Submitted:
08 March 2024
Posted:
11 March 2024
You are already at the latest version
Abstract
Keywords:
1. General Introduction
2. Integrated Stress Response
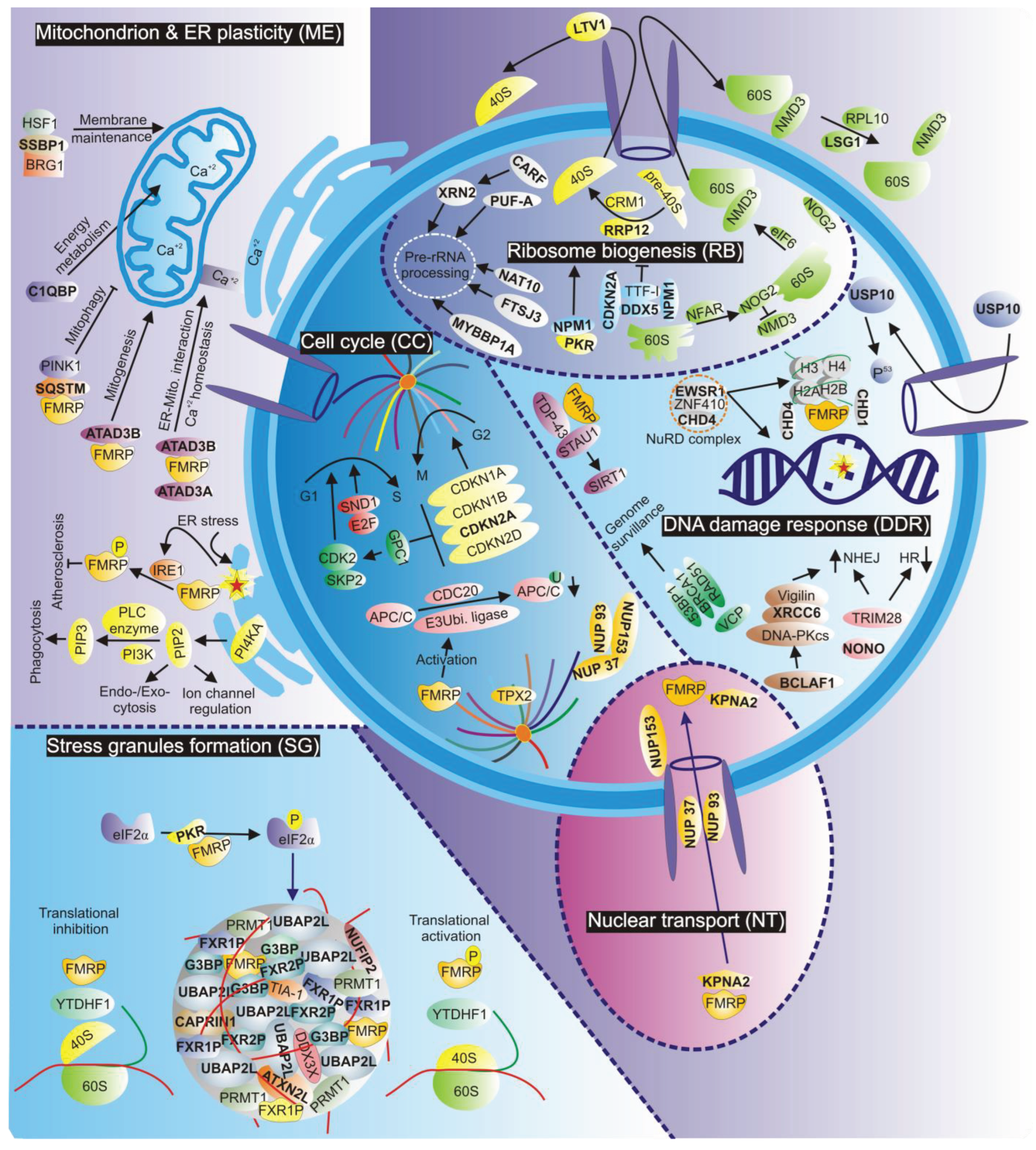
3. Stress Granules Formation
4. Mitochondrion and Endoplasmic Reticulum Plasticity
5. Ribosome Biogenesis
6. Cell Cycle Control
7. DNA Damage Response
8. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclosure Statement
Abbreviations
References
- Maurin, T.; Zongaro, S.; Bardoni, B. Fragile X Syndrome: From molecular pathology to therapy. Neurosci Biobehav Rev 2014, 46 Pt 2, 242–255. [Google Scholar] [CrossRef]
- Ferder, I.; Parborell, F.; Sundblad, V.; Chiauzzi, V.; Gomez, K.; Charreau, E.H.; Tesone, M.; Dain, L. Expression of fragile X mental retardation protein and Fmr1 mRNA during folliculogenesis in the rat. Reproduction 2013, 145, 335–343. [Google Scholar] [CrossRef]
- Tian, H.; Cao, Y.X.; Zhang, X.S.; Liao, W.P.; Yi, Y.H.; Lian, J.; Liu, L.; Huang, H.L.; Liu, W.J.; Yin, M.M.; et al. The targeting and functions of miRNA-383 are mediated by FMRP during spermatogenesis. Cell Death Dis 2013, 4, e617. [Google Scholar] [CrossRef]
- Novak, S.M.; Joardar, A.; Gregorio, C.C.; Zarnescu, D.C. Regulation of Heart Rate in Drosophila via Fragile X Mental Retardation Protein. PLoS ONE 2015, 10, e0142836. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, Y.; Meng, C.; Fang, N. FMRP regulates endothelial cell proliferation and angiogenesis via the miR-181a-CaM-CaMKII pathway. Cell Biol Int 2018, 42, 1432–1444. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, X. Concise review: Fragile X proteins in stem cell maintenance and differentiation. Stem cells (Dayton, Ohio) 2014, 32, 1724–1733. [Google Scholar] [CrossRef]
- Schultz-Pedersen, S.; Hasle, H.; Olsen, J.H.; Friedrich, U. Evidence of decreased risk of cancer in individuals with fragile X. Am J Med Genet 2001, 103, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Ascano, M., Jr.; Mukherjee, N.; Bandaru, P.; Miller, J.B.; Nusbaum, J.D.; Corcoran, D.L.; Langlois, C.; Munschauer, M.; Dewell, S.; Hafner, M.; et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 2012, 492, 382–386. [Google Scholar] [CrossRef]
- Brown, V.; Jin, P.; Ceman, S.; Darnell, J.C.; O’Donnell, W.T.; Tenenbaum, S.A.; Jin, X.; Feng, Y.; Wilkinson, K.D.; Keene, J.D.; et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 2001, 107, 477–487. [Google Scholar] [CrossRef]
- Darnell, J.C.; Klann, E. The translation of translational control by FMRP: Therapeutic targets for FXS. Nat Neurosci 2013, 16, 1530–1536. [Google Scholar] [CrossRef]
- Darnell, J.C.; Van Driesche, S.J.; Zhang, C.; Hung, K.Y.; Mele, A.; Fraser, C.E.; Stone, E.F.; Chen, C.; Fak, J.J.; Chi, S.W.; et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011, 146, 247–261. [Google Scholar] [CrossRef]
- Fernandez, E.; Rajan, N.; Bagni, C. The FMRP regulon: From targets to disease convergence. Front Neurosci 2013, 7, 191. [Google Scholar] [CrossRef]
- Sakano, H.; Zorio, D.A.R.; Wang, X.; Ting, Y.S.; Noble, W.S.; MacCoss, M.J.; Rubel, E.W.; Wang, Y. Proteomic analyses of nucleus laminaris identified candidate targets of the fragile X mental retardation protein. J Comp Neurol 2017, 525, 3341–3359. [Google Scholar] [CrossRef]
- Santoro, M.R.; Bray, S.M.; Warren, S.T. Molecular mechanisms of fragile X syndrome: A twenty-year perspective. Annu Rev Pathol 2012, 7, 219–245. [Google Scholar] [CrossRef] [PubMed]
- Ferron, L.; Nieto-Rostro, M.; Cassidy, J.S.; Dolphin, A.C. Fragile X mental retardation protein controls synaptic vesicle exocytosis by modulating N-type calcium channel density. Nat Commun 2014, 5, 3628. [Google Scholar] [CrossRef]
- Billuart, P.; Chelly, J. From fragile X mental retardation protein to Rac1 GTPase: New insights from Fly CYFIP. Neuron 2003, 38, 843–845. [Google Scholar] [CrossRef]
- Nolze, A.; Schneider, J.; Keil, R.; Lederer, M.; Huttelmaier, S.; Kessels, M.M.; Qualmann, B.; Hatzfeld, M. FMRP regulates actin filament organization via the armadillo protein p0071. RNA 2013, 19, 1483–1496. [Google Scholar] [CrossRef] [PubMed]
