1. Introduction
Entamoeba histolytica (
E. histolytica), the causative agent of human amoebiasis, primarily infects the intestinal tract. However, it can also spread to other organs, giving rise to severe complications such as hepatic abscesses, pneumonia, purulent pericarditis, and in rare cases, cerebral amebiasis [
1,
2]. It is estimated that this pathogen is responsible for approximately 50 million infections worldwide annually, with a resultant morbidity that exceeds 100,000 deaths each year [
3].
Throughout the course of host infection, the parasite encounters a myriad of stressful conditions, including acidic pH levels, the presence of noxious agents, engagement with the host immune defenses, drug exposure, heavy metal toxicity, hypoxia, nutrient scarcity, and thermal fluctuations, among others. Despite these hostile environments, the parasite demonstrates remarkable adaptive capabilities, enabling it to persist and induce amoebiasis [
4,
5,
6,
7]. Therefore, it is important to understand the molecular mechanisms that trophozoites employ to overcome stressful situations and ensure their survival.
In various organisms, including
Homo sapiens (
H. sapiens) [
8],
Saccharomyces cerevisiae (
S. cerevisiae) [
9],
Drosophila melanogaster (
D. melanogaster) [
10], and
Caenorhabditis elegans (
C. elegans) [
11], highly conserved Heat Shock Transcription Factors (HSTFs) have been identified. These HSTFs are crucial in regulating stress responses, thereby safeguarding cell survival under challenging conditions. These discoveries emphasize the evolutionary conservation of stress response mechanisms and underline the significance of HSTFs in the biology of various species.
HSTFs have a structurally conserved DNA-Binding Domain (DBD) featuring three alpha helices and four folded β-sheets, an Oligomerization Domain (OD) with a coiled-coil motif for dimer and trimer formation, and a Carboxyl-terminal Transactivation Domain (CTD). The transcriptional activity of HSTFs is intricately controlled by both intra- and intermolecular interactions, as well as by a variety of Post-translational Modifications (PTMs) [
12,
13].
In their inactive state, HSTFs are present as monomers, but they become active when they transition into trimers, a change facilitated by specific PTMs such as phosphorylation. These modifications allow HSTFs to bind to specific sequences known as Heat Shock Elements (HSE) in the promoters of their target genes during stressful conditions. This complex regulatory mechanism underscores the critical role of HSTFs in modulating cellular responses to stress and highlights their multifaceted function in maintaining cellular homeostasis [
12,
14].
According to the AmoebaDB database (
https://amoebadb.org/amoeba/app, accessed on 15 December 2023) [
15], the
E. histolytica HSFs, referred to as EhHSTFs (Heat Shock Transcription Factors), until now, are considered the second largest family after those in plants. This family consists of 7 members, identified as
Ehhstf1 (EHI_008230),
Ehhstf2 (EHI_049510),
Ehhstf3 (EHI_087630),
Ehhstf4 (EHI_142120),
Ehhstf5 (EHI_137000),
Ehhstf6 (EHI_008660), and
Ehhstf7 (EHI_200020). Currently, it has only been reported that the EhHSTF7 protein can oligomerize and recognize the HSE within the promoter region of the
EhPgp5 gene (HSE_
EhPgp5) [
16]. The binding capabilities of the other EhHSTFs to HSE have not yet been analyzed
in vitro or
in silico. Therefore, the objective of this study was to investigate both the potential oligomerization and the DNA-binding affinity of the recombinant EhHSTF5 protein (rEhHSTF5) to the HSE_
EhPgp5.
3. Discussion
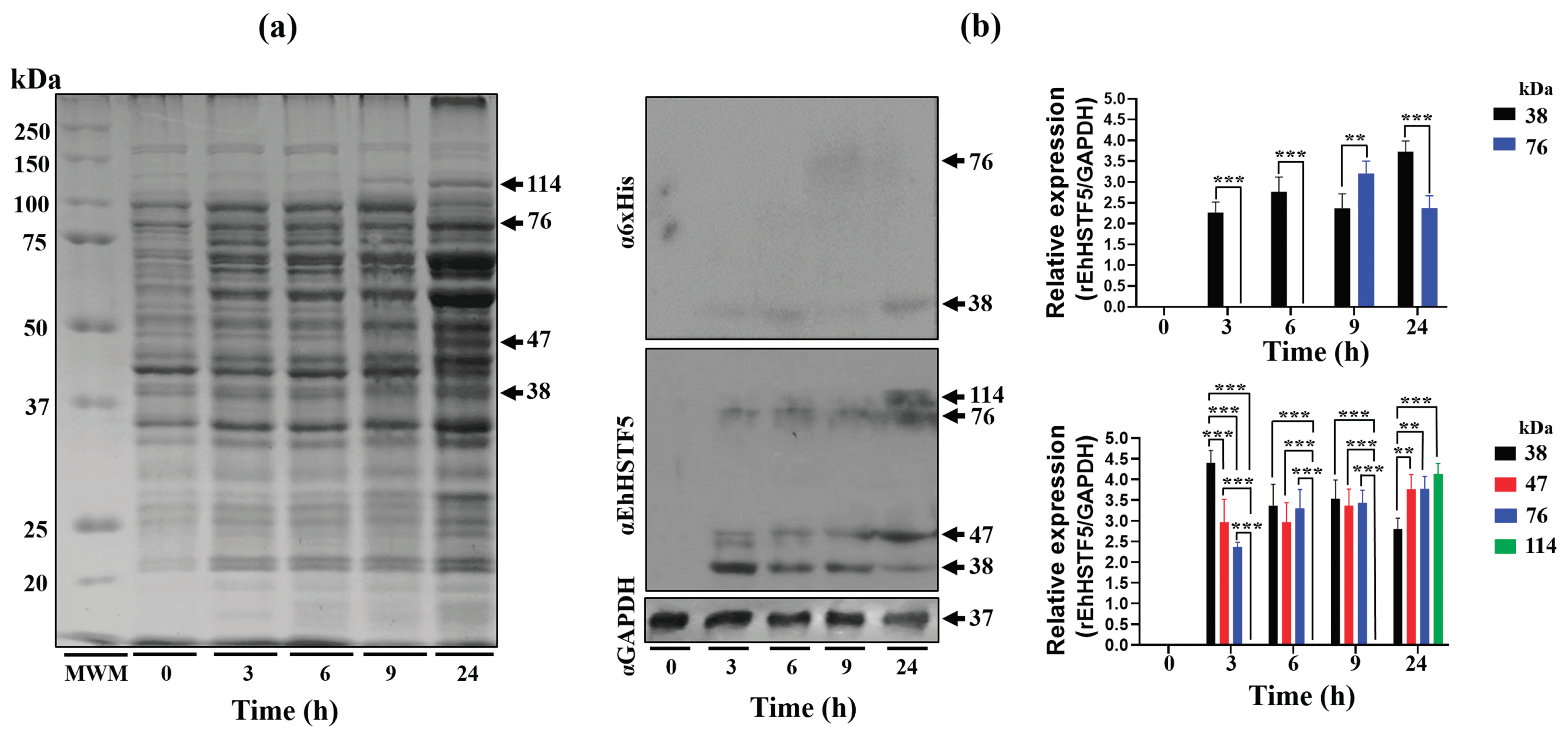
In this study, we demonstrated that the rEhHSTF5 protein undergoes oligomerization, forming dimers and trimers in protein extracts from bacteria harboring the recombinant protein. Induction with IPTG for 3 h enhances the formation of mrEhHSTF5, while induction at 24 h favors the formation of drEhHSTF5 and trEhHSTF5, suggesting that oligomerization is concentration-dependent. Protein oligomerization has been documented in HSTFs from various species. HSTF1[
8], HSTF2 [
17], and HSTF4 [
18] from
H. sapiens; HSTF1 from
A. thaliana [
19], HSTF1 from
D. melanogaster [
20], and EhHSTF7 from
E. histolytica [
16], among others. These transcription factors tend to oligomerize in cellular environments with elevated protein concentrations, while remaining as monomers at lower concentrations.
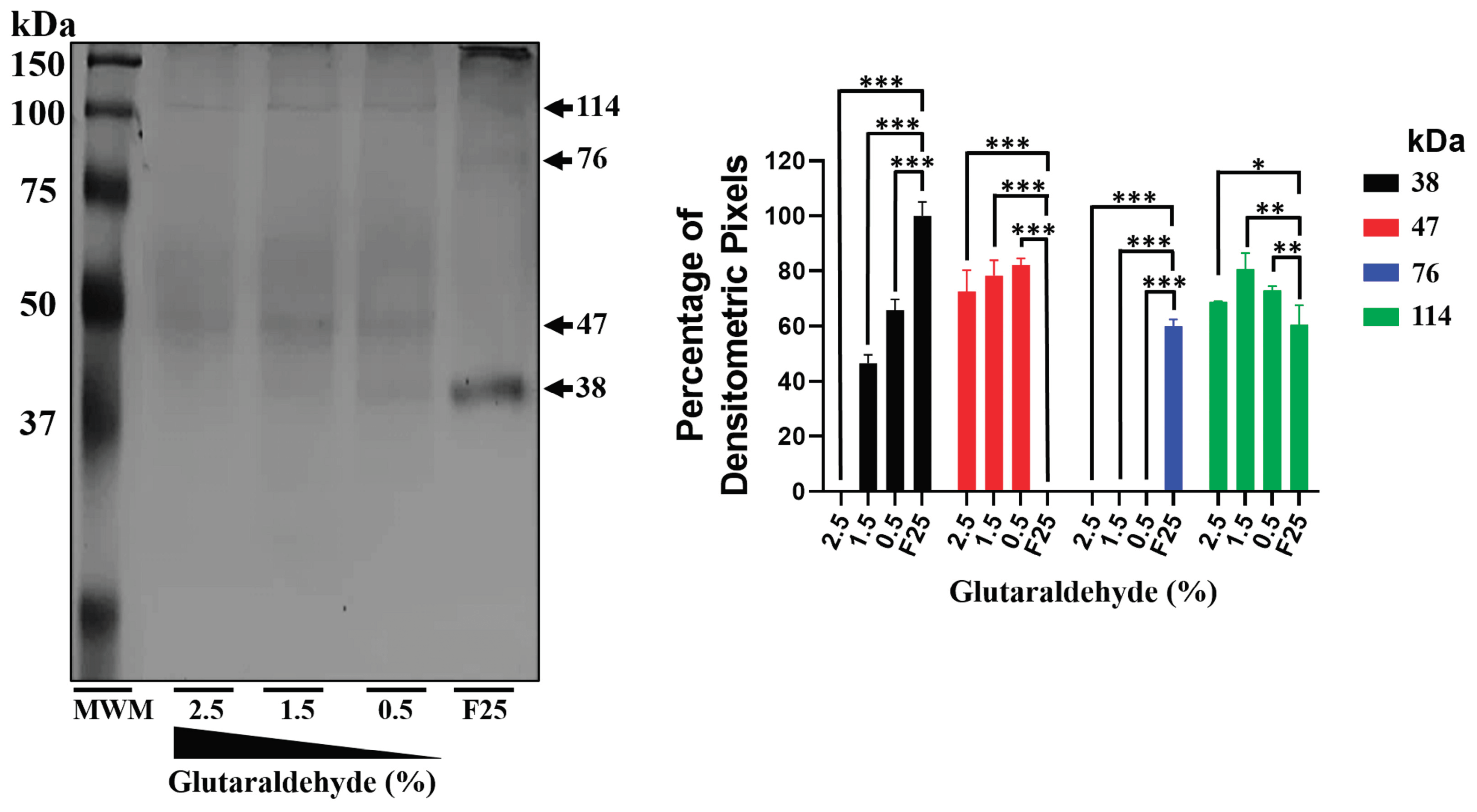
On the other hand, the 47 kDa protein expressed during IPTG induction, which was recognized by the αEhHSTF5 antibody, was also observed upon addition of glutaraldehyde to the purified mrEhHSTF5 protein (fraction 25), suggesting that the 47 kDa protein may represent a stable conformation. This conformation could be an important transient state for dimer and trimer formation, which allows the interaction and coordinated assembly of the subunits, maintaining the stability of the oligomeric structure as occurs in different proteins [
21,
22], including Superoxide Dismutase 1 (SOD1)[
22], or Heat Shock Protein 90 (HSP90) [
23], among others.
