1. Introduction
Penile squamous cell carcinoma (PSCC) is an uncommon neoplasm. Its incidence has marked geographic variability [
1], and it is particularly high (2.5/100,000 inhabitants) in Spain [
2]. Phimosis, absence of circumcision, chronic inflammation, lichen sclerosus, smoking, and human papil-lomavirus (HPV) infection have been described as risk factors for the development of the disease [
3]. According to the World Health Organization (WHO), PSCCs should be classified based on their association with HPV. For this reason, immunohistochemical (IHC) staining for p16, a well-known surrogate of transforming HPV infection, is strongly recommended by the WHO in its last classification of urological tumors [
3].
Genomic instability secondary to the overexpression of the oncoproteins E6 and E7, leading to an uncontrolled progression of the cell cycle is considered a key molecular mechanism involved in HPV-associated PSCC [
4,
5]. Conversely, the molecular background of HPV-independent PSCC is less well understood, with several recent series reporting that
TP53 mutations are highly recurrent in this subset of tumors [
6,
7,
8,
9]. Remarkably, a few studies have suggested that the
TP53 mutational status is associated with nodal metastases and thus with a worse prognosis in PSCC patients [
4,
6,
8,
10,
11,
12,
13].
In a recent study [
14], we showed that a p53 IHC pattern-based framework described initially for vulvar tumors [
15] can also be applied to PSCC and strongly correlates with
TP53 mutational status, suggesting that, if carefully evaluated, a simple p53 IHC technique, available in most pathology laboratories, could be considered a reliable surrogate of
TP53 mutational status. In this scheme, two p53 IHC patterns (scattered and mid-epithelial), defined as “normal”, indicate a wild-type
TP53 protein, whereas four well-defined patterns (diffuse overexpression, basal over-expression, cytoplasmic and null), defined as “abnormal”, strongly correlate with a mutated
TP53. Remarkably, three of the mutant patterns in this scheme, namely, basal overexpression, cytoplasmic and null, have been classified as “normal” according to conventional evaluations in previous studies on PSCC [
14,
16,
17].
We aimed to evaluate the HPV and TP53 statuses in a large series of PSCC from three public institutions in Spain by classifying the tumors based on HPV in situ hybridization and p16 and p53 IHC analysis into three molecular categories: HPV-associated, HPV-independent/p53 normal and HPV-independent/p53 abnormal. We analysed the prognostic implications of this molecular classification system.
2. Methods
2.1. Patients
We retrospectively identified all patients surgically treated for PSCC in two tertiary general hospitals (Hospital Clinic de Barcelona, Hospital Vall d’Hebron) and a monographic urological center (Fundació Puigvert) in Barcelona, Spain, from January 2000 to December 2020. All patients fulfilled the following inclusion criteria: 1) had a primary diagnosis of PSCC, 2) had a follow-up of at least 22 months or until death, and 3) had sufficient available tumor tissue for ancillary IHC studies.
All patients were treated following the guidelines of the European Association of Urology (EAU) [
18] depending on the clinical staging, which was determined on the basis of physical examination plus imaging techniques (ultrasound scan, computed tomography scan and/or positron emission tomography, etc.) when required. All local excisions aimed at organ sparing and reconstructive techniques were used when necessary to minimize the functional impact. Inguinal lymph node evaluation was performed if required. Guided sentinel node biopsy was the first option. Endoscopic inguinal modified lymphadenectomy was performed when sentinel node biopsy was not available. In all patients with positive sentinel node a radical inguinal lymphadenectomy was performed.
The following clinical and pathological variables were retrieved from the electronic files: age at diagnosis, tumor location, type and date/s of treatment/s, margin status, vascular invasion, perineural invasion, stage at diagnosis, date of first cancer recurrence, and patient status at follow-up.
The study was approved by the Healthcare Ethics Committee of the Hospital Clinic of Barcelona, Hospital Vall d’Hebron and Fundació Puigvert (HCB/2020/1207, PR(AG)578/2021, FP2021/05c, respectively). Informed written consent was obtained from all the patients included in the study.
2.2. p16 IHC
IHC for p16 was performed for all samples using the CINtec Histology Kit (clone E6H4; Roche, Heidelberg, Germany). Tumors with strong and diffuse block-type staining were considered positive, whereas patchy or completely negative p16 staining was considered p16 negative [
3,
19,
20]. In each run a p16-positive squamous carcinoma of the vulva was used as positive control. All patients were independently evaluated by two pathologists with expertise in urological pathology and interpretation of p16 staining (I.T. and N.R.).
2.3. ISH
RNA in situ hybridization (ISH) was performed for all samples using the automated Leica Biosystems BOND-III and RNAscope ISH Probe High Risk HPV. The assay qualitatively detects E6 mRNA in 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, and 82 high-risk HPV types. In each run a carcinoma of the uterine cervix with known HPV16 positivity was used as control.
