Submitted:
13 March 2024
Posted:
14 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
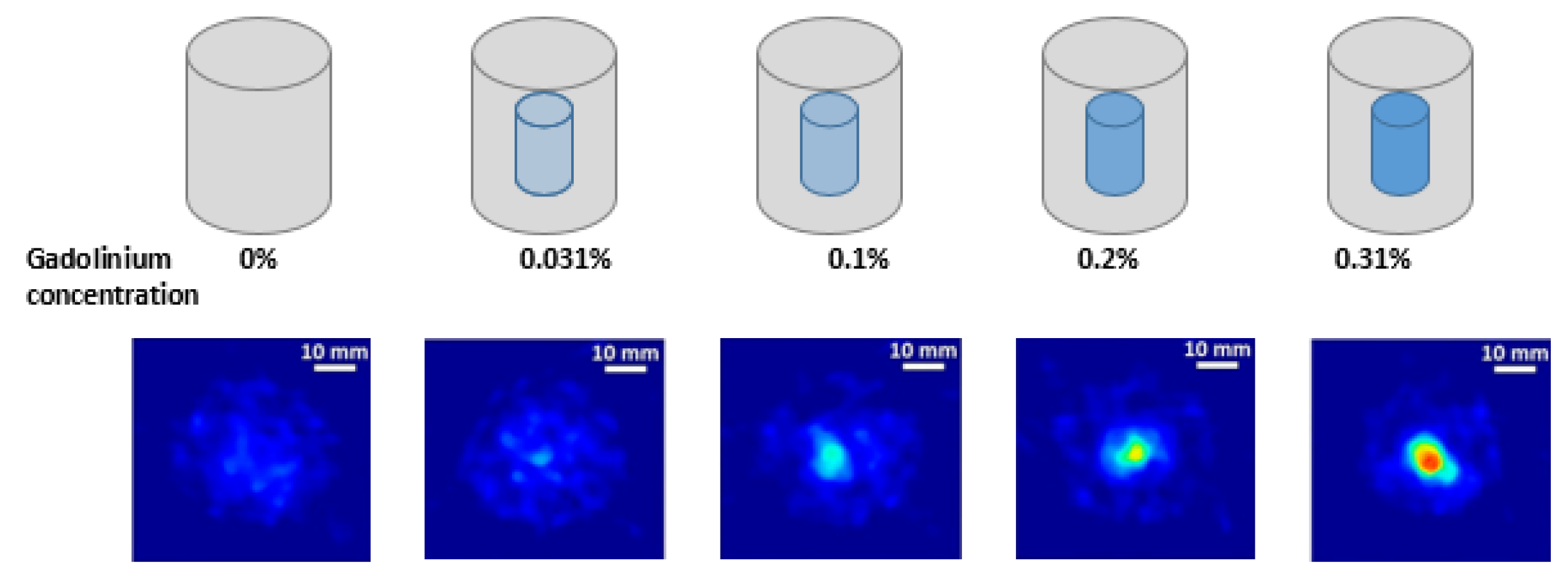
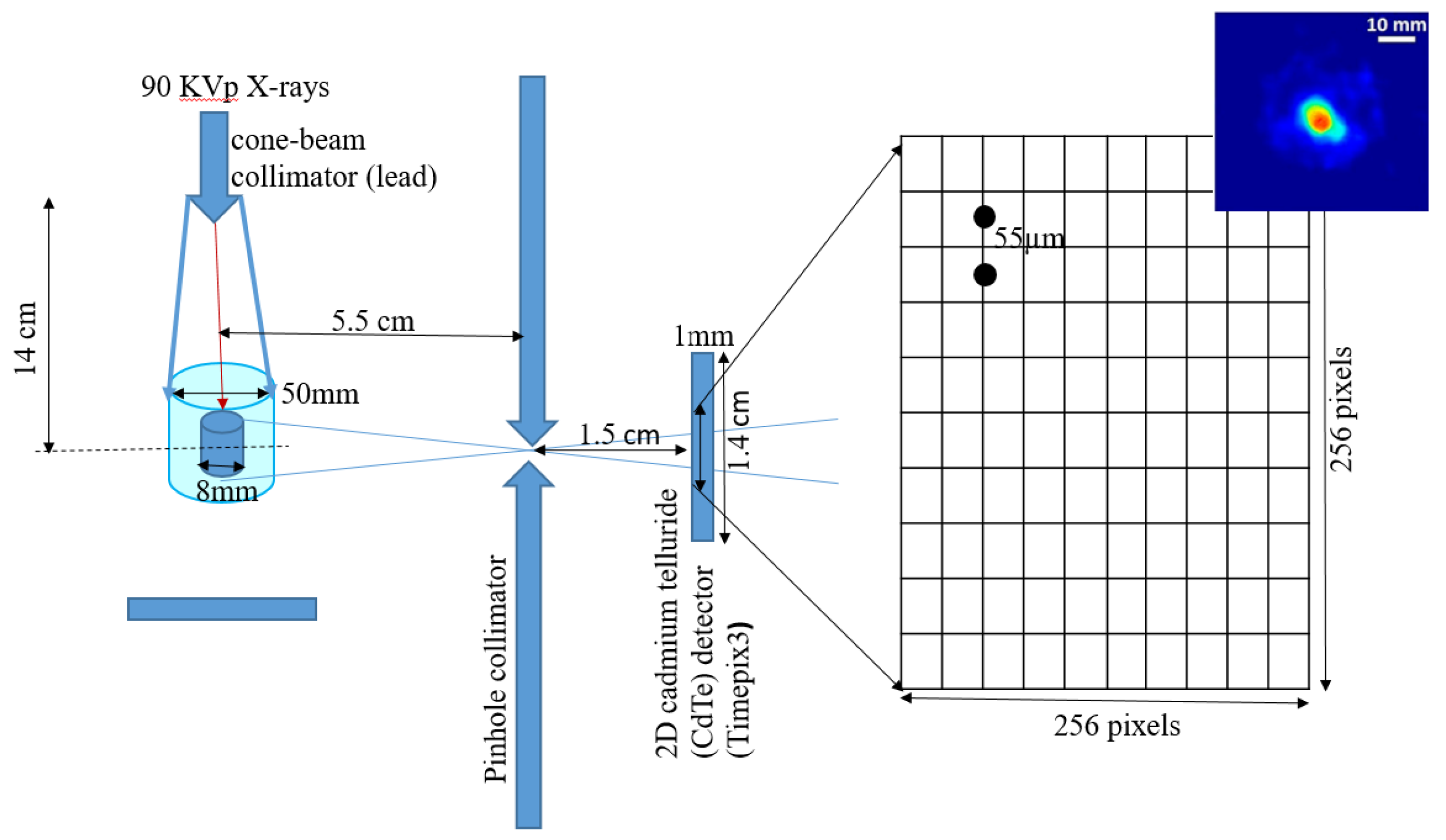
2.1. Experimental Setup
2.2. Deep Blind Image Denoising Model
2.2.1. Dataset
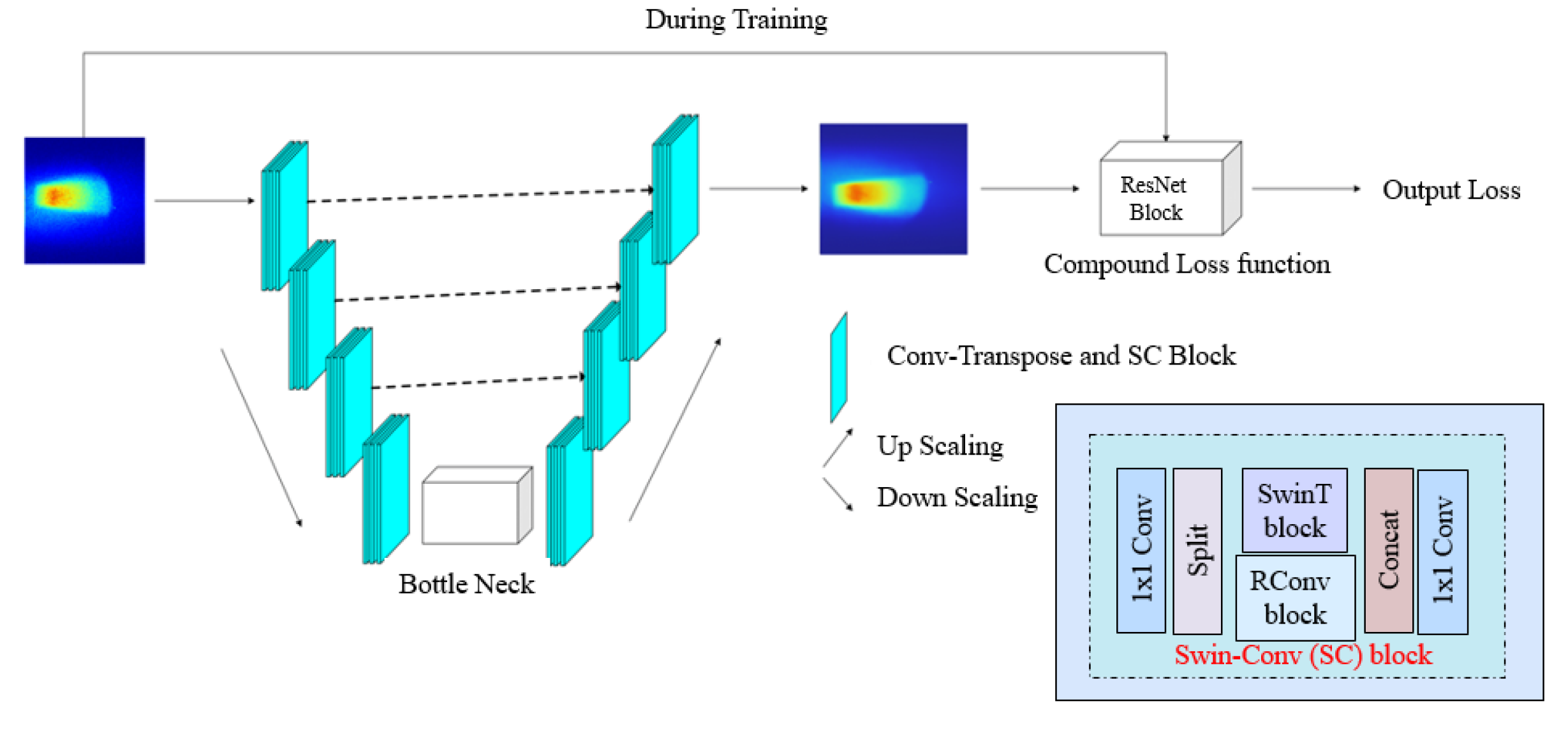
2.2.2. Proposed Model
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| XFCT | X-ray fluorescence computed tomography |
| XRF | X-ray fluorescence |
| AI | Artificial Intelligence |
| DL | Deep Learning |
| SCUNet | Swin-Conv-UNet |
| CT | Computed Tomography |
| PSNR | Peak signal-to-noise ratio |
| SSIM | Structural Similarity Index |
| BM3D | Block-Matching and 3D filtering |
| NLM | Non-local means |
| DnCNN | Denoising Convolutional Neural Network |
| CNN | convolutional neural network |
| GAN | generative adversarial networks |
| Gd | Gadolinium |
| wt | Weight |
| Swin | Shifted window Transformer |
| KeV | Kilo electron volt |
| mA | miliampere |