In WB assays, we found that the oligomerization of the mrEhHSTF5 protein can hide the polyhistidine tag, which makes it difficult to detect using the α6xHis antibody in induced bacteria extracts. Furthermore, after conducting nickel-affinity chromatography, we were unsuccessful in purifying the 114 kDa protein. This reinforces the idea that the polyhistidine region is hidden, making it difficult to bind to the nickel column. This observation is consistent with the behavior of the rEhHSTF7 protein reported by Bello et al. [
16], where, under similar experimental conditions, they were unable neither to detect the trimeric conformation of the protein using the α6xHis antibody nor to purify it through nickel affinity purification. Moreover, it has been previously demonstrated that the oligomerization of recombinant proteins can alter the accessibility of polyhistidine tags, potentially affecting their binding capacity to affinity matrices[
24,
25,
26].
HSTFs are recognized for their role in the heat stress response and their ability to bind to HSEs to regulate gene expression [
27,
28,
29]. The oligomerization and stability of these factors are of vital importance for their proper function, as their ability to bind to HSE and activate transcription depends on the formation of stable oligomeric structures. HSTF1 and HSTF2 from
H. sapiens form homotrimers or heterotrimers with other HSTFs. This oligomer formation increases the structural stability and their affinity for HSE, resulting in an increased ability to regulate gene expression. Therefore, oligomerization is a critical factor to achieve the necessary stability to recognize HSE [
30,
31,
32]. This fact is in concordance with our EMSA assays demonstrating that the dimeric conformation of the rEhHSTF5 protein exhibits a higher affinity for binding to the HSE_
EhPgp5. In contrast, the monomeric conformation under the same experimental conditions did not bind with the HSE_
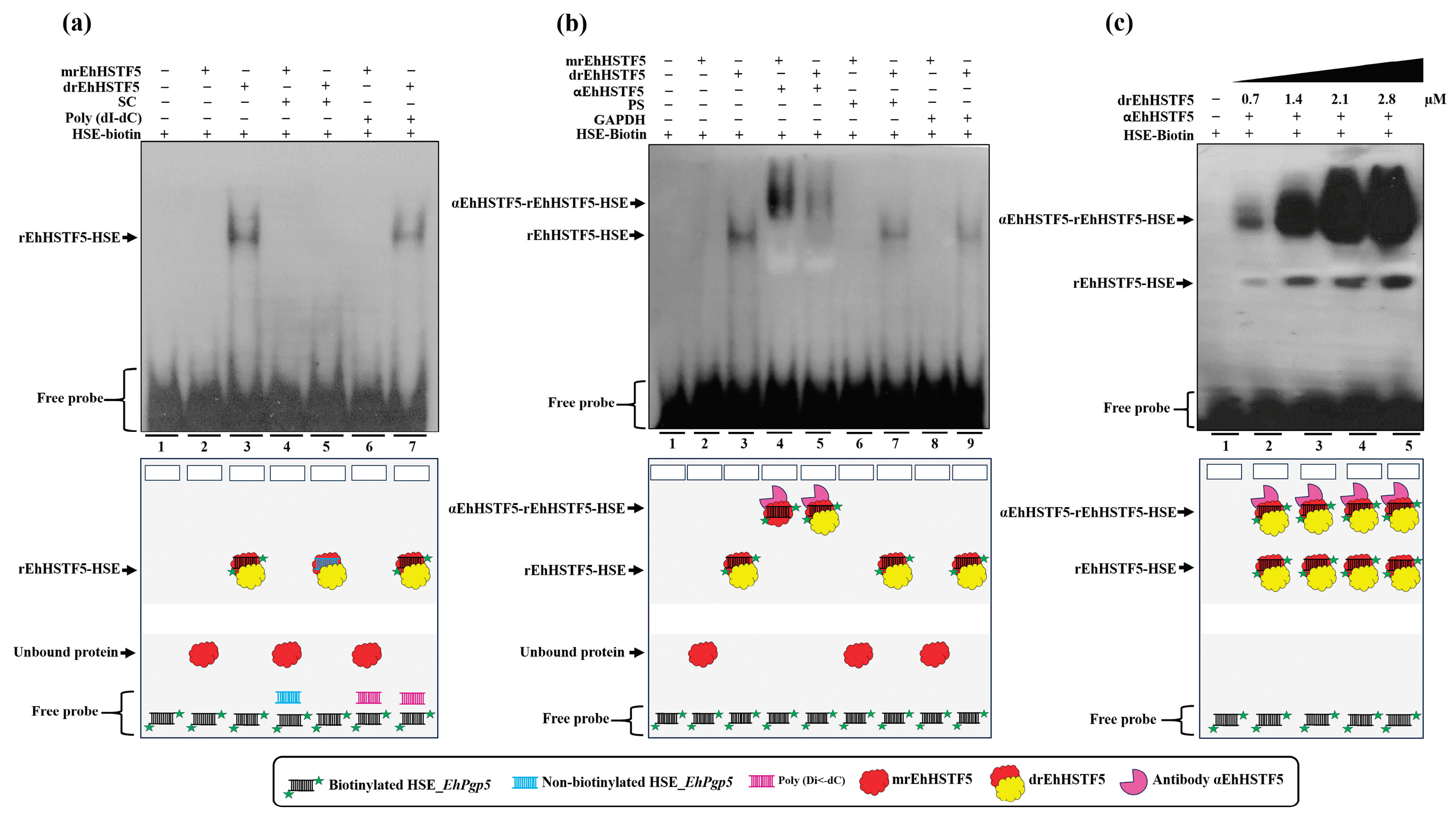
EhPgp5, confirming the necessity of oligomerization to achieve greater stability and, consequently, higher affinity to the HSE_EhPgp5. Supershift assays validated the EhHSTF5-HSE interaction, forming the αEhHSTF5-drEhHSTF5-HSE supercomplex.
The binding of mrEhHSTF5 to the HSE_
EhPgp5 was not detectable by EMSA assays. However, when the protein was preincubated with the αEhHSTF5 antibody, the formation of a highly specific supercomplex was observed. These results suggest that the antibody binding to mrEhHSTF5 induces a conformational change that exposes the DBD and, consequently, enables the binding to the HSE_
EhPgp5. A similar finding was previously reported by Zimarino et al. [
33], who found that the binding activity to the HSE by the inactive form of the HSTF from
D. melanogaster could be induced
in vitro by the addition of a polyclonal antibody against the purified factor; proposing that binding of an anti-HSTF antibody led to a conformational change, exposing its DBD to activate its transcriptional capability. Moreover, the studies performed with HsHSTF1 showed that its DBD is hidden when it is in a monomeric conformation, but is exposed when it is oligomerized, which allows it to bind more easily to HSE [
28,
30,
34]. Similarly, we suggest the possibility that EhDBD5 is hidden in the monomeric conformation of EhHSTF5 and is exposed upon oligomerization or antibody binding, which would more effectively facilitate interaction with HSE_
EhPgp5.
In silico assays is an important tool for understanding protein activity and molecular mechanisms involved [
35].