- Schenck, A.; Bardoni, B.; Langmann, C.; Harden, N.; Mandel, J.L.; Giangrande, A. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron 2003, 38, 887–898. [Google Scholar] [CrossRef]
- Alpatov, R.; Lesch, B.J.; Nakamoto-Kinoshita, M.; Blanco, A.; Chen, S.; Stutzer, A.; Armache, K.J.; Simon, M.D.; Xu, C.; Ali, M.; et al. A chromatin-dependent role of the fragile X mental retardation protein FMRP in the DNA damage response. Cell 2014, 157, 869–881. [Google Scholar] [CrossRef]
- Liu, J.; Koscielska, K.A.; Cao, Z.; Hulsizer, S.; Grace, N.; Mitchell, G.; Nacey, C.; Githinji, J.; McGee, J.; Garcia-Arocena, D.; et al. Signaling defects in iPSC-derived fragile X premutation neurons. Hum Mol Genet 2012, 21, 3795–3805. [Google Scholar] [CrossRef]
- Zhang, W.; Cheng, Y.; Li, Y.; Chen, Z.; Jin, P.; Chen, D. A feed-forward mechanism involving Drosophila fragile X mental retardation protein triggers a replication stress-induced DNA damage response. Hum Mol Genet 2014, 23, 5188–5196. [Google Scholar] [CrossRef]
- Pasciuto, E.; Bagni, C. SnapShot: FMRP interacting proteins. Cell 2014, 159, 218–218.e1. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.S.; Nouri, K.; Milroy, L.G.; Moll, J.M.; Herrmann, C.; Brunsveld, L.; Piekorz, R.P.; Ahmadian, M.R. Subcellular fractionation and localization studies reveal a direct interaction of the fragile X mental retardation protein (FMRP) with nucleolin. PLoS ONE 2014, 9, e91465. [Google Scholar] [CrossRef] [PubMed]
- Bartley, C.M.; O’Keefe, R.A.; Bordey, A. FMRP S499 is phosphorylated independent of mTORC1-S6K1 activity. PLoS ONE 2014, 9, e96956. [Google Scholar] [CrossRef]
- Myrick, L.K.; Hashimoto, H.; Cheng, X.; Warren, S.T. Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain. Hum Mol Genet 2015, 24, 1733–1740. [Google Scholar] [CrossRef]
- Bardoni, B.; Sittler, A.; Shen, Y.; Mandel, J.L. Analysis of domains affecting intracellular localization of the FMRP protein. Neurobiol Dis 1997, 4, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Gutekunst, C.A.; Eberhart, D.E.; Yi, H.; Warren, S.T.; Hersch, S.M. Fragile X mental retardation protein: Nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci 1997, 17, 1539–1547. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Bellini, M.; Ceman, S. Fragile X mental retardation protein FMRP binds mRNAs in the nucleus. Mol Cell Biol 2009, 29, 214–228. [Google Scholar] [CrossRef]
- Tamanini, F.; Bontekoe, C.; Bakker, C.E.; van Unen, L.; Anar, B.; Willemsen, R.; Yoshida, M.; Galjaard, H.; Oostra, B.A.; Hoogeveen, A.T. Different targets for the fragile X-related proteins revealed by their distinct nuclear localizations. Hum Mol Genet 1999, 8, 863–869. [Google Scholar] [CrossRef]
- Hoogeveen, A.T.; Willemsen, R.; Oostra, B.A. Fragile X syndrome, the Fragile X related proteins, and animal models. Microscopy research and technique 2002, 57, 148–155. [Google Scholar] [CrossRef]
- Sakai, Y.; Shaw, C.A.; Dawson, B.C.; Dugas, D.V.; Al-Mohtaseb, Z.; Hill, D.E.; Zoghbi, H.Y. Protein interactome reveals converging molecular pathways among autism disorders. Science translational medicine 2011, 3, 86ra49. [Google Scholar] [CrossRef]
- Schenck, A.; Bardoni, B.; Moro, A.; Bagni, C.; Mandel, J.-L. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proceedings of the National Academy of Sciences 2001, 98, 8844–8849. [Google Scholar] [CrossRef]
- Winograd, C.; Ceman, S. Fragile X family members have important and non-overlapping functions. Biomolecular concepts 2011, 2, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Cheever, A.; Ceman, S. Translation regulation of mRNAs by the fragile X family of proteins through the microRNA pathway. RNA biology 2009, 6, 175–178. [Google Scholar] [CrossRef]
- Wang, T.; Bray, S.M.; Warren, S.T. New perspectives on the biology of fragile X syndrome. Curr Opin Genet Dev 2012, 22, 256–263. [Google Scholar] [CrossRef]
- Chen, E.; Joseph, S. Fragile X mental retardation protein: A paradigm for translational control by RNA-binding proteins. Biochimie 2015, 114, 147–154. [Google Scholar] [CrossRef]