| CBCT | Cone Beam Computed Tomography |
| MAP | Maximum A Posteriori |
| DRUNet | Dilated-Residual U-Net |
| SwinIR | Image Restoration Using Swin Transformer |
| SConv | Strided Convolution |
| TConv | Transposed Convolution |
| RConv | Residual Convolutional |
| SwinT | Swin Transformer |
| BW | bin width |
| AWGN | Additive White Gaussian Noise |
References
- Feng, P.; Luo, Y.; Zhao, R.; Huang, P.; Li, Y.; He, P.; Tang, B.; Zhao, X. Reduction of Compton background noise for X-ray fluorescence computed tomography with deep learning. Photonics 2022, 9, 108. [Google Scholar] [CrossRef]
- Zhang, S.; Li, L.; Chen, J.; Chen, Z.; Zhang, W.; Lu, H. Quantitative imaging of Gd nanoparticles in mice using benchtop cone-beam X-ray fluorescence computed tomography system. International journal of molecular sciences 2019, 20, 2315. [Google Scholar] [CrossRef] [PubMed]
- Manohar, N.; Reynoso, F.J.; Diagaradjane, P.; Krishnan, S.; Cho, S.H. Quantitative imaging of gold nanoparticle distribution in a tumor-bearing mouse using benchtop x-ray fluorescence computed tomography. Scientific reports 2016, 6, 22079. [Google Scholar] [CrossRef] [PubMed]
- Larsson, J.C.; Vogt, C.; Vågberg, W.; Toprak, M.S.; Dzieran, J.; Arsenian-Henriksson, M.; Hertz, H.M. High-spatial-resolution x-ray fluorescence tomography with spectrally matched nanoparticles. Physics in Medicine & Biology 2018, 63, 164001. [Google Scholar]
- Takeda, T.; Yu, Q.; Yashiro, T.; Zeniya, T.; Wu, J.; Hasegawa, Y.; Hyodo, K.; Yuasa, T.; Dilmanian, F.; Akatsuka, T.; others. Iodine imaging in thyroid by fluorescent X-ray CT with 0.05 mm spatial resolution. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 2001, 467, 1318–1321. [Google Scholar] [CrossRef]
- Cheong, S.K.; Jones, B.L.; Siddiqi, A.K.; Liu, F.; Manohar, N.; Cho, S.H. X-ray fluorescence computed tomography (XFCT) imaging of gold nanoparticle-loaded objects using 110 kVp x-rays. Physics in Medicine & Biology 2010, 55, 647. [Google Scholar]