2.4. p53 IHC
p53 IHC was performed in all patients with a monoclonal antibody (clone DO-7; Roche) according to the manufacturer’s protocol. All the IHC staining procedures (for p16 and p53) were performed on an automated staining system (Ventana Benchmark ULTRA, Ventana Medical Systems, Tucson, AZ, USA) on whole slide sections. IHC staining was evaluated for invasive tumors following the recently described p53 pattern-based interpretation framework described for squamous cell carcinomas of the vulva [
21]and confirmed in PSCC [
14]; this method includes two major categories: “normal”, which correlates with wild-type
TP53, and “abnormal staining”, which correlates with mutated
TP53”. The “normal” category included two patterns: 1) occasional positive nuclei in the basal and/or parabasal layer (scattered pattern) and 2) moderate to strong nuclear p53 IHC staining in the parabasal layers with absence of expression in the basal cells (mid-epithelial pattern). The “abnormal” category included four p53 IHC patterns: 1) continuous, strong nuclear staining of the basal layer (basal overexpression pattern); 2) continuous and strong nuclear basal staining with suprabasal extension of the positive cells (diffuse overexpression pattern); 3) cytoplasmic staining with or without nuclear positivity (cytoplasmic pattern); and 4) complete absence of staining in the tumor, with evidence of intrinsic positive control in the adjacent skin, stromal or inflammatory cells (null pattern). All patients were independently evaluated by two pathologists with expertise in urological pathology and interpretation of p53 staining (I.T. and N.R.). All discrepancies were discussed in a consensus meeting, and a final evaluation was achieved. In each run a normal tonsil showing scattered positive staining and a serous carcinoma of the ovary with known
TP53 mutation and diffuse p53 IHC overexpression were used as controls. Thirty-three patients were previously included in a recent study focused on the validation of this pattern-based p53 interpretation framework against
TP53 mutational analysis, in which 95% concordance was observed [
14].
2.5. The Criteria for PSCC Classification into Three Groups
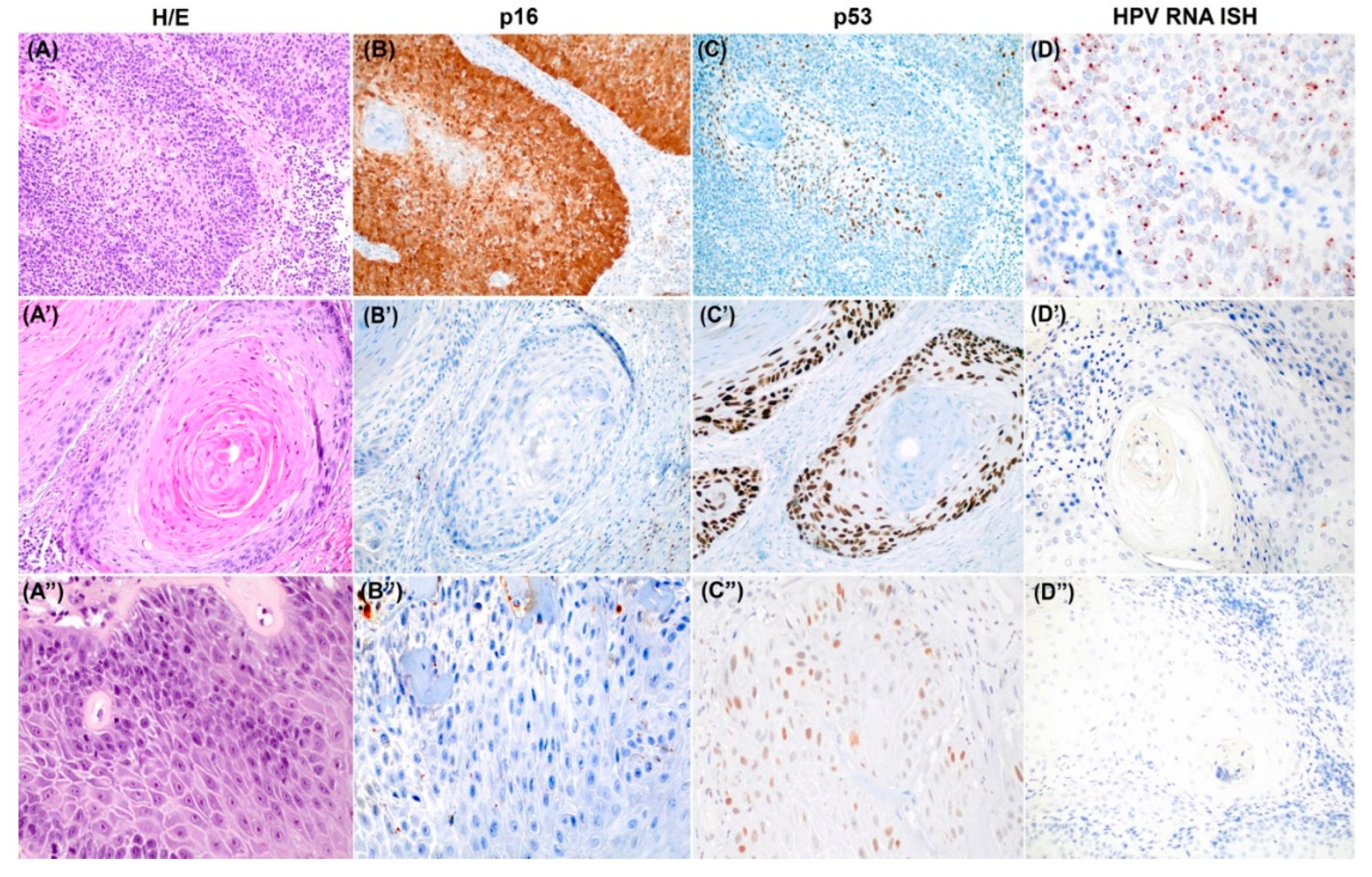
All the study cases were classified into three main categories based on HPV ISH and p16 IHC results and the pattern of p53 IHC expression. The categories included the following: 1) HPV-associated PSCC (positive for HPV ISH and p16 IHC, independent of the p53 IHC pattern); 2) HPV-independent/p53 normal PSCC (negative for HPV ISH and p16 IHC and a scattered or mid-epithelial p53 IHC pattern); and 3) HPV-independent/p53 abnormal PSCC (negative for HPV ISH and p16 IHC and diffuse, basal overexpression, cytoplasmic or null patterns of p53 IHC).