- Irwin, S.A.; Galvez, R.; Greenough, W.T. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex 2000, 10, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Kenny, P.J.; Zhou, H.; Kim, M.; Skariah, G.; Khetani, R.S.; Drnevich, J.; Arcila, M.L.; Kosik, K.S.; Ceman, S. MOV10 and FMRP regulate AGO2 association with microRNA recognition elements. Cell Rep 2014, 9, 1729–1741. [Google Scholar] [CrossRef]
- Alberti, S.; Mateju, D.; Mediani, L.; Carra, S. Granulostasis: Protein Quality Control of RNP Granules. Frontiers in molecular neuroscience 2017, 10, 84. [Google Scholar] [CrossRef] [PubMed]
- Moujaber, O.; Stochaj, U. Cytoplasmic RNA granules in somatic maintenance. Gerontology 2018, 64, 485–494. [Google Scholar] [CrossRef]
- Sfakianos, A.P.; Whitmarsh, A.J.; Ashe, M.P. Ribonucleoprotein bodies are phased in. Biochemical Society transactions 2016, 44, 1411–1416. [Google Scholar] [CrossRef]
- Chyung, E.; LeBlanc, H.F.; Fallon, J.R.; Akins, M.R. Fragile X granules are a family of axonal ribonucleoprotein particles with circuit-dependent protein composition and mRNA cargos. J Comp Neurol 2018, 526, 96–108. [Google Scholar] [CrossRef]
- El Fatimy, R.; Davidovic, L.; Tremblay, S.; Jaglin, X.; Dury, A.; Robert, C.; De Koninck, P.; Khandjian, E.W. Tracking the Fragile X Mental Retardation Protein in a Highly Ordered Neuronal RiboNucleoParticles Population: A Link between Stalled Polyribosomes and RNA Granules. PLoS Genet 2016, 12, e1006192. [Google Scholar] [CrossRef]
- Maziuk, B.; Ballance, H.I.; Wolozin, B. Dysregulation of RNA Binding Protein Aggregation in Neurodegenerative Disorders. Frontiers in molecular neuroscience 2017, 10, 89. [Google Scholar] [CrossRef]
- Galluzzi, L.; Yamazaki, T.; Kroemer, G. Linking cellular stress responses to systemic homeostasis. Nat Rev Mol Cell Biol 2018, 19, 731–745. [Google Scholar] [CrossRef]
- Yang, P.; Mathieu, C.; Kolaitis, R.M.; Zhang, P.; Messing, J.; Yurtsever, U.; Yang, Z.; Wu, J.; Li, Y.; Pan, Q.; et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345.e8. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.S.; Haghighi, F.; Stefanski, A.; Nakhaei-Rad, S.; Kazemein Jasemi, N.S.; Al Kabbani, M.A.; Görg, B.; Fujii, M.; Lang, P.A.; Häussinger, D.; et al. Novel FMRP interaction networks linked to cellular stress. FEBS J 2021, 288, 837–860. [Google Scholar] [CrossRef] [PubMed]
- El Fatimy, R.; Tremblay, S.; Dury, A.Y.; Solomon, S.; De Koninck, P.; Schrader, J.W.; Khandjian, E.W. Fragile X mental retardation protein interacts with the RNA-binding protein Caprin1 in neuronal RiboNucleoProtein complexes [corrected]. PLoS ONE 2012, 7, e39338. [Google Scholar] [CrossRef]
- Bardoni, B.; Castets, M.; Huot, M.E.; Schenck, A.; Adinolfi, S.; Corbin, F.; Pastore, A.; Khandjian, E.W.; Mandel, J.L. 82-FIP, a novel FMRP (fragile X mental retardation protein) interacting protein, shows a cell cycle-dependent intracellular localization. Hum Mol Genet 2003, 12, 1689–1698. [Google Scholar] [CrossRef] [PubMed]
- Ledoux, N.; Lelong, E.I.J.; Simard, A.; Hussein, S.; Adjibade, P.; Lambert, J.P.; Mazroui, R. The Identification of Nuclear FMRP Isoform Iso6 Partners. Cells 2023, 12, 2807. [Google Scholar] [CrossRef]
- Zhuang, Y.; Xu, H.C.; Shinde, P.V.; Warfsmann, J.; Vasilevska, J.; Sundaram, B.; Behnke, K.; Huang, J.; Hoell, J.I.; Borkhardt, A.; et al. Fragile X mental retardation protein protects against tumour necrosis factor-mediated cell death and liver injury. Gut 2020, 69, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, H.; Lu, J.; Chen, Y.; Zhang, Y.; Xiao, L. Accelerated Apoptosis and Down-Regulated FMRP in Human Neuroblastoma Cells with CRISPR/Cas9 Genome Editing. Iran J Public Health 2023, 52, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Mahboubi, H.; Stochaj, U. Nucleoli and stress granules: Connecting distant relatives. Traffic 2014, 15, 1179–1193. [Google Scholar] [CrossRef]
- Kieffer, F.; Hilal, F.; Gay, A.S.; Debayle, D.; Pronot, M.; Poupon, G.; Lacagne, I.; Bardoni, B.; Martin, S.; Gwizdek, C. Combining affinity purification and mass spectrometry to define the network of the nuclear proteins interacting with the N-terminal region of FMRP. Front Mol Biosci 2022, 9, 954087. [Google Scholar] [CrossRef]