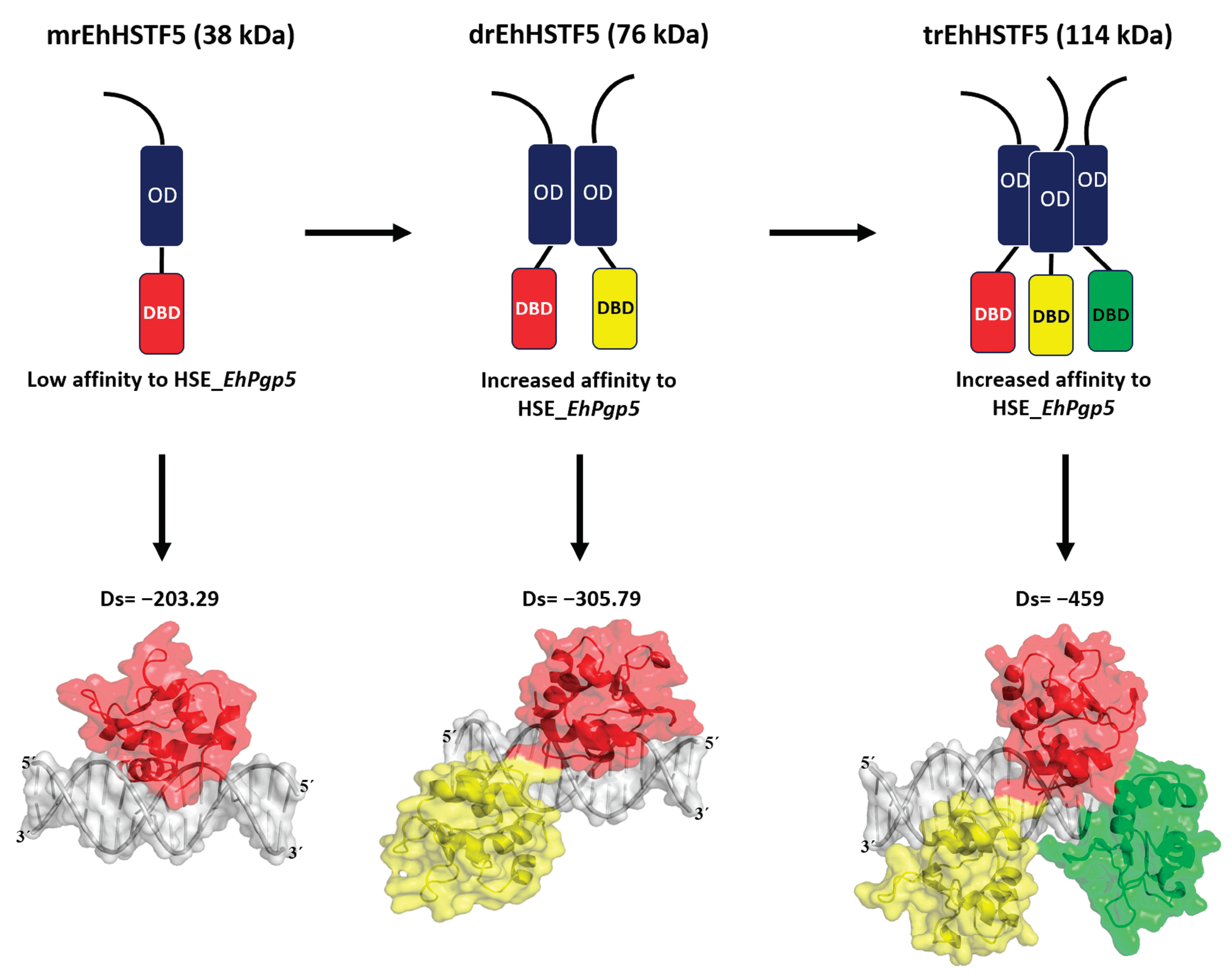
Through our in silico blind molecular docking studies, we identified that mEhDBD5 and dEhDBD5 exhibit binding to HSE_EhPgp5. Notably, dEhDBD5 shows significantly higher binding affinity (−305.79) compared to mEhDBD5 (−203.29).
We previously performed molecular docking under the same conditions as in this study, focusing on the trimeric conformation (tEhDBD5) of the EhHSTF5 protein and discovered its binding to the same HSE with a Docking score of −459 [
36]. This clearly indicates that oligomerization of EhHSTF5 into dimers and trimers progressively improves its stability and affinity towards HSE_
EhPgp5. This behavior has also been demonstrated in other factors such as KlHSTF [
37], HsHSTF1 [
38], and HsHSTF2 [
39], among others.
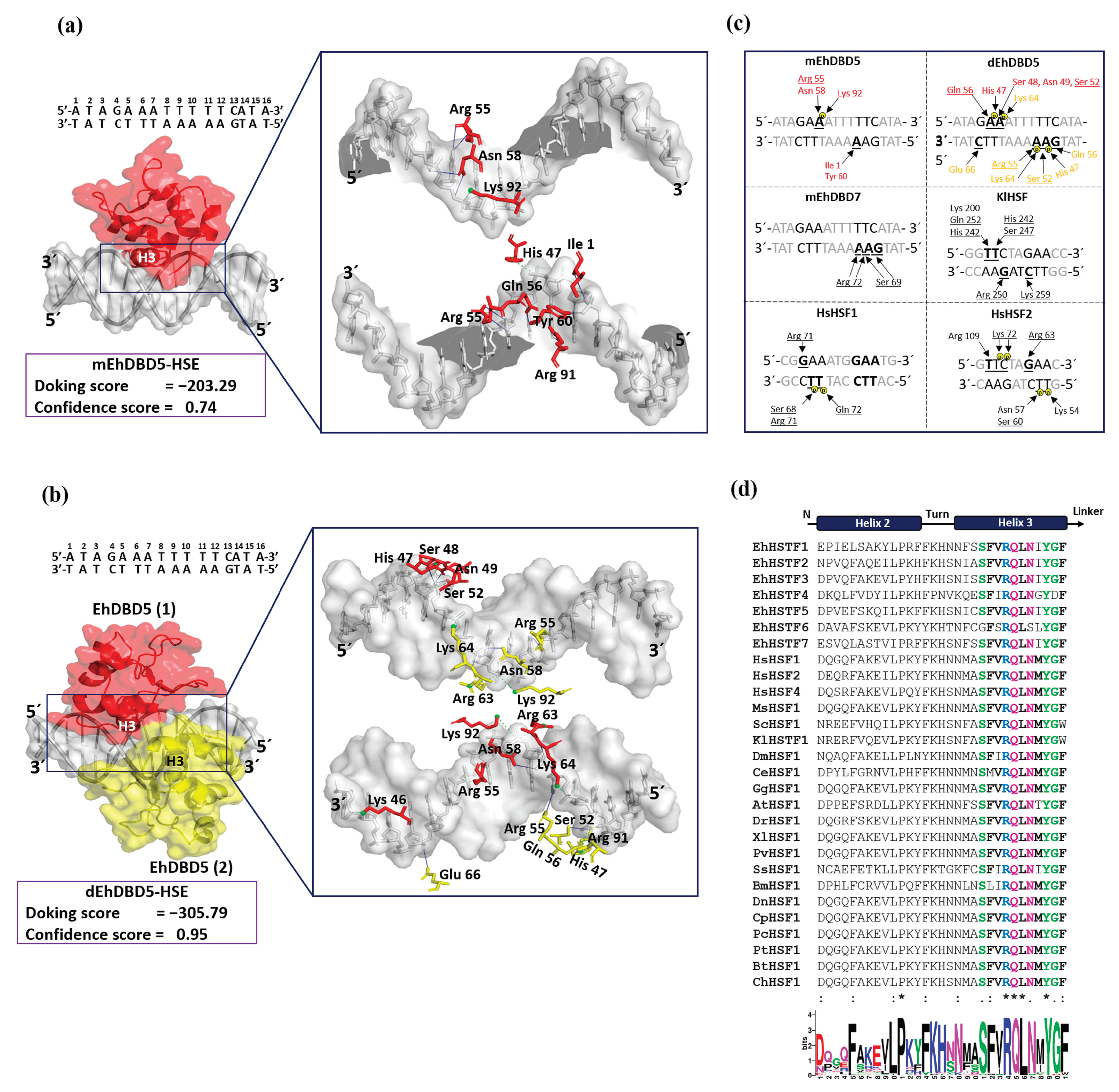
We identified that α3-helix plays a crucial role in the interaction of the DBD domain with the major groove of the HSE, especially in the structural motifs “GAA” and “TTC”, through hydrogen bonds and salt bridges. These results are in agreement with various crystallized complexes (HSTF-HSE) where it has been observed that α3-helix is responsible for direct contact with the major groove of the HSE. In the KlHSTF-HSE complex, for example, amino acids involved in recognizing the structural motifs 5´-TTC-3´, 3´-AAG-5´, and 3´-CTT-5´ are Arg 250, Ser 247, His 242, Gln 252, and Lys 259 [
37]. In the HsHSTF1-HSE crystallographic complex, it has been observed that Arg 71, Ser 68, and Gln 72 recognize the structural motifs 5´-GAA-3´ and 3´-CTT-5´ [
38], while in the HsHSTF2-HSE complex, Arg 109, Arg 63, Ser 60, and Lys 72 participate in recognizing the motifs 5´-TTC-3´, 5´-GAA-3´, and 3´-CTT-5´[
39].
Notably, during the analysis of sequences comprising α3-helix, including EhDBD5, high conservation of amino acids such as Ser, Arg, Gln, Asn, Tyr, and Gly were observed in several species, suggesting their significant relevance in HSE binding. In fact, it has been reported that mutation of amino acids Ser 247, Arg 250, and Asn 253 in α3-helix of
K. lactis leads to a lethal phenotype, further underlining the importance of these amino acids in HSTFs [
37].
An important aspect in the interaction between HSTFs and HSEs is that certain amino acids can interact with the phosphate group of the HSE backbone through salt bridges. This is the case for amino acids Ser 68, Arg 71, and Gln 72 in the HsHSTF1-HSE complex, and Lys 72, Asn 57, Ser 60, and Lys 53 in the HsHSTF2-HSE complex [
38]. This interaction enhances the stability of the DBD, facilitating correct binding of α3-helix to HSE. In our study, we observed a similar interaction mechanism mediated by salt bridges. We identified the presence of two salt bridges in the mEhDBD5_HSE_
EhPgp5 complex, and ten in the dEhDBD5_HSE_
EhPgp5 complex. Hydrogen bonds are also crucial for the stability of the complexes. Interestingly, we observed 12 intermolecular interactions in mEhDBD5_HSE_
EhPgp5 and 17 in dEhDBD5_HSE_
EhPgp5 (
Supplementary Table S1). This suggests that the dimeric conformation, by forming a higher number of salt bridges and hydrogen bonds during its binding to HSE, might be promoting greater stability compared to the monomeric conformation.