- Deng, L.; Ahmed, M.F.; Jayarathna, S.; Feng, P.; Wei, B.; Cho, S.H. A detector’s eye view (DEV)-based OSEM algorithm for benchtop X-ray fluorescence computed tomography (XFCT) image reconstruction. Physics in Medicine & Biology 2019, 64, 08NT02. [Google Scholar]
- Cong, W.; Shen, H.; Cao, G.; Liu, H.; Wang, G. X-ray fluorescence tomographic system design and image reconstruction. Journal of X-ray science and technology 2013, 21, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.L.; Manohar, N.; Reynoso, F.; Karellas, A.; Cho, S.H. Experimental demonstration of benchtop x-ray fluorescence computed tomography (XFCT) of gold nanoparticle-loaded objects using lead-and tin-filtered polychromatic cone-beams. Physics in Medicine & Biology 2012, 57, N457. [Google Scholar]
- Ahmad, M.; Bazalova-Carter, M.; Fahrig, R.; Xing, L. Optimized detector angular configuration increases the sensitivity of x-ray fluorescence computed tomography (XFCT). IEEE transactions on medical imaging 2014, 34, 1140–1147. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Sung, W.; Ye, S.J. Pinhole X-ray fluorescence imaging of gadolinium and gold nanoparticles using polychromatic X-rays: a Monte Carlo study. International Journal of Nanomedicine 2017, 5805–5817. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Kim, T.; Lee, W.; Kim, H.; Kim, H.S.; Im, H.J.; Ye, S.J. Dynamic in vivo X-ray fluorescence imaging of gold in living mice exposed to gold nanoparticles. IEEE transactions on medical imaging 2019, 39, 526–533. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2015; pp. 1–9. [Google Scholar]
- Chen, H.; Zhang, Y.; Kalra, M.K.; Lin, F.; Chen, Y.; Liao, P.; Zhou, J.; Wang, G. Low-dose CT with a residual encoder-decoder convolutional neural network. IEEE transactions on medical imaging 2017, 36, 2524–2535. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zuo, W.; Chen, Y.; Meng, D.; Zhang, L. Beyond a gaussian denoiser: Residual learning of deep cnn for image denoising. IEEE transactions on image processing 2017, 26, 3142–3155. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, J.; Chao, H.; Yang, M. Image blind denoising with generative adversarial network based noise modeling. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2018; pp. 3155–3164. [Google Scholar]
- Guo, S.; Yan, Z.; Zhang, K.; Zuo, W.; Zhang, L. Toward convolutional blind denoising of real photographs. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 2019; pp. 1712–1722. [Google Scholar]
- Krull, A.; Buchholz, T.O.; Jug, F. Noise2void-learning denoising from single noisy images. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 2019; pp. 2129–2137. [Google Scholar]
- Sun, J.; Tappen, M.F. Learning non-local range Markov random field for image restoration. CVPR 2011. IEEE; 2011; pp. 2745–2752.
- Lefkimmiatis, S. Non-local color image denoising with convolutional neural networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2017; pp. 3587–3596. [Google Scholar]
- Liang, J.; Cao, J.; Sun, G.; Zhang, K.; Van Gool, L.; Timofte, R. Swinir: Image restoration using swin transformer. In Proceedings of the IEEE/CVF international conference on computer vision, 2021; pp. 1833–1844. [Google Scholar]
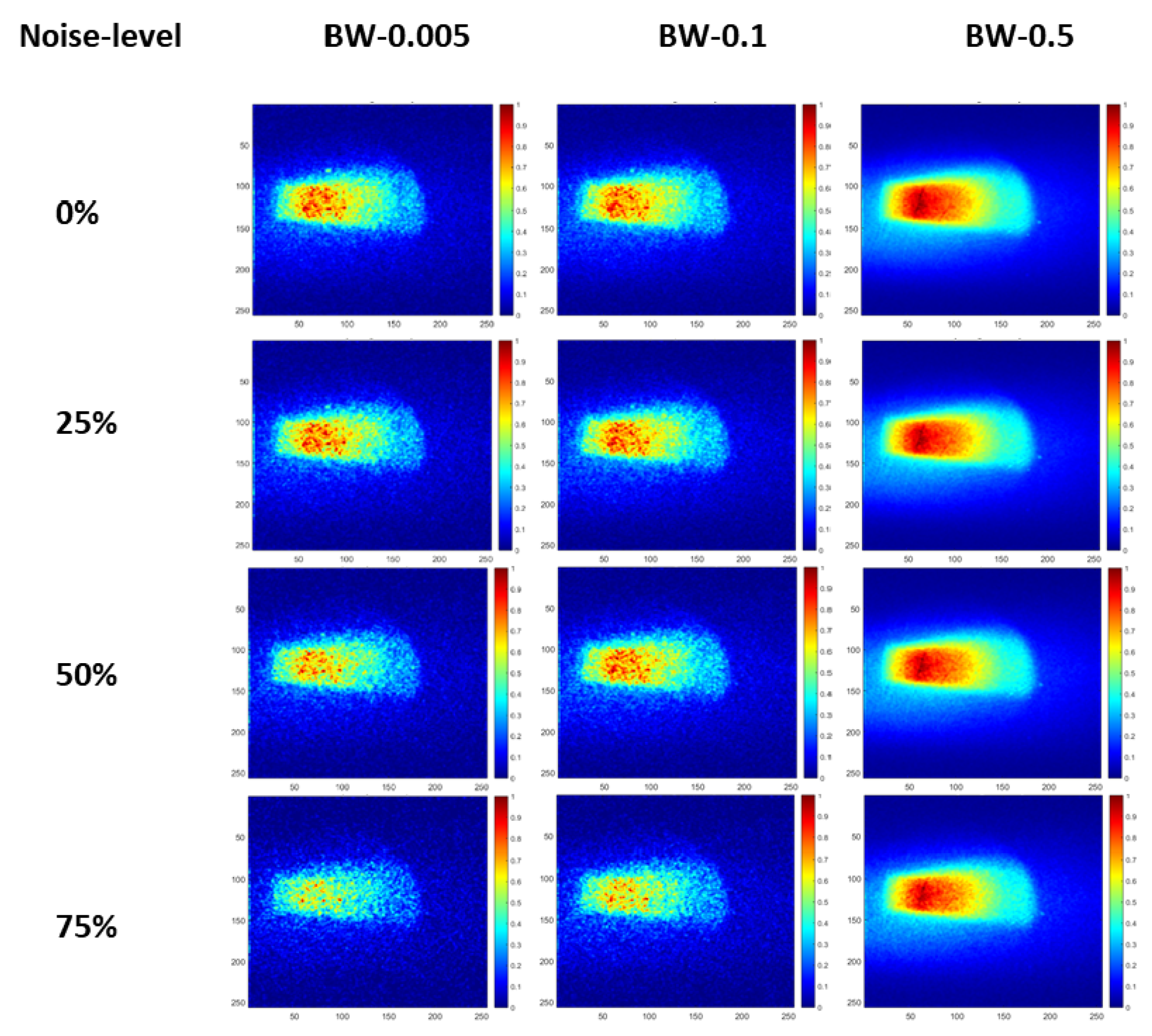
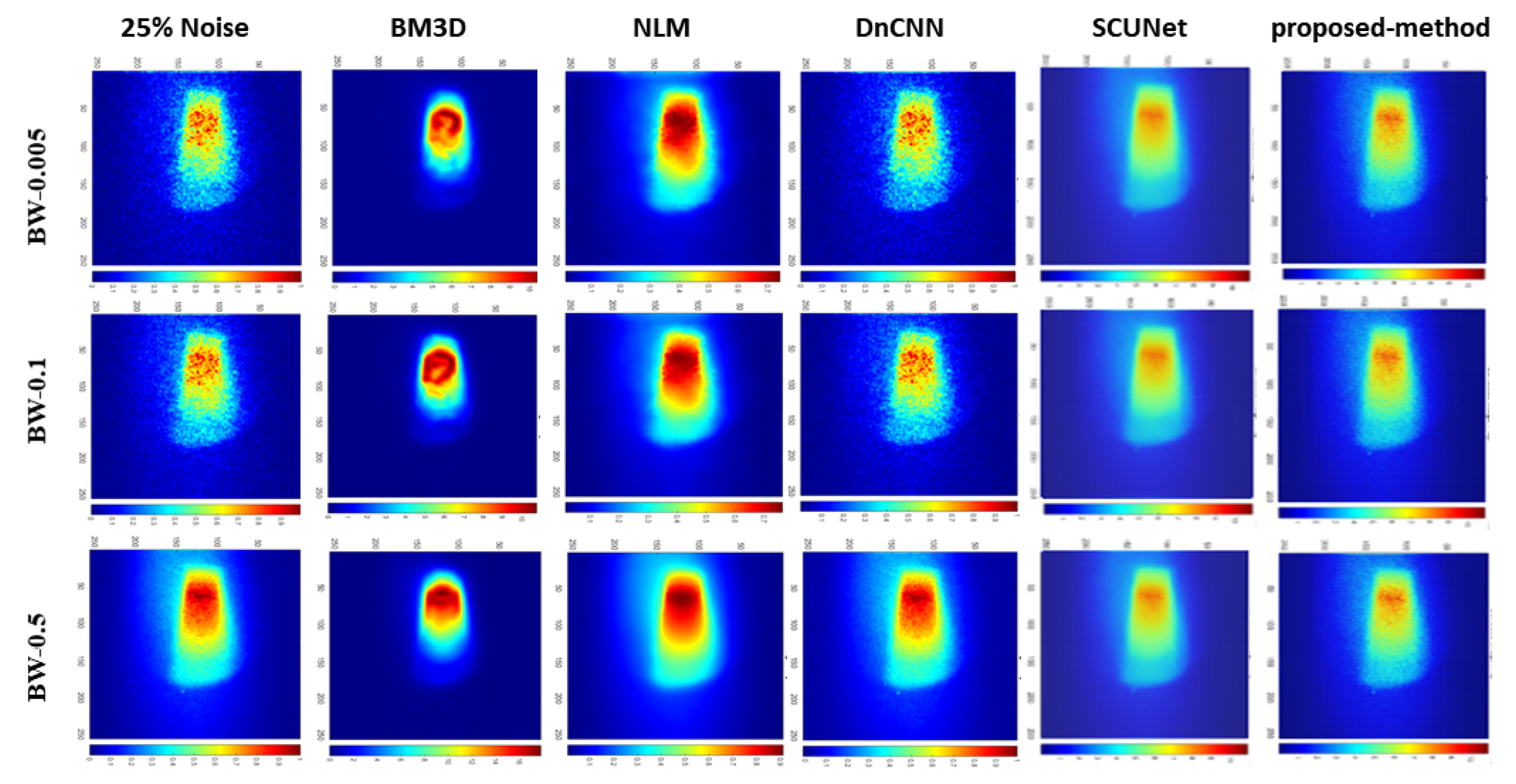
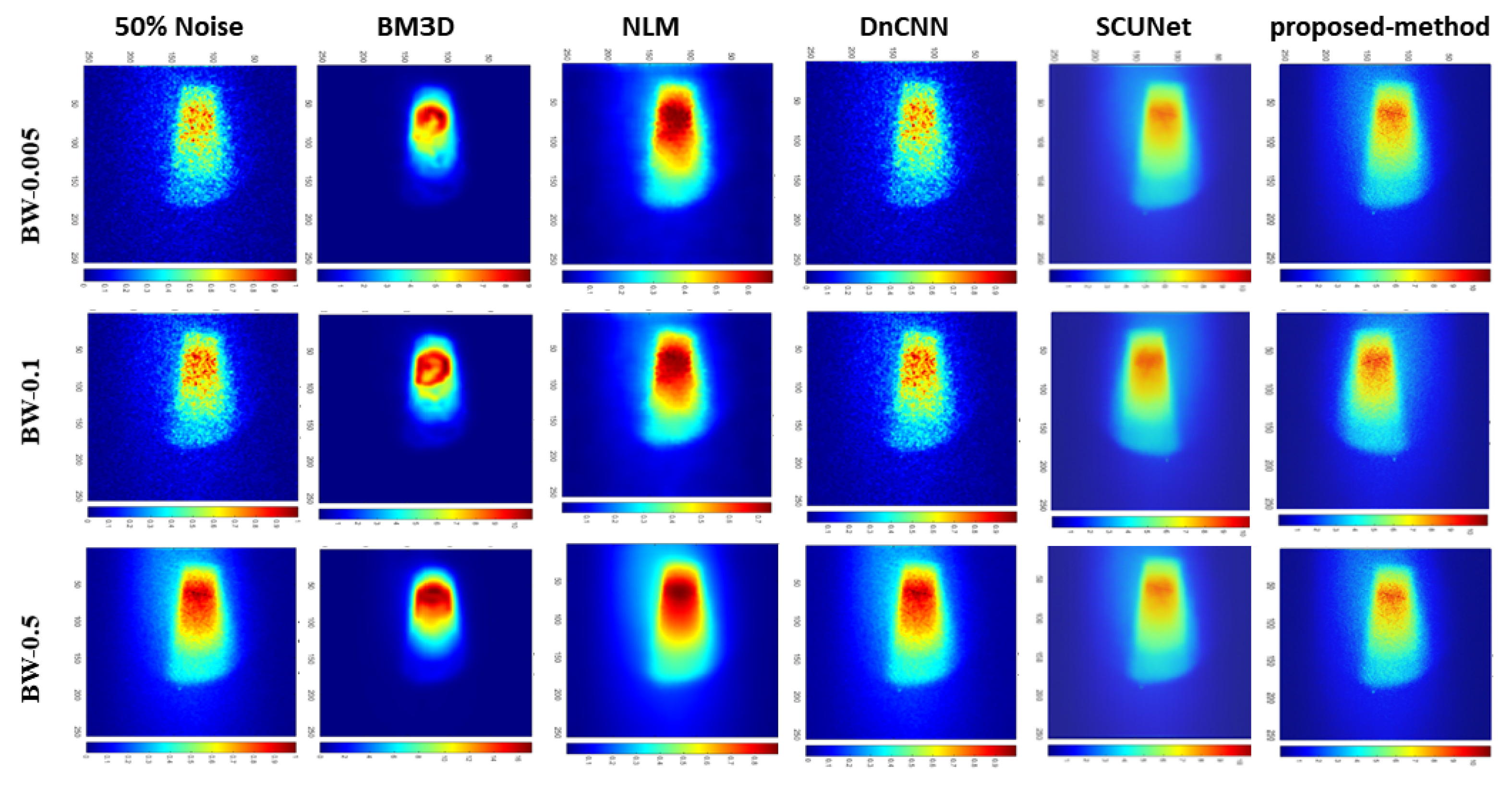
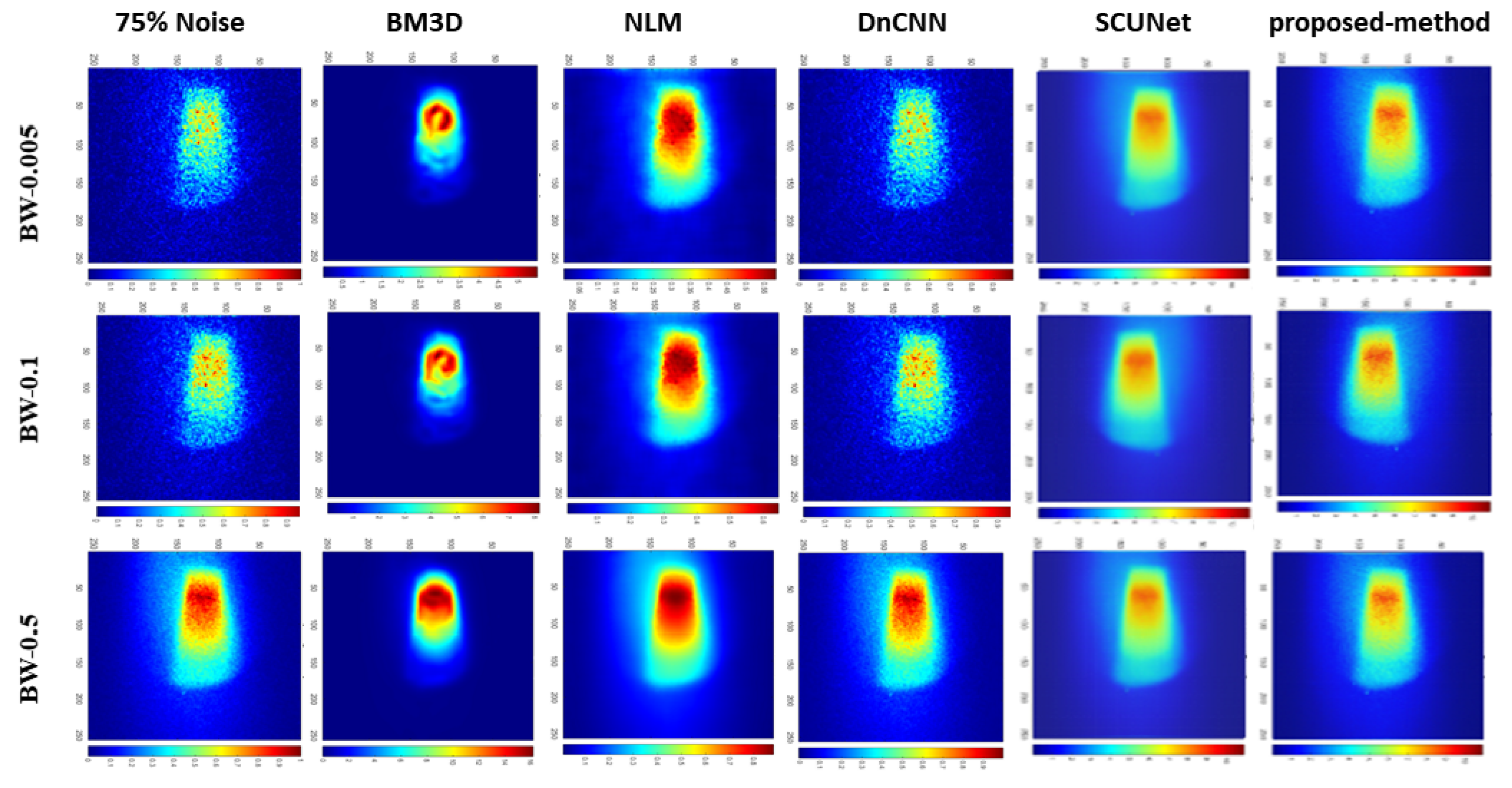
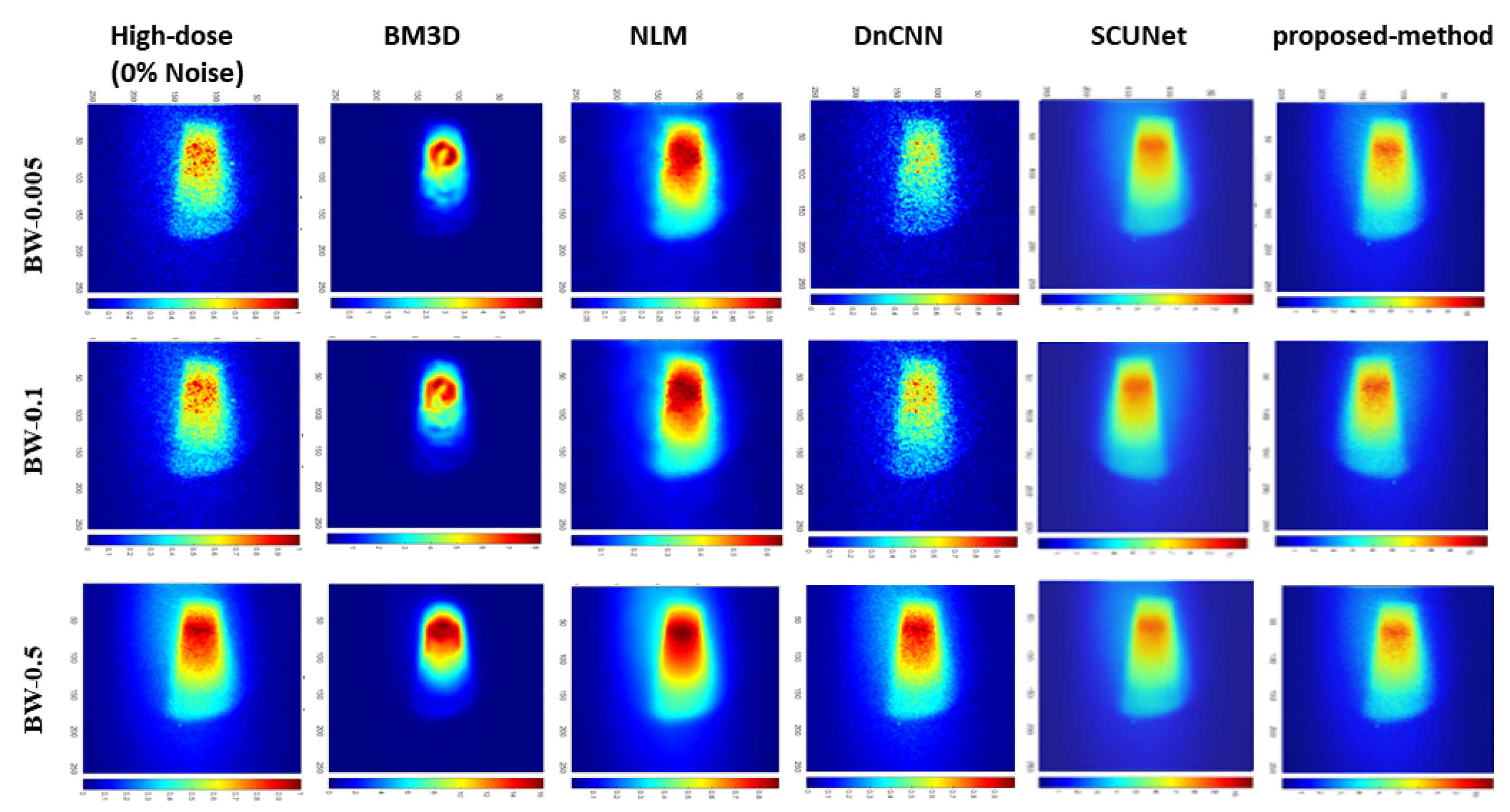
| Bin widths | Noise-level | BM3D | NLM | DnCNN | SCUNet | Proposed model |
|---|---|---|---|---|---|---|
| BW-0.05 | 25% | 13.93 | 27.92 | 36.82 | 22.77 | 29.68 |
| 50% | 13.91 | 25.16 | 31.07 | 24.51 | 26.97 | |
| 75% | 13.89 | 24.58 | 24.75 | 25.29 | 31.44 | |
| BW-0.1 | 25% | 13.70 | 32.42 | 38.88 | 22.87 | 35.48 |