2.6. Statistical Analysis
The statistical analyses were conducted using R Statistical Software (v4.3.2; R Core Team 2021). The chi-square test and Fisher’s exact test were employed for categorical data, while the Wilcoxon rank-sum test was utilized for numerical data, enabling the comparison of clinical and histopathological data.
The endpoints for prognosis were recurrence-free survival (RFS) and disease-specific survival (DSS), which were calculated from the date of treatment (primary surgery) to the date of first recurrence or progression or to death due to the disease, respectively. Cumulative incidences were depicted through plotted curves, and differences between the curves were assessed using Gray’s test. Univariate and adjusted (multivariate) models were obtained using the Cox proportional hazards model. For the multivariate analysis, two models were built, one including the molecular type and the second including the p53 IHC status, due to the collinearity of these two variables. Two-sided tests were used, and a p value less than 0.05 indicated statistical significance.
3. Results
3.1. Clinical Pathological Features of the Overall Series
One hundred twenty-two patients were included in the study. Of these, 43 were from the Hospital Clinic de Barcelona, 35 were from the Hospital Vall d’Hebron and 44 were from the Fundació Puigvert. The mean age at diagnosis was 68.6 years (range 40-96). Fifty-eight patients (47.5%) were stage I at diagnosis, 40 (32.8%) were stage II, 16 (13.1%) were stage III, and 8 (6.6%) were stage IV tumors. The median follow-up period was 56.9 months (range 22-60 months). Sixty-five patients (53.3%) underwent penectomy (partial or radical), 48 (39.3%) glansectomy and in 9 patients (7.4%) circumcision was performed. Metastatic involvement of the lymph nodes was identified in 24 patients (19.7%).
3.2. HPV ISH and p16 and p53 IHC Results and Tumor Classification
HPV ISH was positive in 36/122 tumors (29.5%), and in all of them, p16 IHC was positive. These tumors were classified accordingly as HPV-associated tumors. Thirty-two of the 36 HPV-associated tumors had a normal pattern of p53 IHC expression (88.9%), 31 had a scattered pattern, and one had a mid-epithelial pattern of p53 IHC. Only four (11.1%) HPV-associated tumors presented an abnormal p53: two had diffuse over-expression, one had basal overexpression, and one had a null pattern.
Eighty-six out of the 122 tumors (70.5%) were negative for HPV ISH and p16 IHC and were classified as HPV-independent. Among the 86 tumors, 35 (28.7% of the overall series) showed a normal pattern of p53 IHC expression and were classified as HPV-independent/p53 normal, all of which exhibited a scattered pattern. Fifty-one tumors in this HPV-independent category (41.8% of the overall series) had an abnormal p53 IHC staining pattern and were classified as HPV-independent/p53 abnormal. The most common abnormal p53 IHC pattern in this cohort was diffuse overexpression (27/51, 53.0%), followed by a null pattern (14 patients, 27.4%), basal overexpression (8 patients, 15.7%) and cytoplasmic expression (2/51, 3.9%).
3.3. Characteristics of the Three Molecular Types of PSCC
Table 1 summarizes the clinical and pathological features of the patients and the three molecular categories defined in this study. Patients with HPV-independent/p53 normal tumors were older (mean age of 72 years) than were those with the other two categories (67 years for both HPV-associated and HPV-independent/p53 abnormal tumors [p=0.040]). Patients with HPV-independent/p53 abnormal tumors had a greater risk of lymph node metastases than patients with HPV-independent/p53-normal tumors (p=0.012). The histological variants of the HPV-associated tumors significantly differed from the variants identified in the HPV-independent molecular types. Moreover, there were no differences in terms of histological variants between HPV-independent/p53 normal and HPV-independent/p53 abnormal tumors. There were no differences in terms of anatomical location, vascular or perineural invasion, margin status, or stage at diagnosis.
Figure 1 shows a representative example of each of the three tumor categories, including hematoxylin and eosin staining features as well as HPV ISH and p16 and p53 staining.
