neural Networks (ANN) is used to address the complexity of emergency attendance in a tourist area, where significant seasonal variations occur. In [
25], the Multivariate Exponential Smoothing Recurrent Neural Network (MES-RNN) framework is proposed to improve forecasts. This approach draws from the principles outlined in the works of [
26,
27]. They expand upon earlier research in multivariate time series exponential smoothing to create structural models that can generate optimal forecasts for individual data series. The MES-RNN method demonstrates consistent results when forecasting COVID-19 outbreaks in aggregated disease morbidity datasets, outperforming purely statistical or deep learning models. In the study by [
28] a hybrid model combining linear regression models, often referred to as autoregression-LR, or ARIMA (Auto-Regressive Integrated Moving Average), with nonlinear models based on deep belief networks (DBN) is presented. This blending is employed to effectively capture both linear and nonlinear patterns within time series data, demonstrating that these propositions enhance performance when compared to purely linear or nonlinear models; in contrast to the aforementioned approaches, our CCLR-DL method not only seeks improvement in deep learning forecasting through statistical techniques but also employs these statistical methods in conjunction with Granger causality (GC) for feature selection. This approach aims to select the most influential predictors while enhancing the transparency of the model.
The need to eliminate non-informational variables in the dataset that can worsen the forecasting and the need to understand the underlying relationships between features in multivariate contexts, create an ideal environment for applying feature selection strategies. The gold-standard feature selection methods for enhancing explainability in DL is based on the post-hoc Shapley Additive Explanations (SHAP) method, introduced in [
9]. While the exact computation of Shapley values is computationally challenging, the innovation that SHAP methods bring is that the explanation model is represented as an additive feature attribution method, a linear model. It is considered an agnostic post-hoc method since it is used as a feature importance technique after the training of any model. SHAP method has a limitation based on the need of a strategy for selecting the number of predictors, usually based on a pre-established threshold. Another method of dimensionality reduction widely used in multivariate time-series databases is GC [
29]. In the work of [
30], a dimensionality reduction algorithm is proposed for multivariate time series datasets, which can effectively extract the most significant and discriminative input features for output predictions. In [
31], a variant of RNN known as the Echo State Network (ESN) leverages GC to learn nonlinear models for time series classification. Other studies have employed GC as a feature selection method to enhance model interpretability and improve forecasting [
32,
33]. Our CCLR-DL methodology blends Lagged Multiple regression models and Granger causality to extract significant predictors. This serves a dual purpose: firstly, it refines the feature selection strategy considering lags for identifying predictors, and secondly, it enhances the forecasting accuracy of Deep Learning (DL) models. The objective is to create an autonomous method to select predictors in multivariate time series data and enhance model explicability.
The results of these studies demonstrate the potential of hybrid strategies for predicting time series in both univariate and multivariate scenarios. Our proposal aims to develop a model that can be used in a multivariate temporal database without relying on specific domain knowledge, assuming that predictors are present in the dataset. The method is based on a feature selection strategy based on statistical procedures for both enhancing explainability and improving forecasting accuracy of DL models.