Consistently, upon evaluating the physicochemical properties, we observed that the dEhDBD5-HSE_
EhPgp5 complex, compared to the mEhDBD5-HSE_
EhPgp5 complex, exhibits more aromatic interactions, indicating a higher affinity at the binding interface. It also displays a stronger electric charge, suggesting more robust ionic interactions. Additionally, the complex is more hydrophobic, which enhances its structural stability. Notably, the ionization capacity implies diverse electrostatic interactions. The increased exposure to solvent suggests a more intricate structure and higher solubility. These features indicate a complex network of intramolecular and intermolecular interactions in the dimeric complex, potentially contributing to its greater stability compared to the monomeric conformation, as has been documented in numerous studies for different proteins [
40,
41,
42,
43]
A study that highlights a similar functional and structural behavior to rEhHSTF5 is the report by Bello et al. [
16] which revealed that the rEhHSTF7 factor can oligomerize in a concentration-dependent manner within total extracts of C41 bacteria, and this oligomerization mechanism was verified by αEhHSTF7 and α6xHis antibodies. In addition, successful purification of the protein in its oligomeric states has been achieved through nickel affinity chromatography. Analogously to the current study, EMSA and supershift assays have demonstrated that the oligomeric form of
E. histolytica rEhHSTF7 can recognize the HSE_
EhPgp5 gene. It is interesting to observe that through
in silico tools, it was confirmed that α3-helix also recognizes the 3´-AAG-5´ motif, primarily through the amino acids Arg 72 and Ser 69. Moreover, it has been shown that the expression of the
EhPgp5 gene decreases when a specific siRNA is used to knockdown the expression of the
Ehhstf7 gene in
E. histolytica trophozoites growth with 8 µM of Emetine, underlining the significance of transcriptional modulation by this factor under stress conditions. It is worth highlighting that the EhPGP5 protein of
E. histolytica is recognized as a multidrug resistance protein, acting as a pump to expel drugs, and enhancing amoebic survival when exposed to amoebicidal drugs. In this work, we observed that oligomeric rEhHSTF5 can recognize the HSE_
EhPgp5 gene similarly to rEhHSTF7, suggesting that both factors could collaborate in regulating the gene transcription of
EhPgp5 and, consequently, help the survival of the amoeba by responding to stressors present in its environment.
It is important to emphasize the capability of the rEhHSTF5 factor to recognize the structural motifs “GAA” and “TTC” within the HSE, which expands the potential for recognition not only of the HSE_
EhPgp5 gene [
16,
44], but also, under specific conditions, the HSEs present in other promoters regulated by this element, such as the seven HSEs identified in
EhrabB [
45], three in
Ehhsp100 [
46], and four in the
Ehmlbp gene promoter [
47]. These proteins are of great significance for the parasite, as EhRABB plays a pivotal role in cytoskeleton regulation, cellular motility, potential cyst formation, invasion, and parasitic pathogenicity [
45,
48,
49,
50,
51]. EhHSP100 is involved in amoeba response to heat and oxidative stress, protection against cell damage and modulation of pathogenicity [
46]. Furthermore, EhMLBP is a mRNA-binding protein known to interact specifically with the 3' untranslated region of mRNAs of various genes of the parasite, suggesting its potential role in posttranscriptional regulation and messenger RNA stability [
47,
52,
53,
54].
Recently, Dorantes et al. [
36] conducted an
in silico analysis of HSEs located in the promoter regions (ranging from −500 to +50 bp) of the 8,343 genes present in the
E. histolytica genome. This analysis revealed the presence of 2578 HSEs in total, of which 1412 HSEs were in the promoter regions of 1010 hypothetical genes, and 1,166 HSEs in the promoter regions of 957 coding genes. Remarkably, 24% of the genes could potentially be regulated by HSEs. Furthermore, it was observed that these HSEs are situated in promoters of genes associated with various functions, including ATP-dependent activity, binding, catalytic activity, cytoskeletal motor activity, molecular adaptor activity, molecular function regulator, molecular transducer activity, structural molecule activity, transcription regulator activity, translation regulator activity, and transporter activity, among others.
These findings further underscore the functional importance of the EhHSTF5 factor as a potential regulator of various genes that harbor HSEs in their promoters in E. histolytica. This ability could allow the parasite to dynamically adapt to its environment, thereby ensuring its survival across diverse physiological conditions.
Current in vitro experiments are underway to evaluate the significance of the “GAA” and “TTC” motifs. Our approach involves targeted manipulation and mutation of these nucleotides within the HSE_EhPgp5 sequence to understand their impact on binding affinity with rEhHSTF5. Furthermore, conducting assays on E. histolytica trophozoites will deepen our comprehension of how oligomerization affects the affinity of EhHSTF5 for various HSEs, particularly under stress conditions. Simultaneously, we will proceed to evaluate the relevance of EhHSTF5 in the parasite by specifically blocking its expression through siRNAs.
In summary, as illustrated in
Figure 9, our findings indicate that the mrEhHSTF5 protein, with a molecular weight of 38 kDa, can form dimeric (76 kDa) and trimeric (114 kDa) structures. This oligomerization is crucial, as it increases the stability of the protein, which translates into a higher affinity for HSE_
EhPgp5.
4. Materials and Methods
4.1. Cloning of the Ehhstf5 Gene and Expression of the rEhHSTF5 Protein
The Ehhstf5 gene was cloned into the pET-28a(+) plasmid by GenScript (Piscataway, NJ, USA). The gene was flanked by the restriction sites of the BamHI and XhoI enzymes at the 5' and 3' ends, respectively. Additionally, a 6-histidine tag was incorporated at the 5' end of the plasmid. The insertion of the Ehhstf5 gene was confirmed through gene amplification. The forward primer (5'-ATCCGATCGGATGAGTGAACCACACAAACA-3') and reverse primer (5'-CGACTCGACTTTAAAACTTCCATGGAATTT-3') oligonucleotides were employed. The amplification process started with an initial denaturation step at 94°C for 5 min, followed by 30 cycles. Each cycle involved denaturation at 94°C for 1 min, primer annealing at 49.4°C for 1 min and 30 s, and extension of the new strand at 72°C for 1 min. Following the cycles, there was an incubation at 72°C for 7 min to conclude the amplification process. The resulting PCR product was sequenced using the ABI PRISM 3130 sequencer (Thermo Fisher Scientific, Waltham, MA, USA).
The constructed plasmid pET-28a(+)-
Ehhstf5 was used to transform competent cells of
E. coli C41 strain (Sigma-Aldrich, San Luis, MO, USA). Expression of the recombinant rEhHSTF5 protein was induced using 1 mM Isopropyl-beta-D-thiogalactopyranoside (IPTG) (Thermo Fisher Scientific, Waltham, MA, USA) with agitation at 220 rpm in SOC medium for 24 h at 37°C. The progress of recombinant protein induction was observed at 3, 6, 9, and 24 h. Resulting bacterial cells were resuspended in lysis buffer (20 mM Tris-HCl, pH 8, 1 mg/ml lysozyme, and 1% Triton X-100) and then lysed at 4°C by sonication using an Ultrasonic Processor equipment (MRC, laboratory instruments, Harlow, ESX, UK) at 60% amplitude. Five cycles of 15 s were applied with 15 s rest intervals. The obtained soluble proteins were quantified using the Bradford method [
55]. Integrity and expression of the rEhHSTF5 protein were assessed through 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using 20 μg of protein.
4.2. Immunodetection of rEhHSTF5 Protein by Western Blotting
The proteins separated by electrophoresis were transferred to PVDF membranes with 0.45 μm pores (Sigma-Aldrich, Basel, Switzerland). The membranes were incubated for 30 min with the primary antibody anti-6His (α6xHis) (Sigma-Aldrich, Basel, Switzerland) at a dilution of 1:3000, or with a polyclonal antibody anti-EhHSTF5 (αEhHSTF5) at a dilution of 1:1000, generated in Balb/c mice by SPIDDSNNVELP (GLBiochem, Shanghai, China) EhHSTF5 peptide immunization. The membranes were subsequently incubated for 30 min with the secondary antibody goat anti-mouse IgG (H+L) conjugated to horseradish peroxidase (Thermo Fisher Scientific, Waltham, MA, USA) at a dilution of 1:20,000. Detection was performed through chemiluminescence using Immobilon™ Western chemiluminescent HRP substrate (Millipore, Burlington, MA, USA).
4.3. Purification of Recombinant Protein rEhHSTF5
Bacteria containing the overexpressed rEhHSTF5 protein were suspended in buffer I (20 mM Tris-HCl, 500 mM NaCl, and 25 mM Imidazole, pH 8.5) and lysed by sonication. The soluble fraction was obtained through centrifugation at 20,000 xg for 45 min at 4°C, followed by filtration using 0.22 μm pore size filters. The purification of the recombinant protein was conducted through nickel affinity chromatography using the ÄKTA pure™ chromatography system (Cytiva, Marlborough, MA, USA). Briefly, the protein lysate was loaded onto a 5 mL HisTrap FF column (Cytiva, Marlborough, MA, USA), which was subsequently washed with 5 column volumes (25 ml) of buffer I, and the protein was eluted from the column with buffer II (20 mM Tris-HCl, 500 mM NaCl, 500 mM Imidazole, pH 8.5). The purification of the rEhHSTF5 protein was evaluated by Western Blot following the methodology described previously.
4.4. Oligomerization States of the mrEhHSTF5 Protein
The oligomerization of the monomeric rEhHSTF5 protein (mrEhHSTF5) was assessed by incubating 20 µg of purified protein at 25°C in the presence of glutaraldehyde (Sigma-Aldrich, St. Louis, MO, USA) at concentrations of 0.5%, 1.5%, and 2.5% for 15 min. The oligomerization reaction was stopped by adding 10 µl of a loading buffer (2X) containing 100 mM Tris-Cl at pH 6.8, 4% SDS (sodium dodecyl sulfate; electrophoresis-grade), 0.2% bromophenol blue, 20% glycerol, 200 mM DTT (dithiothreitol), and 200 mM β-mercaptoethanol. Oligomerization states were evaluated through 12% SDS-PAGE gel electrophoresis.
4.5. Biotinylation and Hybridization of the HSE_EhPgp5 Sequence
The sense (5'-ATAGAAATTTTTCATA-3') and antisense (3'-TATCTTTAAAAAGTAT-5') oligonucleotides of the HSE_EhPgp5 were synthesized at Integrated DNA Technologies (IDT) (Coralville, IA, USA). The 3' end of the single-stranded oligonucleotides was biotinylated using the Biotin 3' End DNA Labeling Kit (Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer's instructions. Subsequently, the oligonucleotides were hybridized in equimolar ratios in an Axygen-Maxigene thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA) at 95°C for 5 min, followed by sequential descending incubations from 10°C to 25°C, holding each temperature for one min.