3.4. Survival Analysis
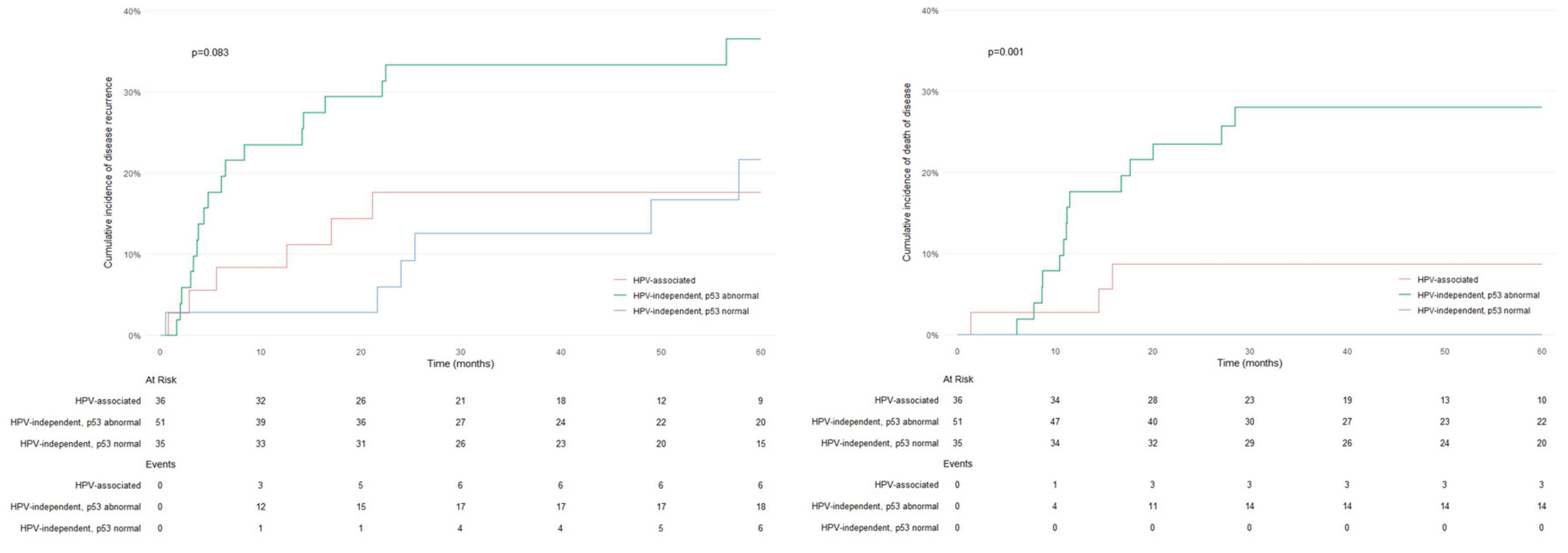
Thirty patients (24.6%) experienced disease recurrence during follow-up, but no differences were observed among the three molecular types in terms of recurrence rate (p=0.067). Seventeen patients (13.9%) died due to PSCC, and 13 (10.6%) died due to other causes. Disease-related death was observed in 3/36 (8.3%) patients with HPV-associated PSCC and 0/35 (0.0%) patients with HPV-independent/p53 normal PSCC, with the highest number of events occurring in patients with HPV-independent/p53 abnormal PSCC (14/51 (27.5%) (p<0.001)). The patterns of p53 IHC expression in the HPV-independent tumors of the patients who died due to the tumor were diffuse overexpression (7/27; 25.9%), null pattern (5/14; 35.7%), basal overexpression (1/8; 12.5%) and cytoplasmic expression (1/2; 50%).
Figure 2 shows the cumulative incidence curves for RFS and DSS for the three molecular categories. No differences in RFS were observed between the three molecular types (p=0.083); however, significant differences in DSS were detected (p=0.001), with patients with HPV-independent/p53 abnormal PSCC having the worst prognosis. In terms of tumor staging, no differences in RFS were observed between patients with early-stage tumors and patients with advanced-stage tumors (p=0.073); however, significant differences in DSS were identified (p<0.001). Remarkably, 10/14 (71.4%) HPV-independent/p53 abnormal PSCC patients diagnosed with stage III or IV died from the disease compared to only 2/8 (25.0%) of the patients with HPV-associated tumors in stages III or IV and 0/2 (0.0%) of the HPV-independent/p53 normal patients in stages III or IV (p=0.048).
Table 2 shows the Cox regression analysis for RFS. Vascular invasion, perineural invasion, lymph node metastasis and advanced disease stage were associated with impaired RFS in the univariate analysis. According to the multivariate analysis, only vascular invasion reached statistical significance. The results of the Cox regression analysis for DSS are shown in
Table 3. The molecular type (HPV-independent/p53 abnormal), p53 IHC abnormal pattern, vascular invasion, perineural invasion, lymph node metastases and advanced stages were associated with impaired DSS in the univariate analysis. According to the two multivariate models, HPV-independent/p53 abnormal molecular type (p=0.001) or p53 IHC expression (p=0.001), in addition to vascular and perineural invasion, lymph node metastases, and advanced stage, were significantly associated with impaired DSS.
4. Discussion
The most remarkable finding of our study, which included a large series of patients with PSCC treated at three different institutions in Barcelona, Spain, was the difference in prognosis observed between the three molecular types of PSCC defined according to their association with HPV and p53 IHC: HPV-associated, HPV-independent with normal p53, and HPV-independent with abnormal p53 PSCC. Remarkably, the classification of the tumors based on HPV status and p53 IHC patterns had a stronger impact on DSS in the multivariate analysis than did the staging system, suggesting that not only HPV status but also p53 IHC should be routinely evaluated in all PSCC patients.
The good prognosis of patients with HPV-associated tumors (over 90% DSS at 5 years) strongly supports the current 2022 WHO classification of PSCC, which separates tumors based on HPV status [
22]. Studies evaluating the prognostic impact of HPV status in PSCC patients have shown controversial results, with some reporting no differences in DSS [
23], whereas others showing longer DSS in HPV-associated PSCC [
9,
13], similar to what occurs in HPV-associated carcinomas in other anatomical sites, such as the head and neck [
24] and the lower female genital tract [
25]. The good DSS of patients with HPV-associated tumors in our study is remarkable, especially considering that these patients were frequently diagnosed in advanced stages and had metastatic involvement of the lymph nodes in almost 25% of the patients. These results suggest that, as shown in HPV-associated tumors from other anatomical areas, HPV-associated PSCC is highly sensitive to radiation and chemotherapy [
26]. In accordance with the findings of other European series [
27], HPV-associated tumors represented a small percentage (29.5%) of all PSCC, which is in contrast with the findings of studies from sub-Saharan Africa or the Caribbean, which showed rates of HPV-associated PSCC as high as 80% [
28]. As previously reported, p16 IHC results have shown excellent correlation with HPV ISH [
29], reinforcing the validity of the recent WHO 2022 recommendation of using p16 IHC as a surrogate for the presence of high-risk HPV [
22]. Our study revealed that the second type of PSCC defined by the WHO, HPV-independent tumors, includes at least two categories with different clinical and pathological features and, most importantly, a different prognosis. In the first category, HPV-independent PSCC with normal p53 expression is associated with several specific clinical pathological features. These tumors arise in older men, have a very low rate of lymph node metastases and are rarely diagnosed at stage III or IV. Remarkably, although these patients had similar rates of recurrence compared to those in the other two groups, they had excellent DSS, with no tumor-related deaths. This category of HPV-independent tumors with normal p53 IHC has previously been described in the vulva [
30,
31], where they show similar behavior to that observed in our series of PSCC, with frequent recurrences but extremely good DSS [
32].