- Dolicka, D.; Foti, M.; Sobolewski, C. The Emerging Role of Stress Granules in Hepatocellular Carcinoma. Int J Mol Sci 2021, 22, 9428. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, Y. Stress granules: Potential therapeutic targets for infectious and inflammatory diseases. Front Immunol 2023, 14, 1145346. [Google Scholar] [CrossRef]
- Glineburg, M.R.; Yildirim, E.; Gomez, N.; Li, X.; Pak, J.; Altheim, C.; Waksmacki, J.; McInerney, G.; Barmada, S.J.; Todd, P.K. Stress granule formation helps to mitigate neurodegeneration. bioRxiv 2023. [Google Scholar]
- Mahboubi, H.; Stochaj, U. Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochim Biophys Acta Mol Basis Dis 2017, 1863, 884–895. [Google Scholar] [CrossRef]
- Lamichhane, P.P.; Samir, P. Cellular Stress: Modulator of Regulated Cell Death. Biology (Basel) 2023, 12, 1172. [Google Scholar] [CrossRef]
- Ryan, L.; Rubinsztein, D.C. The autophagy of stress granules. FEBS Lett 2024, 598, 59–72. [Google Scholar] [CrossRef]
- Protter, D.S.W.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol 2016, 26, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Buddika, K.; Ariyapala, I.S.; Hazuga, M.A.; Riffert, D.; Sokol, N.S. Canonical nucleators are dispensable for stress granule assembly in Drosophila intestinal progenitors. J Cell Sci 2020, 133, jcs243451. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.E.; Blackford, A.N.; Chapman, J.R.; Baskcomb, L.; Gravel, S.; Rusch, A.; Thomas, A.; Blundred, R.; Smith, P.; Kzhyshkowska, J.; et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Molecular cell 2012, 45, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Luo, K.; Zhang, L.; Cheville, J.C.; Lou, Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 2010, 140, 384–396. [Google Scholar] [CrossRef]
- Cirillo, L.; Cieren, A.; Barbieri, S.; Khong, A.; Schwager, F.; Parker, R.; Gotta, M. UBAP2L Forms Distinct Cores that Act in Nucleating Stress Granules Upstream of G3BP1. Curr Biol 2020, 30, 698–707.e6. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Chen, Y.; Dai, H.; Zhang, H.; Xie, M.; Chen, F.; Kang, X.; Bai, X.; Chen, Z. UBAP2L arginine methylation by PRMT1 modulates stress granule assembly. Cell Death Differ 2020, 27, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Asano-Inami, E.; Yokoi, A.; Sugiyama, M.; Hyodo, T.; Hamaguchi, T.; Kajiyama, H. The association of UBAP2L and G3BP1 mediated by small nucleolar RNA is essential for stress granule formation. Commun Biol 2023, 6, 415. [Google Scholar] [CrossRef]
- Zou, Z.; Wei, J.; Chen, Y.; Kang, Y.; Shi, H.; Yang, F.; Shi, Z.; Chen, S.; Zhou, Y.; Sepich-Poore, C.; et al. FMRP phosphorylation modulates neuronal translation through YTHDF1. Molecular cell 2023, 83, 4304–4317.e8. [Google Scholar] [CrossRef]
- Geng, J.; Khaket, T.P.; Pan, J.; Li, W.; Zhang, Y.; Ping, Y.; Cobos Sillero, M.I.; Lu, B. Deregulation of ER-mitochondria contact formation and mitochondrial calcium homeostasis mediated by VDAC in fragile X syndrome. Dev Cell 2023, 58, 597–615.e10. [Google Scholar] [CrossRef]
- Bülow, P.; Zlatic, S.A.; Wenner, P.A.; Bassell, G.J.; Faundez, V. FMRP attenuates activity dependent modifications in the mitochondrial proteome. Mol Brain 2021, 14, 75. [Google Scholar] [CrossRef]
- Angelova, P.R.; Abramov, A.Y. Role of mitochondrial ROS in the brain: From physiology to neurodegeneration. FEBS Lett 2018, 592, 692–702. [Google Scholar] [CrossRef]
- Nissanka, N.; Moraes, C.T. Mitochondrial DNA damage and reactive oxygen species in neurodegenerative disease. FEBS Lett 2018, 592, 728–742. [Google Scholar] [CrossRef]
- Liu, H.; Dai, C.; Fan, Y.; Guo, B.; Ren, K.; Sun, T.; Wang, W. From autophagy to mitophagy: The roles of P62 in neurodegenerative diseases. Journal of bioenergetics and biomembranes 2017, 49, 413–422. [Google Scholar] [CrossRef]
- Baudier, J. ATAD3 proteins: Brokers of a mitochondria-endoplasmic reticulum connection in mammalian cells. Biol Rev Camb Philos Soc 2018, 93, 827–844. [Google Scholar] [CrossRef]