4.6. Electrophoretic Mobility Shift Assays (EMSA)
The interaction between the rEhHSTF5 factor and HSE_EhPgp5 was evaluated using 0.7 μM of monomeric protein (mrEhHSTF5) or dimeric protein (drEhHSTF5). The proteins were incubated for 30 min at room temperature with the components of the Lightshift EMSA kit (Thermo Fisher Scientific, Waltham, MA, USA), including 2 µl of binding buffer, 1 µl of Poly (dI-dC) (50 ng/µl), 1 µl of glycerol (50%), 1 µl of MgCl2 (100 mM), and 300 µg of BSA. For competition assays, a non-biotinylated probe (specific competitor) or Poly (dI-dC) was used as an unspecific competitor at molar excesses of 150 and 350 times, respectively. Prior to incubation with the binding mixture and the biotinylated probe, competitors were preincubated for 30 min at 4°C with mrEhHSTF5 and drEhHSTF5. The samples were subjected to electrophoresis on non-denaturing 0.5X-TBE polyacrylamide gels and then transferred to PVDF membranes (Sigma-Aldrich, Basel, Switzerland). The formation of the rEhHSTF5-HSE complex was detected through chemiluminescence using the provided kit solution.
4.7. Electrophoretic Mobility Supershift Assay
The αEhHSTF5 antibody was preincubated at a 1:3000 dilution with mrEhHSTF5 or drEhHSTF5 protein for 30 min. This was followed by another 30 min incubation with the binding mixture and biotinylated probe under the same conditions. Preimmune serum (serum from mice not immunized with the immunogenic peptide of EhHSTF5) or an unrelated commercial antibody (αGAPDH, 1:3000) were used as controls. The procedure for detecting complexes was carried out following the EMSA conditions described previously. Furthermore, a kinetics study involving an increase in drEhHSTF5 concentration (from 0.7 to 2.8 µM) was conducted to confirm the consistency and correspondence of the formed complexes with the drEhHSTF5-HSE_EhPgp5 complex.
4.8. Three-Dimensional Modeling of the HSE_EhPgp5
We utilized UCSF Chimera version 1.15 software (San Francisco, CA, USA) to construct the three-dimensional model of HSE_EhPgp5, using the sense sequence 5'-ATAGAAATTTTTCATA-3' and antisense sequence 3'-TATCTTTAAAAAGTAT-5'. The resulting double-stranded structure in conformation b was saved in .pdb format for subsequent bioinformatics analysis.
4.9. 3D Modeling, Structural Validation, and Stability Analysis of mEhDBD5 and dEhDBD5
The amino acid sequence 46-137, corresponding to the DNA-binding domain (DBD) of the EhHSTF5 factor (EhDBD5), was obtained from the AmoebaDB platform (
https://amoebadb.org/amoeba/app, accessed on 2 November 2023) with the ID EHI_137000 to generate the 3D models of the DNA-binding domain in monomeric (mEhDBD5) and dimeric (dEhDBD5) conformations. The SWISS-MODEL server (
https://swissmodel.expasy.org/, accessed on 2 November 2023) was utilized to model the 3D structures of mEhDBD5 and dEhDBD5, using the DBD of human HSTF2 in complex with HSE as a template, deposited with the ID 5D8L in the Protein Data Bank (PDB) (
https://www.rcsb.org, accessed on 2 November 2023). The obtained models were structurally validated using various servers, including PDBsum for Ramachandran analysis (
http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=index.html, accessed on 5 November 2023), ProSA-web (
https://prosa.services.came.sbg.ac.at/prosa.php, accessed on 10 November 2023), ERRAT and VERIFY 3D contained in UCLA-DOE LAB-SAVES v6.0 (
https://saves.mbi.ucla.edu, accessed on 8 December 2023), and VADAR (
http://vadar.wishartlab.com/index.html?, accessed on 10 December 2023). Additionally, the structural stability of mEhDBD5 was predicted using the normalized B-factor or temperature factor calculated by the I-Tasser server (
https://zhanggroup.org/I-TASSER/, accessed on 15 December 2023).
4.10. In Silico Molecular Docking
Blind molecular docking simulations were conducted using the HDOCK server (
http://hdock.phys.hust.edu.cn/, accessed on 20 December 2023) to assess the intermolecular interactions between HSE_
EhPgp5 and the mEhDBD5 and dEhDBD5 domains of the EhHSTF5 factor. During this process, the .pdb file corresponding to HSE_
EhPgp5 was used as ligand, while the mEhDBD5 and dEhDBD5 domains were used as receptors. The selection of dockings was based on the most negative values obtained in the Docking Score, representing binding affinity. Additionally, selections were made based on the Confidence score.
4.11. Analysis of the Physicochemical Properties of the mEhDBD5-HSE_EhPgp5 and dEhDBD5-HSE_EhPgp5 Complexes
The obtained complexes, mEhDBD5-HSE_EhPgp5 and dEhDBD5-HSE_EhPgp5, were subjected to a comprehensive analysis of physicochemical properties. This analysis covered Aromatics, H-bonds, Charge, Hydrophobicity, Ionizability, and Solvent Accessible Surface (SAS) and was conducted using BIOVIA Discovery Studio, version v.21 (San Diego, CA, USA).
4.12. In Silico Analysis of Intermolecular Interactions in the EhDBD5-HSE_EhPgp5 Complex
To identify the intermolecular interactions within the mEhDBD5-HSE_
EhPgp5 and dEhDBD5-HSE_
EhPgp5 complexes, the Protein-Ligand Interaction Profiler (PLIP) server (
https://plip-tool.biotec.tu-dresden.de/plip-web/plip/index, accessed on 29 December 2023) was used. Interactions were assessed with a bond length cutoff of 4 Å, following the recommendations of the server.
4.13. Structural Similarity of 3D Proteins
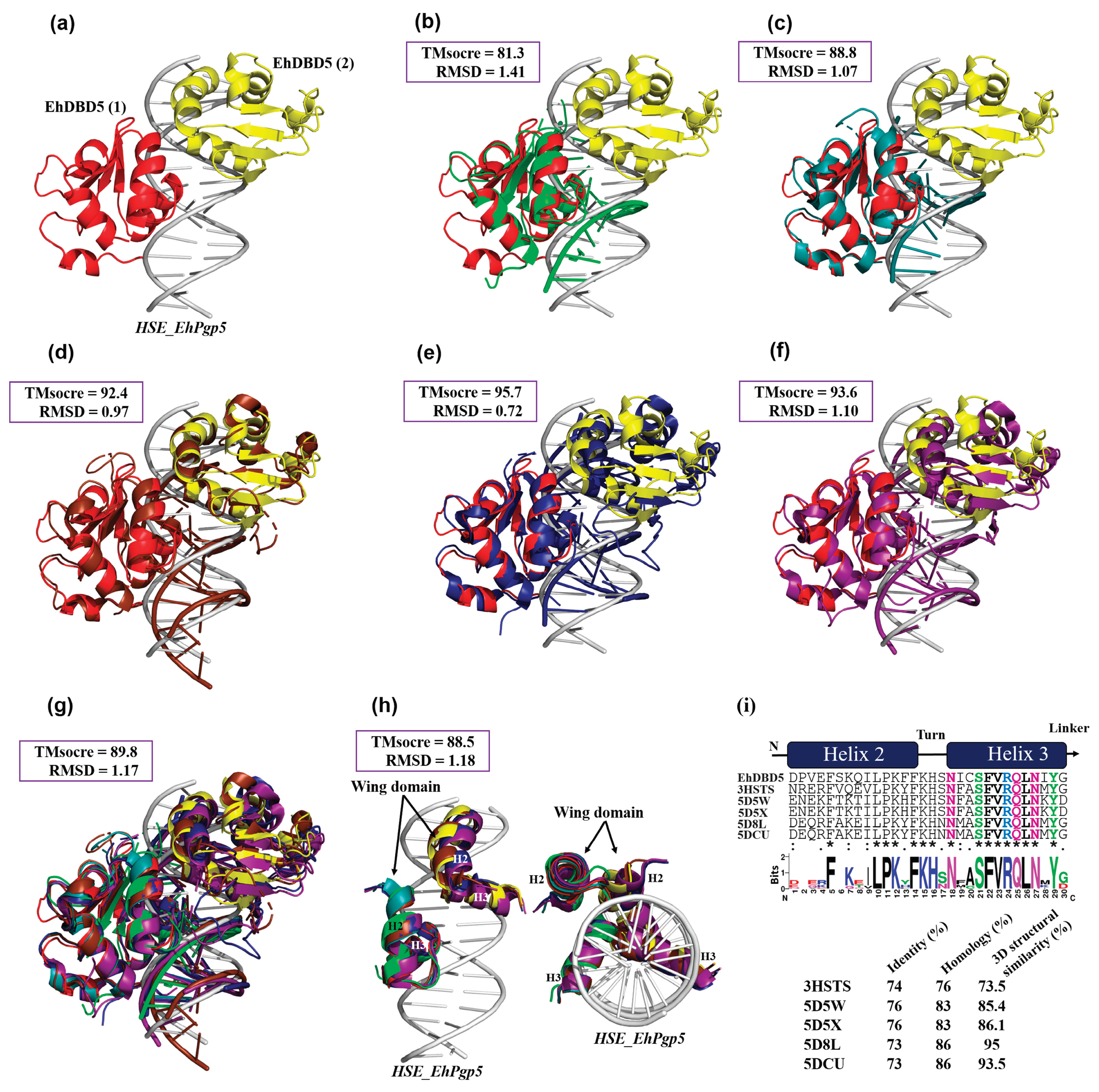
The RaptorX server (
http://raptorx6.uchicago.edu/, accessed on 2 January 2024) was used to perform 3D structure superposition and obtain structural similarity and root mean square error (RMSD). Among the structures deposited on the PDB platform used for the superposition, the DBD of HSTF in complex with an HSE was used, including that of
Kluyveromyces lactis (
K. lactis) (ID; 3HTS),
Saccharomyces cerevisiae-
Chaetomium thermophilum (
S. cerevisiae-
C. thermophilum, chimera) (ID; 5D5X),
Homo sapiens-
Chaetomium thermophilum (
H. sapiens-C.
thermophilum, chimera) (ID; 5D5W),
H. sapiens (ID; 7DCU) and the synthetic
H. sapiens construct (ID; 5D8L). We were also able to obtain only DBD α2-helix and α3-helix in .pdb format from these structures, which were used to calculate the percentage of structural similarity.