The most frequent category of PSCC in our study (60% of all HPV-independent tumors and 40% of all tumors) was HPV-independent/p53 abnormal PSCC. This percentage of abnormal p53 IHC results is similar to the percentage reported by other studies in HPV-independent PSCC [
6,
9]. In contrast with the favorable DSS of patients with HPV-associated tumors and HPV-independent/p53 normal tumors, the prognosis of patients with HPV-independent/p53 abnormal PSCC is poor, with a 27% 5-year mortality. Importantly, the impaired DSS of this subgroup was confirmed via multivariate analysis. Our study confirmed that the pattern-based framework of p53 IHC interpretation, previously described as having an excellent correlation with the
TP53 mutational status in PSCC [
14], significantly improved the conventional evaluation of p53 IHC. In addition to the diffuse overexpression classically accepted by previous researchers [
33], this framework recognizes additional patterns (null, cytoplasmic and basal overexpression) usually neglected in previous studies [
34]. As shown in this study, these patterns correlated with an adverse prognosis, as 7/14 deaths in the HPV-independent/p53 abnormal group were related to tumors showing p53 patterns not recognized via classical p53 IHC. These differences in p53 IHC results may explain the differences observed in previous studies regarding the prognostic impact of p53 IHC [
35].Four out of the 36 HPV-associated PSCC (11%) in our series had abnormal p53 IHC (one null, one basal and two diffuse overexpression).
Although the vast majority of HPV-associated PSCC showed a normal pattern of p53 expression [
14], a small percentage of patients in our series (11%) exhibited an abnormal p53 IHC pattern. This finding has been previously described [
17]. Moreover,
TP53 mutations have been detected in small percentages of HPV-associated PSCC [
36], in HPV-associated tumors of the head and neck [
37] and in the vulva [
38,
39], indicating that abnormal
TP53 (or abnormal p53 IHC staining) is not an exclusive finding of HPV-independent carcinomas. Interestingly, none of the patients with HPV-associated PSCC with an abnormal p53 IHC expression pattern died due to the disease, suggesting that
TP53 mutation does not impair prognosis in this molecular category, although further studies including a greater number of HPV-associated tumors with abnormal p53 IHC staining are needed to reach strong conclusions. Finally, it should be emphasized that although in this study the correlation between p16 IHC overexpression and HPV detection was 100%, a percentage of around 10% of discrepant results has been reported in other studies focused on head and neck and vulvar tumors [
32], and that this phenomenon should also be expected as probably occurring in PSCC.
Interestingly, the presence of vascular invasion was the only factor associated with disease recurrence according to the multivariate analysis. This association is not surprising considering the previously reported association between vascular invasion and lymph node metastases in PSCC [
40].
Our study has several limitations. Due to the large time frame of inclusion, a significant number of patients, mainly from the initial period of inclusion, did not undergo inguinal staging using sentinel lymph node analysis; thus, some inguinal lymph node microscopic metastases could have been missed. In addition, the results could not be corrected for the different treatments due to the small number of patients requiring adjuvant therapies. Finally, the small number of disease-related deaths in the series might have affected the strength of the statistical estimations.
5. Conclusions
Our study showed that patients with HPV-independent/p53 abnormal PSCC have adverse clinical outcomes than patients with HPV-associated and HPV-independent/p53 normal PSCC. p53 IHC defines two prognostic categories in HPV-independent PSCC: HPV-independent/p53 normal PSCC are low-risk tumors, whereas HPV-independent/p53 abnormal tumors can be considered aggressive neoplasms. Our study suggests that PSCC be stratified into three molecular types with distinct clinicopathological features and behaviors based on p16 (as a surrogate of HPV status) and p53 IHC (as a surrogate of TP53 mutational status) status. If these results are confirmed in prospective studies, they could help to refine the staging work-up, treatment schemes and follow-up strategies for patients with PSCC.
Author Contributions
I.T.: study design, investigation, methodology, writing of the first draft, writing and editing; F.A.: investigation, methodology, writing of the first draft, writing and editing; I.d.T.: investigation, writing and editing; A.S.: investigation, writing of the first draft, writing and editing; L.M.: investigation, methodology, writing and editing; N.P.: investigation, methodology, writing and editing; L.D.-A.: investigation, methodology, writing and editing; L.S.: investigation, methodology, writing and editing; K.D.: investigation, methodology, writing and editing; A.M.: statistical analysis, writing and editing; M.d.P.: statistical analysis, writing of the first draft, writing and editing; C.F-P.: recruitment of patients, clinical follow-up, writing and editing; M.J.R.: recruitment of patients, clinical follow-up, writing and editing; T.A.: recruitment of patients, clinical follow-up, writing and editing; J.M.C.: recruitment of patients, clinical follow-up, writing and editing; J.M.G.: recruitment of patients, clinical follow-up, writing and editing; O.R.: recruitment of patients, clinical follow-up, writing and editing; O.O.: data analysis; writing and editing; I.R-C.: study design, investigation, methodology, writing of the first draft, writing and editing; A.G-H.: study design, investigation, methodology, writing of the first draft, writing and editing; N.R. study design, investigation, methodology, writing of the first draft, writing and editing, project management. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The institutional ethical approval for this study was obtained (registry reference HCB/2020/1207).