- Geisler, S.; Holmstrom, K.M.; Skujat, D.; Fiesel, F.C.; Rothfuss, O.C.; Kahle, P.J.; Springer, W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature cell biology 2010, 12, 119–131. [Google Scholar] [CrossRef]
- Narendra, D.; Kane, L.A.; Hauser, D.N.; Fearnley, I.M.; Youle, R.J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 2010, 6, 1090–1106. [Google Scholar] [CrossRef]
- Hoffmann, M.; Bellance, N.; Rossignol, R.; Koopman, W.J.; Willems, P.H.; Mayatepek, E.; Bossinger, O.; Distelmaier, F.C. elegans ATAD-3 is essential for mitochondrial activity and development. PLoS ONE 2009, 4, e7644. [Google Scholar] [CrossRef]
- Santini, E.; Huynh, T.N.; Longo, F.; Koo, S.Y.; Mojica, E.; D’Andrea, L.; Bagni, C.; Klann, E. Reducing eIF4E-eIF4G interactions restores the balance between protein synthesis and actin dynamics in fragile X syndrome model mice. Sci Signal 2017, 10. [Google Scholar] [CrossRef]
- Feichtinger, R.G.; Olahova, M.; Kishita, Y.; Garone, C.; Kremer, L.S.; Yagi, M.; Uchiumi, T.; Jourdain, A.A.; Thompson, K.; D’Souza, A.R.; et al. Biallelic C1QBP Mutations Cause Severe Neonatal-, Childhood-, or Later-Onset Cardiomyopathy Associated with Combined Respiratory-Chain Deficiencies. American journal of human genetics 2017, 101, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Yagi, M.; Uchiumi, T.; Sagata, N.; Setoyama, D.; Amamoto, R.; Matsushima, Y.; Kang, D. Neural-specific deletion of mitochondrial p32/C1qbp leads to leukoencephalopathy due to undifferentiated oligodendrocyte and axon degeneration. Scientific reports 2017, 7, 15131. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Fujimoto, M.; Takii, R.; Takaki, E.; Hayashida, N.; Nakai, A. Mitochondrial SSBP1 protects cells from proteotoxic stresses by potentiating stress-induced HSF1 transcriptional activity. Nat Commun 2015, 6, 6580. [Google Scholar] [CrossRef]
- Yao, A.; Jin, S.; Li, X.; Liu, Z.; Ma, X.; Tang, J.; Zhang, Y.Q. Drosophila FMRP regulates microtubule network formation and axonal transport of mitochondria. Hum Mol Genet 2011, 20, 51–63. [Google Scholar] [CrossRef]
- Weisz, E.D.; Towheed, A.; Monyak, R.E.; Toth, M.S.; Wallace, D.C.; Jongens, T.A. Loss of Drosophila FMRP leads to alterations in energy metabolism and mitochondrial function. Hum Mol Genet 2018, 27, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, Z.; Baboo, S.; Hamid, S.M.; Dogan, A.E.; Tufanli, O.; Robichaud, S.; Emerton, C.; Diedrich, J.K.; Vatandaslar, H.; Nikolos, F.; et al. Intercepting IRE1 kinase-FMRP signaling prevents atherosclerosis progression. EMBO Mol Med 2022, 14, e15344. [Google Scholar] [CrossRef] [PubMed]
- Bojjireddy, N.; Botyanszki, J.; Hammond, G.; Creech, D.; Peterson, R.; Kemp, D.C.; Snead, M.; Brown, R.; Morrison, A.; Wilson, S.; et al. Pharmacological and genetic targeting of the PI4KA enzyme reveals its important role in maintaining plasma membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate levels. J Biol Chem 2014, 289, 6120–6132. [Google Scholar] [CrossRef] [PubMed]
- Czech, M.P. PIP2 and PIP3: Complex roles at the cell surface. Cell 2000, 100, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Pfister, A.S. Emerging Role of the Nucleolar Stress Response in Autophagy. Front Cell Neurosci 2019, 13, 156. [Google Scholar] [CrossRef] [PubMed]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.M.; Lamond, A.I. The nucleolus under stress. Molecular cell 2010, 40, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Thomson, E.; Ferreira-Cerca, S.; Hurt, E. Eukaryotic ribosome biogenesis at a glance. J Cell Sci 2013, 126 Pt 21, 4815–4821. [Google Scholar] [CrossRef] [PubMed]
- van Riggelen, J.; Yetil, A.; Felsher, D.W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nature reviews. Cancer 2010, 10, 301–309. [Google Scholar] [CrossRef]
- Saporita, A.J.; Chang, H.C.; Winkeler, C.L.; Apicelli, A.J.; Kladney, R.D.; Wang, J.; Townsend, R.R.; Michel, L.S.; Weber, J.D. RNA helicase DDX5 is a p53-independent target of ARF that participates in ribosome biogenesis. Cancer research 2011, 71, 6708–6717. [Google Scholar] [CrossRef]