4.14. Conservation of DBD α2-Helix and α3-Helix
To evaluate the conservation degree of α-helices 2 and 3 within the DBDs of HS
TFs, an amino acid sequence analysis was conducted using the Clustal Omega platform (
https://www.ebi.ac.uk/jdispatcher/msa/clustalo, accessed on 6 November 2023). This analysis included the sequences of the seven EhHSTFs of
E. histolytica previously mentioned, as well as sequences obtained from the National Center for Biotechnology Information platform (NCBI) (
https://www.ncbi.nlm.nih.gov/, accessed on 6 November 2023) of HSTF1, HSTF2 and HSTF4 from
H. sapiens (HsHSTF1, 2 and 4 , IDs: 3297, 3298 and 3299), as well as sequences from
Mus musculus (MmHSTF1, ID: 15499),
S. cerevisiae (ScHSTF1, ID: 1322586),
K. lactis (KlHSTF1, ID: 1751387658),
Drosophila melanogaster (DmHSTF1, ID: 37068),
Caenorhabditis elegans (CeHSTF1, ID: 45643032),
Gallus gallus (GgHSTF1, ID: 76523668686),
Arabidopsis thaliana (AtHSTF1, ID: 827496),
Danio rerio (DrHSTF1, ID: 134026298),
Xenopus laevis (XlHSTF1, ID: 148222464
), Pteropus vampyrus (PvHSTF1, ID: 759162997),
Sus scrofa (SsHSTF1, ID: 100511321),
Brugia malayi (BmHSTF1, ID: CRZ24125. 1),
Dasypus novemcinctus (DnHSTF1, ID: 101424131),
Cavia porcellus (CpHSTF1, ID: 101787582),
Phascolarctos cinereus (PcHSTF1, ID: 1190400766),
Pan troglodytes (PtHSTF1, ID: 741396),
Bos taurus (BtHSTF1, ID: 506235) and
Capra hircus (ChHSTF1, ID: 102178552).
Simultaneously, amino acid sequences were extracted from the crystals deposited in the PDB database for K. lactis (ID: 3HTS), S. cerevisiae-C. thermophilum (ID: 5D5X), H. sapiens-C. thermophilum (ID: 5D5W), synthetic H. sapiens construct (ID: 5D8L), and H. sapiens (ID: 7DCU) to perform a sequence alignment and determine the percentage of identity and homology.
Complementarily, logos were constructed using the amino acids from the aforementioned sequences through the WebLogo platform (
https://weblogo.berkeley.edu/logo.cgi, accessed on 7 November 2023).
4.15. 3D Structure Viewer
PyMol, version 2.5.0, (Nueva York, NY, USA) and BIOVIA Discovery Studio were used to visualize and generate images for different bioinformatics analyses.
4.16. Relative Expression and Graphics
The ImageJ software, version 1.54, (Bethesda, MD, USA) was used to quantify the relative protein expression through pixel analysis. For the statistical analysis, a two-way ANOVA (Analysis of Variance) was utilized, followed by Tukey's multiple comparison test, in accordance with the American Psychological Association (APA) format for adjusted p-value analysis, detailed as follows: *p ≤ 0.033, **p ≤ 0.002, and ***p ≤ 0.001. The data are expressed as the mean ± standard deviation, derived from a minimum of three independent experiments. For graph creation, the GraphPad Prism software, version 8.0.1 (Boston, MA, USA), was used, allowing for a precise and detailed graphical representation of the results.
Author Contributions
Conceptualization, S.P.-M., M.d.C.G.-G, and D.G.P.-I.; methodology, S.P.-M., M.d.C.G.-G., D.G.P.-I., S.V.S-H., M.O.M-F., C.A.R-L., M.A. R., and V. S-M.; software, S.P.-M.; S.V.S-H., and C.A.R-L.; validation, D.G.P.-I., and M.d.C.G.-G.; formal analysis, S.P.-M., D.G.P.-I., M.d.C.G.-G., C.A.R-L., and M.A. R.; investigation, S.P.-M., M.d.C.G.-G., and D.G.P.-I.; resources, M.d.C.G.-G., and D.G.P.-I.; writing—original draft preparation, S.P.-M.; writing—review and editing, S.P.-M., M.d.C.G.-G., D.G.P.-I., C.A.R-L., and M.A. R.; visualization, S.P.-M., S.V.S-H., M.d.C.G.-G., D.G.P.-I., C.A.R-L., and M.A. R.; supervision, M.d.C.G.-G., and D.G.P.-I.; project administration, M.d.C.G.-G., D.G.P.-I., and S.P.-M.; funding acquisition, M.d.C.G.-G., and D.G.P.-I. All authors have read and agreed to the published version of the manuscript.
Figure 1.
Induction and immunodetection of the oligomeric states of the rEhHSTF5 protein. The integrity of total proteins extracted from transformed and IPTG-induced bacteria was evaluated by SDS-PAGE. A Molecular Weight Marker (MWM) and IPTG induction times ranging from 0 to 24 h were included. The gel was stained with Coomassie Brilliant Blue G-250 to visualize protein bands (a). Subsequently, immunodetection was performed by Western blot using α6xHis and αEhHSTF5 specific antibodies to detect the distinct conformations of rEhHSTF5 (b). The graphs display the relative protein expression, calculated as the average of pixel values obtained from three independent samples. Expression was normalized using the endogenous control, GAPDH. The statistical analysis was performed using a two-way ANOVA, followed by the Tukey's multiple comparison test. Significant differences between experimental groups are denoted with (*). The significance levels used were **p≤ 0.002, and ***p≤ 0.001.
Figure 1.
Induction and immunodetection of the oligomeric states of the rEhHSTF5 protein. The integrity of total proteins extracted from transformed and IPTG-induced bacteria was evaluated by SDS-PAGE. A Molecular Weight Marker (MWM) and IPTG induction times ranging from 0 to 24 h were included. The gel was stained with Coomassie Brilliant Blue G-250 to visualize protein bands (a). Subsequently, immunodetection was performed by Western blot using α6xHis and αEhHSTF5 specific antibodies to detect the distinct conformations of rEhHSTF5 (b). The graphs display the relative protein expression, calculated as the average of pixel values obtained from three independent samples. Expression was normalized using the endogenous control, GAPDH. The statistical analysis was performed using a two-way ANOVA, followed by the Tukey's multiple comparison test. Significant differences between experimental groups are denoted with (*). The significance levels used were **p≤ 0.002, and ***p≤ 0.001.
Figure 2.
Purification and immunodetection of oligomeric conformations of the rEhHSTF5 protein. The oligomeric conformations of the rEhHSTF5 protein were monitored before and after loading onto the HisTrap FF column. The total proteins obtained from bacteria transformed with the plasmid construct and induced with IPTG (24 h) were passed through filters with a pore size of 0.22 μm (flowthrough, FT), and the proteins that did not bind to the column (waste, W) are shown (a). The chromatogram and SDS-PAGE assays revealed that the drEhHSTF5 protein was eluted in fractions 7 to 12, while the mrEhHSTF5 protein was eluted in fractions 24 to 26 (b-d). These proteins were detected using α6xHis and αEhHSTF5 antibodies in WB assays (e, f). In the chromatogram (b), the stepped black lines represent the absorbance reading at 280 nm during the elution time, indicating the presence of proteins being eluted from the affinity column. Panel (c) to (f), “F” denotes the fraction number obtained from the chromatography, and “MWM” indicates molecular weight marker.
Figure 2.
Purification and immunodetection of oligomeric conformations of the rEhHSTF5 protein. The oligomeric conformations of the rEhHSTF5 protein were monitored before and after loading onto the HisTrap FF column. The total proteins obtained from bacteria transformed with the plasmid construct and induced with IPTG (24 h) were passed through filters with a pore size of 0.22 μm (flowthrough, FT), and the proteins that did not bind to the column (waste, W) are shown (a). The chromatogram and SDS-PAGE assays revealed that the drEhHSTF5 protein was eluted in fractions 7 to 12, while the mrEhHSTF5 protein was eluted in fractions 24 to 26 (b-d). These proteins were detected using α6xHis and αEhHSTF5 antibodies in WB assays (e, f). In the chromatogram (b), the stepped black lines represent the absorbance reading at 280 nm during the elution time, indicating the presence of proteins being eluted from the affinity column. Panel (c) to (f), “F” denotes the fraction number obtained from the chromatography, and “MWM” indicates molecular weight marker.