Informed Consent Statement
Written study consent was obtained from all the patients enrolled in the study.
Data Availability Statement
All data used in this study are available and can be accessed upon reasonable request.
Acknowledgments
ISGlobal receives support from the Spanish Ministry of Science and Innovation through the “Centro de Excelencia Severo Ochoa 2019-2023” Program (CEX2018-000806-S) and support from the Generalitat de Catalunya through the CERCA Program.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Emmanuel, A.; Nettleton, J.; Watkin, N.; Berney, D.M. The Molecular Pathogenesis of Penile Carcinoma—Current Developments and Understanding. Virchows Archiv 2019, 475, 397–405. [Google Scholar] [CrossRef]
- Borque-Fernando, Á.; Gaya, J.M.; Esteban-Escaño, L.M.; Gómez-Rivas, J.; García-Baquero, R.; Agreda-Castañeda, F.; Gallioli, A.; Verri, P.; Ortiz-Vico, F.J.; Amir-Nicolau, B.F.; et al. Epidemiology, Diagnosis and Management of Penile Cancer: Results from the Spanish National Registry of Penile Cancer. Cancers (Basel) 2023, 15, 1–12. [Google Scholar] [CrossRef]
- Amin, M.B. , Berney D.M., Comperat, E.M., Hartmann, A., Menon S., Netto G.J., Raspollini, M. R., Rubin, M. A., Tickoo, S. and Tujaric, S. Who Classification of Tumours. 5th Edition. Urinary and Male Genital Tumours; Board, W.C. of T.E., Ed.; 5th ed.IARC: Lyon (France), 2022; ISBN 9789283245124. [Google Scholar]
- Chaux, A.; Velazquez, E.; Algaba, F.; Ayala, G.; Cubilla, A. Developments in the Pathology of Penile Squamous Cell Carcinomas. Urology 2010, 76. [Google Scholar] [CrossRef]
- Gross G; Pfister H Role of Human Papillomavirus in Penile Cancer, Penile Intraepithelial Squamous Cell Neoplasias and in Genital Warts. Med Microbiol Immunol 2004, 193, 35–44. [CrossRef]
- Kashofer, K.; Winter, E.; Halbwedl, I.; Thueringer, A.; Kreiner, M.; Sauer, S.; Regauer, S. HPV-Negative Penile Squamous Cell Carcinoma: Disruptive Mutations in the TP53 Gene Are Common. Modern Pathology 2017, 30, 1013–1020. [Google Scholar] [CrossRef]
- Feber, A.; Worth, D.C.; Chakravarthy, A.; de Winter, P.; Shah, K.; Arya, M.; Saqib, M.; Nigam, R.; Malone, P.R.; Tan, W.S.; et al. CSN1 Somatic Mutations in Penile Squamous Cell Carcinoma. Cancer Res 2016, 76, 4720–4727. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, K.; Chen, Y.; Zhou, J.; Liang, Y.; Yang, X.; Li, X.; Cao, Y.; Wang, D.; Luo, L.; et al. Mutational Landscape of Penile Squamous Cell Carcinoma in a Chinese Population. Int J Cancer 2019, 145, 1280–1289. [Google Scholar] [CrossRef]
- McDaniel, A.S.; Hovelson, D.H.; Cani, A.K.; Liu, C.-J.; Zhai, Y.; Zhang, Y.; Weizer, A.Z.; Mehra, R.; Feng, F.Y.; Alva, A.S.; et al. Genomic Profiling of Penile Squamous Cell Carcinoma Reveals New Opportunities for Targeted Therapy. Cancer Res 2015, 75, 5219–5227. [Google Scholar] [CrossRef]
- Stoehr, R.; Weisser, R.; Wendler, O.; Giedl, J.; Daifalla, K.; Gaisa, N.T.; Richter, G.; Campean, V.; Burger, M.; Wullich, B.; et al. P53 Codon 72 Polymorphism and Risk for Squamous Cell Carcinoma of the Penis: A Caucasian Case-Control Study. J Cancer 2018, 9, 4234–4241. [Google Scholar] [CrossRef]
- Lopes A; Bezerra AL; Pinto CA; Serrano SV; de Mello CA; Villa LL. P53 as a New Prognostic Factor for Lymph Node Metastasis in Penile Carcinoma: Analysis of 82 Patients Treated with Amputation and Bilateral Lymphadenectomy. J Urol. 2002, 168, 81–86. [CrossRef]
- Jacob, J.; Ferry, E.; Gay, L.; Elvin, J.A.; Vergilio, J.A.; Ramkissoon, S.; Severson, E.; Necchi, A.; Killian, J.K.; Ali, S.M.; et al. Comparative Genomic Profiling of Refractory and Metastatic Penile and Nonpenile Cutaneous Squamous Cell Carcinoma: Implications for Selection of Systemic Therapy. J Urol 2019, 201, 541–548. [Google Scholar] [CrossRef]
- Ferrandiz-Pulido, C.; Masferrer, E.; Toll, A.; Hernandez-Losa, J.; Mojal, S.; Pujol, R.; Ramon y Cajal, S.; de Torres, I.; Garcia-Patos, V. MTOR Signaling Pathway in Penile Squamous Cell Carcinoma: PmTOR and PeIF4E over Expression Correlate with Aggressive Tumor Behavior. J Urol 2013, 190, 2288–2295. [Google Scholar] [CrossRef]