- Sato, S.; Ishikawa, H.; Yoshikawa, H.; Izumikawa, K.; Simpson, R.J.; Takahashi, N. Collaborator of alternative reading frame protein (CARF) regulates early processing of pre-ribosomal RNA by retaining XRN2 (5′-3′ exoribonuclease) in the nucleoplasm. Nucleic acids research 2015, 43, 10397–10410. [Google Scholar] [CrossRef]
- Qiu, C.; McCann, K.L.; Wine, R.N.; Baserga, S.J.; Hall, T.M. A divergent Pumilio repeat protein family for pre-rRNA processing and mRNA localization. Proceedings of the National Academy of Sciences of the United States of America 2014, 111, 18554–18559. [Google Scholar] [CrossRef]
- Morello, L.G.; Coltri, P.P.; Quaresma, A.J.; Simabuco, F.M.; Silva, T.C.; Singh, G.; Nickerson, J.A.; Oliveira, C.C.; Moore, M.J.; Zanchin, N.I. The human nucleolar protein FTSJ3 associates with NIP7 and functions in pre-rRNA processing. PLoS ONE 2011, 6, e29174. [Google Scholar] [CrossRef] [PubMed]
- Hochstatter, J.; Holzel, M.; Rohrmoser, M.; Schermelleh, L.; Leonhardt, H.; Keough, R.; Gonda, T.J.; Imhof, A.; Eick, D.; Langst, G.; et al. Myb-binding protein 1a (Mybbp1a) regulates levels and processing of pre-ribosomal RNA. J Biol Chem 2012, 287, 24365–24377. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Horikawa, S.; Suzuki, T.; Kawauchi, H.; Tanaka, Y.; Suzuki, T.; Suzuki, T. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). J Biol Chem 2014, 289, 35724–35730. [Google Scholar] [CrossRef] [PubMed]
- Blalock, W.L.; Piazzi, M.; Bavelloni, A.; Raffini, M.; Faenza, I.; D’Angelo, A.; Cocco, L. Identification of the PKR nuclear interactome reveals roles in ribosome biogenesis, mRNA processing and cell division. Journal of cellular physiology 2014, 229, 1047–1060. [Google Scholar] [CrossRef] [PubMed]
- Moriggi, G.; Nieto, B.; Dosil, M. Rrp12 and the Exportin Crm1 participate in late assembly events in the nucleolus during 40S ribosomal subunit biogenesis. PLoS Genet 2014, 10, e1004836. [Google Scholar] [CrossRef] [PubMed]
- Seiser, R.M.; Sundberg, A.E.; Wollam, B.J.; Zobel-Thropp, P.; Baldwin, K.; Spector, M.D.; Lycan, D.E. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics 2006, 174, 679–691. [Google Scholar] [CrossRef] [PubMed]
- Strunk, B.S.; Novak, M.N.; Young, C.L.; Karbstein, K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 2012, 150, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Wandrey, F.; Montellese, C.; Koos, K.; Badertscher, L.; Bammert, L.; Cook, A.G.; Zemp, I.; Horvath, P.; Kutay, U. The NF45/NF90 Heterodimer Contributes to the Biogenesis of 60S Ribosomal Subunits and Influences Nucleolar Morphology. Mol Cell Biol 2015, 35, 3491–3503. [Google Scholar] [CrossRef] [PubMed]
- Miluzio, A.; Beugnet, A.; Volta, V.; Biffo, S. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep 2009, 10, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y.; Granneman, S.; Thoms, M.; Manikas, R.G.; Tollervey, D.; Hurt, E. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 2014, 505, 112–116. [Google Scholar] [CrossRef] [PubMed]
- West, M.; Hedges, J.B.; Chen, A.; Johnson, A.W. Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol Cell Biol 2005, 25, 3802–3813. [Google Scholar] [CrossRef]
- Rhind, N.; Russell, P. Signaling pathways that regulate cell division. Cold Spring Harb Perspect Biol 2012, 4, a005942. [Google Scholar] [CrossRef]
- Qiao, D.; Meyer, K.; Friedl, A. Glypican-1 stimulates Skp2 autoinduction loop and G1/S transition in endothelial cells. J Biol Chem 2012, 287, 5898–5909. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.A.; Chu, K.B.; Moon, E.K.; Quan, F.S. Histone Deacetylase Inhibitor-Induced CDKN2B and CDKN2D Contribute to G2/M Cell Cycle Arrest Incurred by Oxidative Stress in Hepatocellular Carcinoma Cells via Forkhead Box M1 Suppression. J Cancer 2021, 12, 5086–5098. [Google Scholar] [CrossRef]
- Fagundes, R.; Teixeira, L.K. Cyclin E/CDK2: DNA Replication, Replication Stress and Genomic Instability. Front Cell Dev Biol 2021, 9, 774845. [Google Scholar] [CrossRef]
- Yang, H.; Zhao, X.; Zhao, L.; Liu, L.; Li, J.; Jia, W.; Liu, J.; Huang, G. PRMT5 competitively binds to CDK4 to promote G1-S transition upon glucose induction in hepatocellular carcinoma. Oncotarget 2016, 7, 72131–72147. [Google Scholar] [CrossRef]