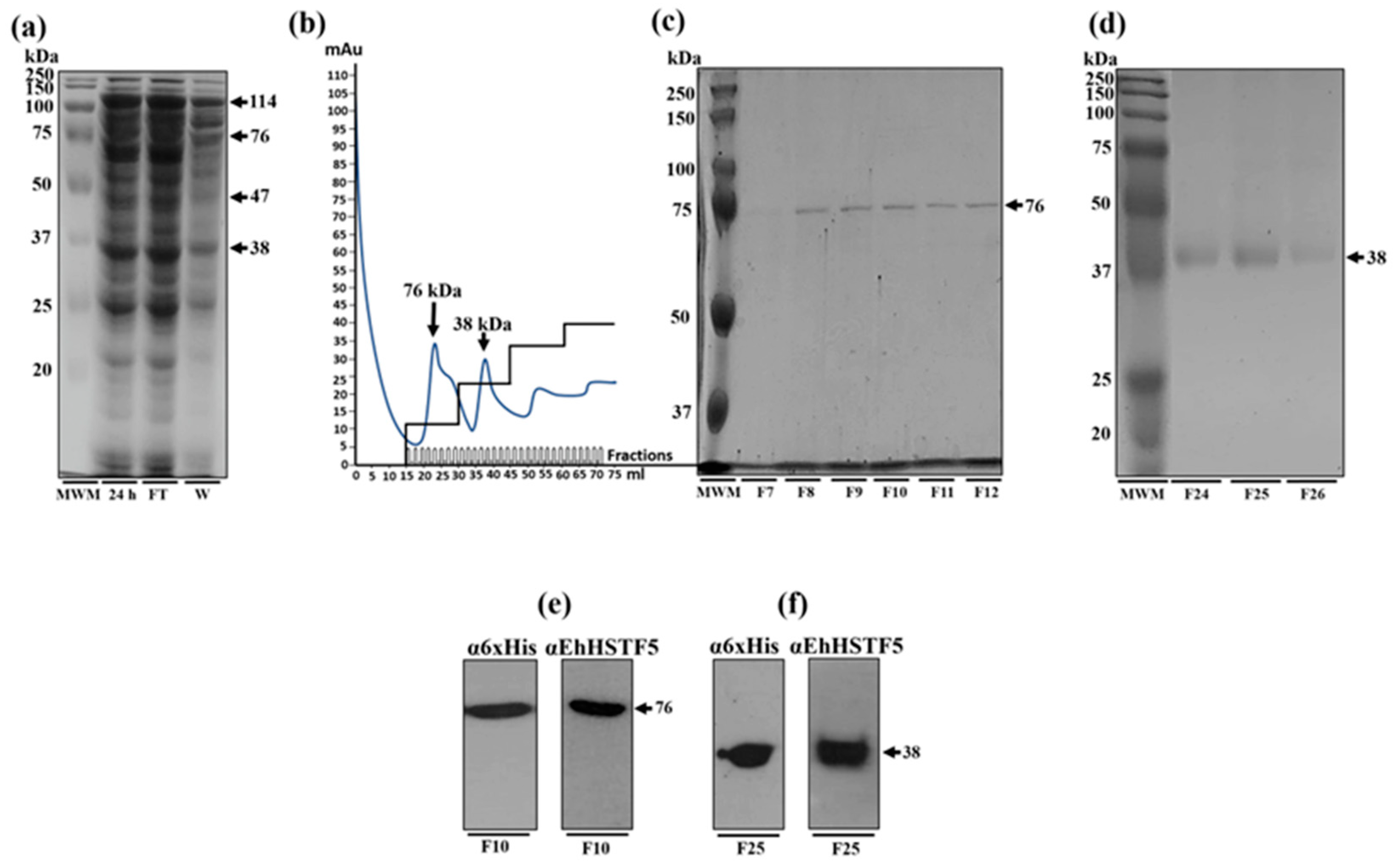
Figure 3.
mrEhHSTF5 oligomerization induction with increasing concentrations of glutaraldehyde. Oligomerization states were evaluated by 12% SDS-PAGE gel electrophoresis stained with Coomassie Brilliant Blue G-250. The statistical analysis was performed using a two-way ANOVA, followed by the Tukey's multiple comparison test. Significant differences between experimental groups are denoted with (*). The significance levels used were *p≤ 0.033, **p≤ 0.002, and ***p≤ 0.001.
Figure 3.
mrEhHSTF5 oligomerization induction with increasing concentrations of glutaraldehyde. Oligomerization states were evaluated by 12% SDS-PAGE gel electrophoresis stained with Coomassie Brilliant Blue G-250. The statistical analysis was performed using a two-way ANOVA, followed by the Tukey's multiple comparison test. Significant differences between experimental groups are denoted with (*). The significance levels used were *p≤ 0.033, **p≤ 0.002, and ***p≤ 0.001.
Figure 4.
HSE recognition of the EhPgp5 gene by the rEhHSTF5 protein. EMSA assays was performed using the monomeric and dimeric conformations of the rEhHSTF5 protein. A specific competitor (non-biotinylated HSE_EhPgp5) and a nonspecific competitor (Poly(dI-dC)) were used (a). For the supershift assays, the specific antibody αEhHSTF5 was utilized for both conformations of the rEhHSTF5 protein. Unrelated antibodies, including mouse preimmune serum (PS) and αGAPDH antibody, were also used as negative controls (b). Likewise, a kinetic study with increasing drEhHSTF5 concentration (from 0.7 to 2.8 µM) drEhHSTF5 was performed in supershift assays (c). In the image legend, the plus sign (+) indicates the presence of a component in the reaction, while the minus sign (−) denotes its absence. A graphical illustration of each condition used in the EMSA and supershift tests is provided at the bottom. .
Figure 4.
HSE recognition of the EhPgp5 gene by the rEhHSTF5 protein. EMSA assays was performed using the monomeric and dimeric conformations of the rEhHSTF5 protein. A specific competitor (non-biotinylated HSE_EhPgp5) and a nonspecific competitor (Poly(dI-dC)) were used (a). For the supershift assays, the specific antibody αEhHSTF5 was utilized for both conformations of the rEhHSTF5 protein. Unrelated antibodies, including mouse preimmune serum (PS) and αGAPDH antibody, were also used as negative controls (b). Likewise, a kinetic study with increasing drEhHSTF5 concentration (from 0.7 to 2.8 µM) drEhHSTF5 was performed in supershift assays (c). In the image legend, the plus sign (+) indicates the presence of a component in the reaction, while the minus sign (−) denotes its absence. A graphical illustration of each condition used in the EMSA and supershift tests is provided at the bottom. .
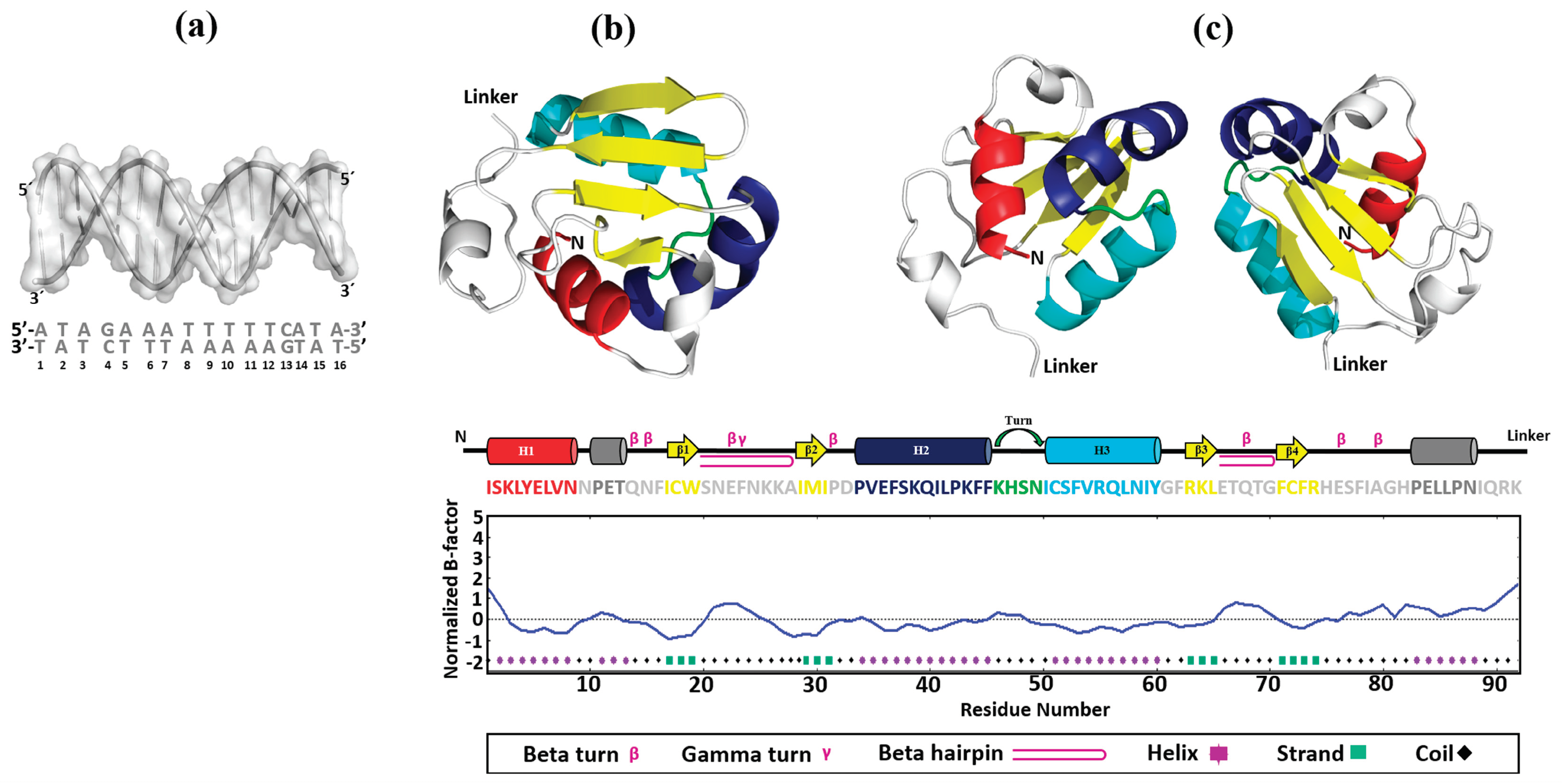
Figure 5.
Construction of 3D models. The 3D structures of HSE_EhPgp5 (a), mEhDBD5 (b), and dEhDBD5 (c) were obtained. At the bottom of the EhDBDs, the secondary structure of a monomer is displayed using a color code, while the graph illustrates its stability, assessed in silico through the normalized B-factor.
Figure 5.
Construction of 3D models. The 3D structures of HSE_EhPgp5 (a), mEhDBD5 (b), and dEhDBD5 (c) were obtained. At the bottom of the EhDBDs, the secondary structure of a monomer is displayed using a color code, while the graph illustrates its stability, assessed in silico through the normalized B-factor.
Figure 6.
Validation of 3D structures. The 3D models mEhDBD5 (a) and dEhDBD5 (b) were validated using Ramachandran analysis, which provides detailed information about the distribution of dihedral angles of amino acids in favorable and unfavorable regions. The capital letters A, B, and L are used to designate the Most Favoured Regions, while the lowercase letters a, b, l, and p represent the Additional Allowed Regions. The tilde (~a, ~b, ~l, ~p) is utilized to indicate the Generously Allowed Regions. The label 'XX' is employed for the Disallowed Regions. Furthermore, a value of −1 is assigned to a torsion angle located within a non-permissible region. The models were subjected to the ProSA-web server for a validation based on Z-score and global energy, where values ≤0 indicate absence of structural errors. ERRAT was used for error quantification, where an asterisk (*) indicates the error value. White bars in the graph represent instances without structural errors, while yellow bars indicate amino acids with structural errors falling within the 95-99% error range, and red bars identify errors exceeding 99%. Verify 3D validates protein structures by evaluating their 3D conformation through comparison with a set of experimental structures. It assigns a score, suggesting an acceptable average 3D−1D score of ≥ 0.1 for validation.