- Trias, I.; Saco, A.; Marimon, L.; López del Campo, R.; Manzotti, C.; Ordi, O.; del Pino, M.; Pérez, F.M.; Vega, N.; Alós, S.; et al. P53 in Penile Squamous Cell Carcinoma: A Pattern-Based Immunohistochemical Framework with Molecular Correlation. Cancers (Basel) 2023, 15. [Google Scholar] [CrossRef]
- Kortekaas KE, Solleveld-Westerink N, Tessier-Cloutier B, Rutten TA, Poelgeest MIE, Gilks CB, Hoang LN, B. T. Performance of the Pattern-Based Interpretation of P53 Immunohistochemistry as a Surrogate for TP53 Mutations in Vulvar Squamous Cell Carcinoma. Histopathology 2020, 77, 92–99. [Google Scholar] [CrossRef]
- Liu, J.Y.; Li, Y.H.; Zhang, Z.L.; Yao, K.; Ye, Y.L.; Xie, D.; Han, H.; Liu, Z.W.; Qin, Z.K.; Zhou, F.J. The Risk Factors for the Presence of Pelvic Lymph Node Metastasis in Penile Squamous Cell Carcinoma Patients with Inguinal Lymph Node Dissection. World J Urol 2013, 31, 1519–1524. [Google Scholar] [CrossRef]
- Zargar-Shoshtari, K.; Spiess, P.E.; Berglund, A.E.; Sharma, P.; Powsang, J.M.; Giuliano, A.; Magliocco, A.M.; Dhillon, J. Clinical Significance of P53 and P16ink4a Status in a Contemporary North American Penile Carcinoma Cohort. Clin Genitourin Cancer 2016, 14, 346–351. [Google Scholar] [CrossRef]
- Brouwer, O.R.; Albersen, M.; Parnham, A.; Protzel, C.; Pettaway, C.A.; Ayres, B.; Antunes-Lopes, T.; Barreto, L.; Campi, R.; Crook, J.; et al. European Association of Urology-American Society of Clinical Oncology Collaborative Guideline on Penile Cancer: 2023 Update. Eur Urol 2023, 83, 548–560. [Google Scholar] [CrossRef]
- Cheng, A.S.; Karnezis, A.N.; Jordan, S.; Singh, N.; McAlpine, J.N.; Gilks, C.B. P16 Immunostaining Allows for Accurate Subclassification of Vulvar Squamous Cell Carcinoma Into HPV-Associated and HPV-Independent Cases. International Journal of Gynecological Pathology 2016, 35, 385–393. [Google Scholar] [CrossRef]
- Santos, M.; Landolfi, S.; Olivella, A.; Lloveras, B.; Klaustermeier, J.; Suárez, H.; Alòs, L.; Puig-Tintoré, L.M.; Campo, E.; Ordi, J. P16 Overexpression Identifies HPV-Positive Vulvar Squamous Cell Carcinomas. Am J Surg Pathol 2006, 30, 1347–1356. [Google Scholar] [CrossRef]
- Tessier-Cloutier, B.; Kortekaas, K.E.; Thompson, E.; Pors, J.; Chen, J.; Ho, J.; Prentice, L.M.; McConechy, M.K.; Chow, C.; Proctor, L.; et al. Major P53 Immunohistochemical Patterns in in Situ and Invasive Squamous Cell Carcinomas of the Vulva and Correlation with TP53 Mutation Status. Modern Pathology 2020, 33, 1595–1605. [Google Scholar] [CrossRef]
- Menon, S.; Moch, H.; Berney, D.M.; Cree, I.A.; Srigley, J.R.; Tsuzuki, T.; Compérat, E.; Hartmann, A.; Netto, G.; Rubin, M.A.; et al. WHO 2022 Classification of Penile and Scrotal Cancers: Updates and Evolution. Histopathology 2023, 82, 508–520. [Google Scholar] [CrossRef]
- Kidd, L.C.; Chaing, S.; Chipollini, J.; Giuliano, A.R.; Spiess, P.E.; Sharma, P. Relationship between Human Papillomavirus and Penile Cancer-Implications for Prevention and Treatment. Transl Androl Urol 2017, 6, 791–802. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, X. Radioimmunotherapy in HPV-Associated Head and Neck Squamous Cell Carcinoma. Biomedicines 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Rakislova, N.; Saco, A.; Sierra, A.; Del Pino, M.; Ordi, J. Role of Human Papillomavirus in Vulvar Cancer. Adv Anat Pathol 2017, 24, 201–214. [Google Scholar] [CrossRef]
- Szymonowicz, K.A.; Chen, J. Biological and Clinical Aspects of HPV-Related Cancers. Cancer Biol Med 2020, 17, 864–878. [Google Scholar] [CrossRef]
- Visser, O.; Adolfsson, J.; Rossi, S.; Verne, J.; Gatta, G.; Maffezzini, M.; Franks, K.N.; Zielonk, N.; Van Eycken, E.; Sundseth, H.; et al. Incidence and Survival of Rare Urogenital Cancers in Europe. Eur J Cancer 2012, 48, 456–464. [Google Scholar] [CrossRef]