- Su, C.; Zhang, C.; Tecle, A.; Fu, X.; He, J.; Song, J.; Zhang, W.; Sun, X.; Ren, Y.; Silvennoinen, O.; et al. Tudor staphylococcal nuclease (Tudor-SN), a novel regulator facilitating G1/S phase transition, acting as a co-activator of E2F-1 in cell cycle regulation. J Biol Chem 2015, 290, 7208–7220. [Google Scholar] [CrossRef]
- Yu, Y.; Gao, R.; Kaul, Z.; Li, L.; Kato, Y.; Zhang, Z.; Groden, J.; Kaul, S.C.; Wadhwa, R. Loss-of-function screening to identify miRNAs involved in senescence: Tumor suppressor activity of miRNA-335 and its new target CARF. Scientific reports 2016, 6, 30185. [Google Scholar] [CrossRef]
- Cheung, C.T.; Singh, R.; Kalra, R.S.; Kaul, S.C.; Wadhwa, R. Collaborator of ARF (CARF) regulates proliferative fate of human cells by dose-dependent regulation of DNA damage signaling. J Biol Chem 2014, 289, 18258–18269. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zheng, M.; Lu, R.; Du, J.; Zhao, Q.; Li, Z.; Li, Y.; Zhang, S. The role of CDC25C in cell cycle regulation and clinical cancer therapy: A systematic review. Cancer Cell Int 2020, 20, 213. [Google Scholar] [CrossRef]
- Zhang, N.; Kaur, R.; Akhter, S.; Legerski, R.J. Cdc5L interacts with ATR and is required for the S-phase cell-cycle checkpoint. EMBO Rep 2009, 10, 1029–1035. [Google Scholar] [CrossRef]
- Bai, D.; Zhang, J.; Xiao, W.; Zheng, X. Regulation of the HDM2-p53 pathway by ribosomal protein L6 in response to ribosomal stress. Nucleic acids research 2014, 42, 1799–1811. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Chen, H.; Zhan, Y.Q.; Yin, R.H.; Li, C.Y.; Ge, C.H.; Yu, M.; Yang, X.M. EWSR1 regulates mitosis by dynamically influencing microtubule acetylation. Cell cycle (Georgetown, Tex.) 2016, 15, 2202–2215. [Google Scholar] [CrossRef] [PubMed]
- Wadsworth, P. TPX2. Curr Biol 2015, 25, R1156–1158. [Google Scholar] [CrossRef] [PubMed]
- Pickard, M.R.; Mourtada-Maarabouni, M.; Williams, G.T. Candidate tumour suppressor Fau regulates apoptosis in human cells: An essential role for Bcl-G. Biochimica et biophysica acta 2011, 1812, 1146–1153. [Google Scholar] [CrossRef]
- Wozniak, M.; Hotowy, K.; Czapinska, E.; Dus-Szachniewicz, K.; Szczuka, I.; Gamian, E.; Gamian, A.; Terlecki, G.; Ziolkowski, P. Early induction of stress-associated Src activator/Homo sapiens chromosome 9 open reading frame 10 protein following photodynamic therapy. Photodiagnosis and photodynamic therapy 2014, 11, 27–33. [Google Scholar] [CrossRef]
- Ren, B.; Burkovetskaya, M.; Jung, Y.; Bergdolt, L.; Totusek, S.; Martinez-Cerdeno, V.; Stauch, K.; Korade, Z.; Dunaevsky, A. Dysregulated cholesterol metabolism, aberrant excitability and altered cell cycle of astrocytes in fragile X syndrome. Glia 2023, 71, 1176–1196. [Google Scholar] [CrossRef]
- Wang, X.; Li, F.; Zhu, J.; Feng, D.; Shi, Y.; Qu, L.; Li, Y.; Guo, K.; Zhang, Y.; Wang, Q.; et al. Up-regulation of cell division cycle 20 expression alters the morphology of neuronal dendritic spines in the nucleus accumbens by promoting FMRP ubiquitination. J Neurochem 2022, 162, 166–189. [Google Scholar] [CrossRef]
- Agote-Arán, A.; Lin, J.; Sumara, I. Fragile X-Related Protein 1 Regulates Nucleoporin Localization in a Cell Cycle-Dependent Manner. Front Cell Dev Biol 2021, 9, 755847. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, J.; Lin, L.; Zhang, Y.; Zhong, F.; Liu, Y.; Yu, Y.; Shen, H.; Han, M.; He, F.; et al. Proteomic study explores AGR2 as pro-metastatic protein in HCC. Mol Biosyst 2012, 8, 2710–2718. [Google Scholar] [CrossRef] [PubMed]
- Ledoux, N.; Gauthier-Naud, W.; Lavoie, O.; Watters, V.; Hussein, S.; Adjibade, P.; Mazroui, R. The nuclear isoforms of the Fragile X mental retardation RNA-binding protein associate with genomic DNA bridges. Mol Biol Cell 2023, 34, ar36. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.N.; Boysen, G.; Sumanasuriya, S.; Seed, G.; Marzo, A.M.; de Bono, J. The molecular underpinnings of prostate cancer: Impacts on management and pathology practice. J Pathol 2017, 241, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.J.; Bin, P.J.; Luo, C.W.; Chai, C.Y. CHD4 plays a critical role in arsenite-induced oxidative damage in human urothelial carcinoma. Pathol Res Pract 2022, 240, 154173. [Google Scholar] [CrossRef]