| 50% | 13.70 | 28.19 | 34.50 | 27.82 | 29.06 | |
| 75% | 13.67 | 27.71 | 27.75 | 26.77 | 29.33 | |
| BW-0.5 | 25% | 11.76 | 39.94 | 49.35 | 22.54 | 31.81 |
| 50% | 11.75 | 38.77 | 43.66 | 22.74 | 32.67 | |
| 75% | 11.75 | 36.75 | 38.40 | 25.58 | 39.05 |
| Bin Widths | Noise-level | BM3D | NLM | DnCNN | SCUNet | Proposed model |
|---|---|---|---|---|---|---|
| BW-0.05 | 25% | 0.073 | 0.7435 | 0.9430 | 0.4661 | 0.7868 |
| 50% | 0.0720 | 0.739 | 0.872 | 0.5177 | 0.8369 | |
| 75% | 0.071 | 0.7156 | 0.7431 | 0.6472 | 0.8654 | |
| BW-0.1 | 25% | 0.0716 | 0.7985 | 0.8594 | 0.6153 | 0.8284 |
| 50% | 0.0715 | 0.7984 | 0.8023 | 0.6206 | 0.8658 | |
| 75% | 0.0712 | 0.7801 | 0.7867 | 0.6453 | 0.8218 | |
| BW-0.5 | 25% | 0.0617 | 0.8029 | 0.9139 | 0.5773 | 0.8786 |
| 50% | 0.0616 | 0.7626 | 0.9014 | 0.4972 | 0.8466 | |
| 75% | 0.0615 | 0.7611 | 0.7582 | 0.5290 | 0.8638 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