- Manzotti, C.; Chulo, L.; López del Campo, R.; Trias, I.; del Pino, M.; Saúde, O.; Basílio, I.; Tchamo, N.; Lovane, L.; Lorenzoni, C.; et al. Penile Squamous Cell Carcinomas in Sub-Saharan Africa and Europe: Differential Etiopathogenesis. Cancers (Basel) 2022, 14. [Google Scholar] [CrossRef]
- Mohanty, S.K.; Mishra, S.K.; Bhardwaj, N.; Sardana, R.; Jaiswal, S.; Pattnaik, N.; Pradhan, D.; Sharma, S.; Kaushal, S.; Baisakh, M.R.; et al. P53 and P16ink4a As Predictive and Prognostic Biomarkers for Nodal Metastasis and Survival in A Contemporary Cohort of Penile Squamous Cell Carcinoma. Clin Genitourin Cancer 2021, 19, 510–520. [Google Scholar] [CrossRef]
- Nooij, L.S.; Ter Haar, N.T.; Ruano, D.; Rakislova, N.; van Wezel, T.; Smit, V.T.H.B.M.; Trimbos, B.J.B.M.Z.; Ordi, J.; van Poelgeest, M.I.E.; Bosse, T. Genomic Characterization of Vulvar (Pre)Cancers Identifies Distinct Molecular Subtypes with Prognostic Significance. Clin Cancer Res 2017, 23, 6781–6789. [Google Scholar] [CrossRef]
- Kortekaas, K.E.; Bastiaannet, E.; van Doorn, H.C.; de Vos van Steenwijk, P.J.; Ewing-Graham, P.C.; Creutzberg, C.L.; Akdeniz, K.; Nooij, L.S.; van der Burg, S.H.; Bosse, T.; et al. Vulvar Cancer Subclassification by HPV and P53 Status Results in Three Clinically Distinct Subtypes. Gynecol Oncol 2020, 159, 649–656. [Google Scholar] [CrossRef]
- Carreras-Dieguez, N.; Saco, A.; del Pino, M.; Marimon, L.; López del Campo, R.; Manzotti, C.; Fusté, P.; Pumarola, C.; Torné, A.; Garcia, A.; et al. Human Papillomavirus and P53 Status Define Three Types of Vulvar Squamous Cell Carcinomas with Distinct Clinical, Pathological, and Prognostic Features. Histopathology 2023, 17–30. [Google Scholar] [CrossRef]
- Martins, A.C.P.; Faria, S.M.; Cologna, A.J.; Suaid, H.J.; Tucci, S. Immunoexpression of P53 Protein and Proliferating Cell Nuclear Antigen in Penile Carcinoma. Journal of Urology 2002, 167, 89–93. [Google Scholar] [CrossRef]
- Bowie, J.; Singh, S.; O’hanlon, C.; Shiatis, V.; Brunckhorst, O.; Muneer, A.; Ahmed, K. A Systematic Review of Non-HPV Prognostic Biomarkers Used in Penile Squamous Cell Carcinoma. Turk J Urol 2021, 47, 358–365. [Google Scholar] [CrossRef]
- Yanagawa, N.; Osakabe, M.; Hayashi, M.; Tamura, G.; Motoyama, T. Detection of HPV-DNA, P53 Alterations, and Methylation in Penile Squamous Cell Carcinoma in Japanese Men. Pathol Int 2008, 58, 477–482. [Google Scholar] [CrossRef]
- Ermakov, M.S.; Kashofer, K.; Regauer, S. Different Mutational Landscapes in Human Papillomavirus–Induced and Human Papillomavirus–Independent Invasive Penile Squamous Cell Cancers. Modern Pathology 2023, 36, 100250. [Google Scholar] [CrossRef]
- Westra, W.H.; Taube, J.M.; Poeta, M.L.; Begum, S.; Sidransky, D.; Koch, W.M. Inverse Relationship between Human Papillomavirus-16 Infection and Disruptive P53 Gene Mutations in Squamous Cell Carcinoma of the Head and Neck. Clinical Cancer Research 2008, 14, 366–369. [Google Scholar] [CrossRef]
- Zięba, S.; Kowalik, A.; Zalewski, K.; Rusetska, N.; Goryca, K.; Piaścik, A.; Misiek, M.; Bakuła-Zalewska, E.; Kopczyński, J.; Kowalski, K.; et al. Somatic Mutation Profiling of Vulvar Cancer: Exploring Therapeutic Targets. Gynecol Oncol 2018, 150, 552–561. [Google Scholar] [CrossRef]
- Yang, H.; Almadani, N.; Thompson, E.F.; Tessier-Cloutier, B.; Chen, J.; Ho, J.; Senz, J.; McConechy, M.K.; Chow, C.; Ta, M.; et al. Classification of Vulvar Squamous Cell Carcinoma and Precursor Lesions by P16 and P53 Immunohistochemistry: Considerations, Caveats, and an Algorithmic Approach. Modern Pathology 2023, 36, 100145. [Google Scholar] [CrossRef]
- Slaton, J.W.; Morgenstern, N.; Levy, D.A.; Santos, M.W.; Tamboli, P.; Ro, J.Y.; Ayala, A.G.; Pettaway, C.A. Tumor Stage, Vascular Invasion and the Percentage of Poorly Differentiated Cancer: Independent Prognosticators for Inguinal Lymph Node Metastasis in Penile Squamous Cancer. J Urol 2001, 165, 1138–1142. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).