- Vinjamur, D.S.; Yao, Q.; Cole, M.A.; McGuckin, C.; Ren, C.; Zeng, J.; Hossain, M.; Luk, K.; Wolfe, S.A.; Pinello, L.; et al. ZNF410 represses fetal globin by singular control of CHD4. Nat Genet 2021, 53, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Graca Marques, J.; Pavlovic, B.; Ngo, Q.A.; Pedot, G.; Roemmele, M.; Volken, L.; Kisele, S.; Perbet, R.; Wachtel, M.; Schäfer, B.W. The Chromatin Remodeler CHD4 Sustains Ewing Sarcoma Cell Survival by Controlling Global Chromatin Architecture. Cancer research 2024, 84, 241–257. [Google Scholar] [CrossRef] [PubMed]
- Luca, R.; Averna, M.; Zalfa, F.; Vecchi, M.; Bianchi, F.; La Fata, G.; Del Nonno, F.; Nardacci, R.; Bianchi, M.; Nuciforo, P.; et al. The fragile X protein binds mRNAs involved in cancer progression and modulates metastasis formation. EMBO Mol Med 2013, 5, 1523–1536. [Google Scholar] [CrossRef]
- Gleicher, N.; McAlpine, J.N.; Gilks, C.B.; Kushnir, V.A.; Lee, H.J.; Wu, Y.G.; Lazzaroni-Tealdi, E.; Barad, D.H. Absence of BRCA/FMR1 correlations in women with ovarian cancers. PLoS ONE 2014, 9, e102370. [Google Scholar] [CrossRef]
- Cabart, P.; Chew, H.K.; Murphy, S. BRCA1 cooperates with NUFIP and P-TEFb to activate transcription by RNA polymerase II. Oncogene 2004, 23, 5316–5329. [Google Scholar] [CrossRef]
- Kawai, S.; Amano, A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J Cell Biol 2012, 197, 201–208. [Google Scholar] [CrossRef]
- Nicol, S.M.; Bray, S.E.; Black, H.D.; Lorimore, S.A.; Wright, E.G.; Lane, D.P.; Meek, D.W.; Coates, P.J.; Fuller-Pace, F.V. The RNA helicase p68 (DDX5) is selectively required for the induction of p53-dependent p21 expression and cell-cycle arrest after DNA damage. Oncogene 2013, 32, 3461–3469. [Google Scholar] [CrossRef]
- Tago, K.; Funakoshi-Tago, M.; Itoh, H.; Furukawa, Y.; Kikuchi, J.; Kato, T.; Suzuki, K.; Yanagisawa, K. Arf tumor suppressor disrupts the oncogenic positive feedback loop including c-Myc and DDX5. Oncogene 2015, 34, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Kar, A.; Kaur, M.; Ghosh, T.; Khan, M.M.; Sharma, A.; Shekhar, R.; Varshney, A.; Saxena, S. RPA70 depletion induces hSSB1/2-INTS3 complex to initiate ATR signaling. Nucleic acids research 2015, 43, 4962–4974. [Google Scholar] [CrossRef] [PubMed]
- Salton, M.; Lerenthal, Y.; Wang, S.Y.; Chen, D.J.; Shiloh, Y. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell cycle (Georgetown, Tex.) 2010, 9, 1568–1576. [Google Scholar] [CrossRef] [PubMed]
- Krietsch, J.; Caron, M.C.; Gagne, J.P.; Ethier, C.; Vignard, J.; Vincent, M.; Rouleau, M.; Hendzel, M.J.; Poirier, G.G.; Masson, J.Y. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic acids research 2012, 40, 10287–10301. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Z.; Blackwell, K.; Carmichael, G.G. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr Biol 2005, 15, 384–391. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Yu, Y.B.; Gunawardena, H.P.; Xie, L.; Chen, X. BCLAF1 is a radiation-induced H2AX-interacting partner involved in gammaH2AX-mediated regulation of apoptosis and DNA repair. Cell Death Dis 2012, 3, e359. [Google Scholar] [CrossRef] [PubMed]
- Marin-Vicente, C.; Domingo-Prim, J.; Eberle, A.B.; Visa, N. RRP6/EXOSC10 is required for the repair of DNA double-strand breaks by homologous recombination. J Cell Sci 2015, 128, 1097–1107. [Google Scholar] [CrossRef]
- Huertas, P. DNA resection in eukaryotes: Deciding how to fix the break. Nature structural & molecular biology 2010, 17, 11–16. [Google Scholar]
- Wang, Y.; Zhu, W.G.; Zhao, Y. Autophagy substrate SQSTM1/p62 regulates chromatin ubiquitination during the DNA damage response. Autophagy 2017, 13, 212–213. [Google Scholar] [CrossRef] [PubMed]
- Meerang, M.; Ritz, D.; Paliwal, S.; Garajova, Z.; Bosshard, M.; Mailand, N.; Janscak, P.; Hubscher, U.; Meyer, H.; Ramadan, K. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nature cell biology 2011, 13, 1376–1382. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