Figure 6.
Validation of 3D structures. The 3D models mEhDBD5 (a) and dEhDBD5 (b) were validated using Ramachandran analysis, which provides detailed information about the distribution of dihedral angles of amino acids in favorable and unfavorable regions. The capital letters A, B, and L are used to designate the Most Favoured Regions, while the lowercase letters a, b, l, and p represent the Additional Allowed Regions. The tilde (~a, ~b, ~l, ~p) is utilized to indicate the Generously Allowed Regions. The label 'XX' is employed for the Disallowed Regions. Furthermore, a value of −1 is assigned to a torsion angle located within a non-permissible region. The models were subjected to the ProSA-web server for a validation based on Z-score and global energy, where values ≤0 indicate absence of structural errors. ERRAT was used for error quantification, where an asterisk (*) indicates the error value. White bars in the graph represent instances without structural errors, while yellow bars indicate amino acids with structural errors falling within the 95-99% error range, and red bars identify errors exceeding 99%. Verify 3D validates protein structures by evaluating their 3D conformation through comparison with a set of experimental structures. It assigns a score, suggesting an acceptable average 3D−1D score of ≥ 0.1 for validation.
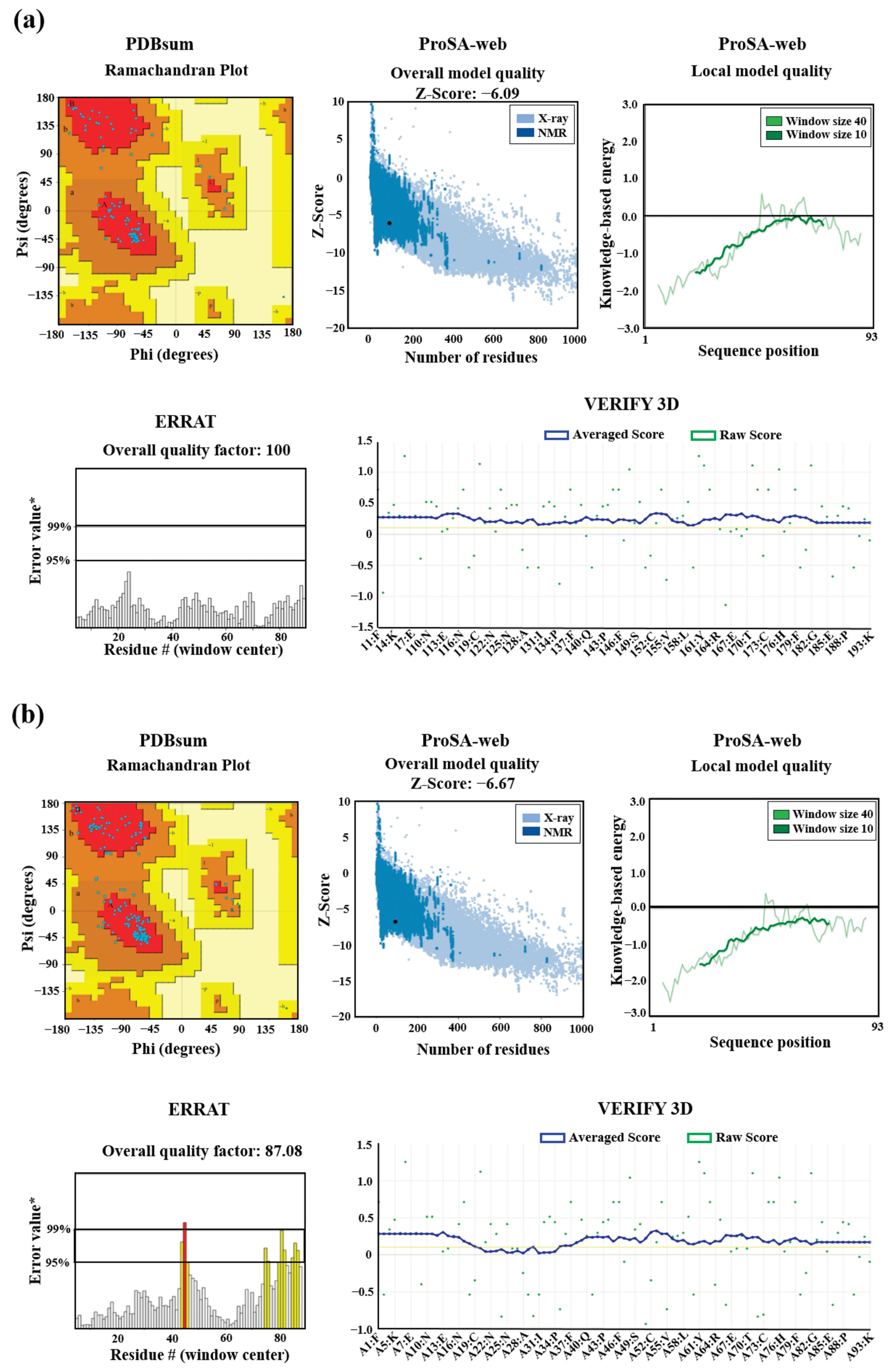
Figure 7.
Molecular docking and intermolecular evaluation of the mEhDBD5-HSE_EhPgp5 and dEhDBD5-HSE_ EhPgp5 complexes. Blind dockings were performed with the domains mEhDBD5 or dEhDBD5 and the HSE_EhPgp5. Using the PLIP server, intermolecular interactions of amino acids involved in the interaction with the sense and antisense strands of HSE were estimated. A close-up of the area of interaction is shown in the boxes, where amino acids from mEhDBD5 (EhDBD5(1)) are illustrated in red, while the analogue of the second EhDBD5 domain from dEhDBD5 (EhDBD5(2)) is shown in yellow (a) and (b). The amino acids involved in the interaction with the amino acids of the mEhDBD7-HSE, KlHSTF-HSE, HsHSTF1-HSE, and HsHSTF2-HSE complexes were compared (c). The high conservation degree of α2-helix and α3-helix of DBDs from 28 species, including the EhHSTFs family, is shown (d). .
Figure 7.
Molecular docking and intermolecular evaluation of the mEhDBD5-HSE_EhPgp5 and dEhDBD5-HSE_ EhPgp5 complexes. Blind dockings were performed with the domains mEhDBD5 or dEhDBD5 and the HSE_EhPgp5. Using the PLIP server, intermolecular interactions of amino acids involved in the interaction with the sense and antisense strands of HSE were estimated. A close-up of the area of interaction is shown in the boxes, where amino acids from mEhDBD5 (EhDBD5(1)) are illustrated in red, while the analogue of the second EhDBD5 domain from dEhDBD5 (EhDBD5(2)) is shown in yellow (a) and (b). The amino acids involved in the interaction with the amino acids of the mEhDBD7-HSE, KlHSTF-HSE, HsHSTF1-HSE, and HsHSTF2-HSE complexes were compared (c). The high conservation degree of α2-helix and α3-helix of DBDs from 28 species, including the EhHSTFs family, is shown (d). .
Figure 8.
3D structural similarity between DBDs-HSEs complexes. Based on the obtained dEhDBD5-HSE_EhPgp5 complex (a), overlay comparisons were performed with crystallographic structures deposited in the PDB database. These included the K. lactis DBD-HSE complex (ID: 3HTS) (b), S. cerevisiae-C. thermophilum complex (ID: 5D5X) (c), H. sapiens-C. thermophilum complex (ID: 5D5W) (d), synthetic H. sapiens construction (ID: 5D8L) (e), and H. sapiens (ID: 7DCU) (f). A joint overlay of all these structures was performed (g) and focusing solely on the wing domain (h). In addition, a significant degree of conservation was identified through the alignment of amino acid sequences of helices 2 and 3 (i). The squares illustrate the TM-scores in percentages, while the RMSD is in Angstroms (Å).
Figure 8.
3D structural similarity between DBDs-HSEs complexes. Based on the obtained dEhDBD5-HSE_EhPgp5 complex (a), overlay comparisons were performed with crystallographic structures deposited in the PDB database. These included the K. lactis DBD-HSE complex (ID: 3HTS) (b), S. cerevisiae-C. thermophilum complex (ID: 5D5X) (c), H. sapiens-C. thermophilum complex (ID: 5D5W) (d), synthetic H. sapiens construction (ID: 5D8L) (e), and H. sapiens (ID: 7DCU) (f). A joint overlay of all these structures was performed (g) and focusing solely on the wing domain (h). In addition, a significant degree of conservation was identified through the alignment of amino acid sequences of helices 2 and 3 (i). The squares illustrate the TM-scores in percentages, while the RMSD is in Angstroms (Å).
Figure 9.
Our
in vitro findings demonstrate that the mrEhHSTF5 protein is capable of oligomerizing, forming dimers and trimers. Furthermore, through EMSA assays and
in silico molecular docking, we discovered that the dimeric conformation exhibits greater affinity for HSE_
EhPgp5 compared to the monomeric conformation. Dorantes et al. [
36] reported higher affinity (-459) binding of tEhHSTF5 to the same HSE, supporting our results and indicating that the degree of oligomerization increases the affinity towards HSE_
EhPgp5. OD; oligomerization domain, DBD; DNA binding domain, Ds; Docking score.
Figure 9.
Our
in vitro findings demonstrate that the mrEhHSTF5 protein is capable of oligomerizing, forming dimers and trimers. Furthermore, through EMSA assays and
in silico molecular docking, we discovered that the dimeric conformation exhibits greater affinity for HSE_
EhPgp5 compared to the monomeric conformation. Dorantes et al. [
36] reported higher affinity (-459) binding of tEhHSTF5 to the same HSE, supporting our results and indicating that the degree of oligomerization increases the affinity towards HSE_
EhPgp5. OD; oligomerization domain, DBD; DNA binding domain, Ds; Docking score.