Submitted:
04 March 2024
Posted:
06 March 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Materials and Methods
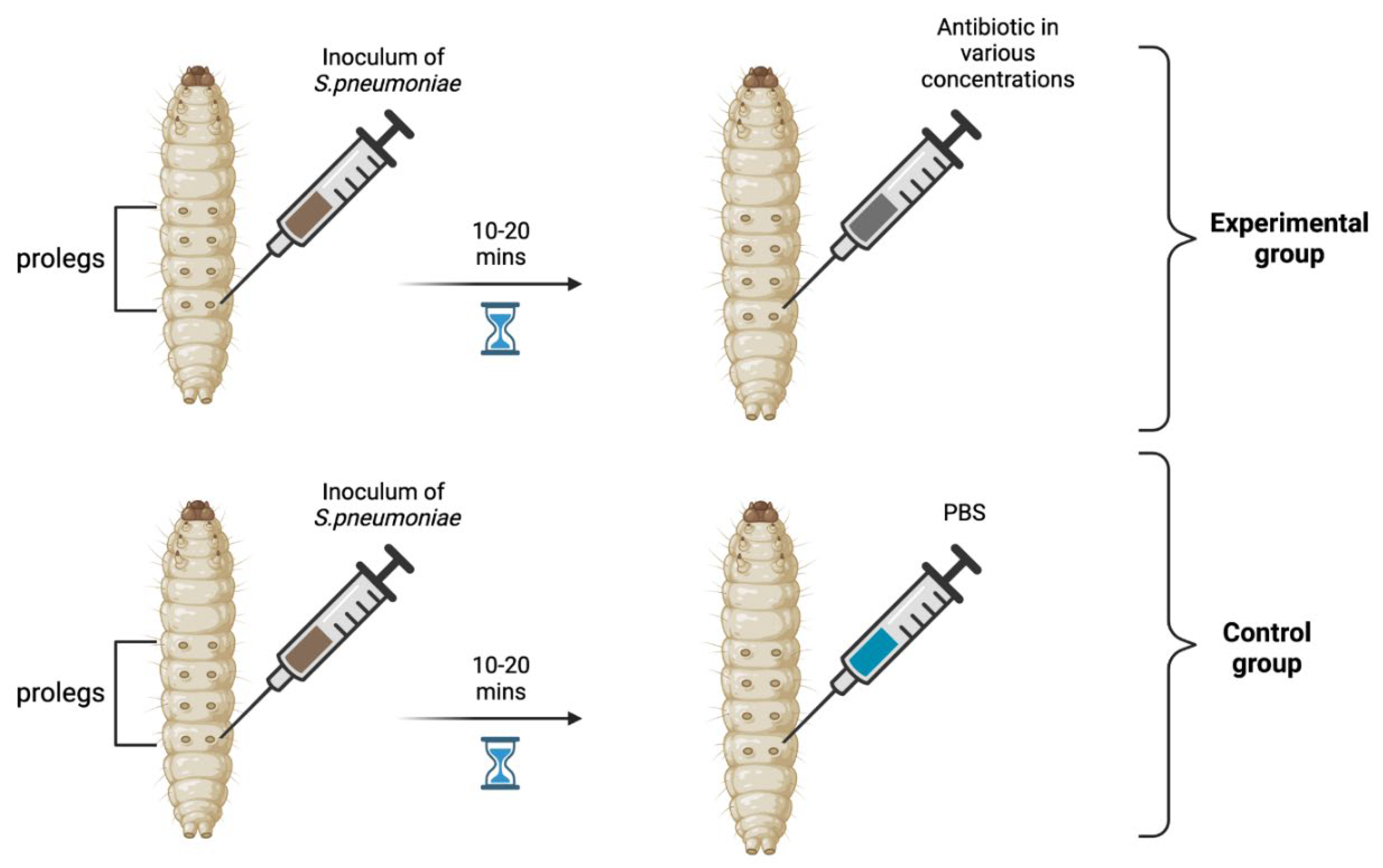
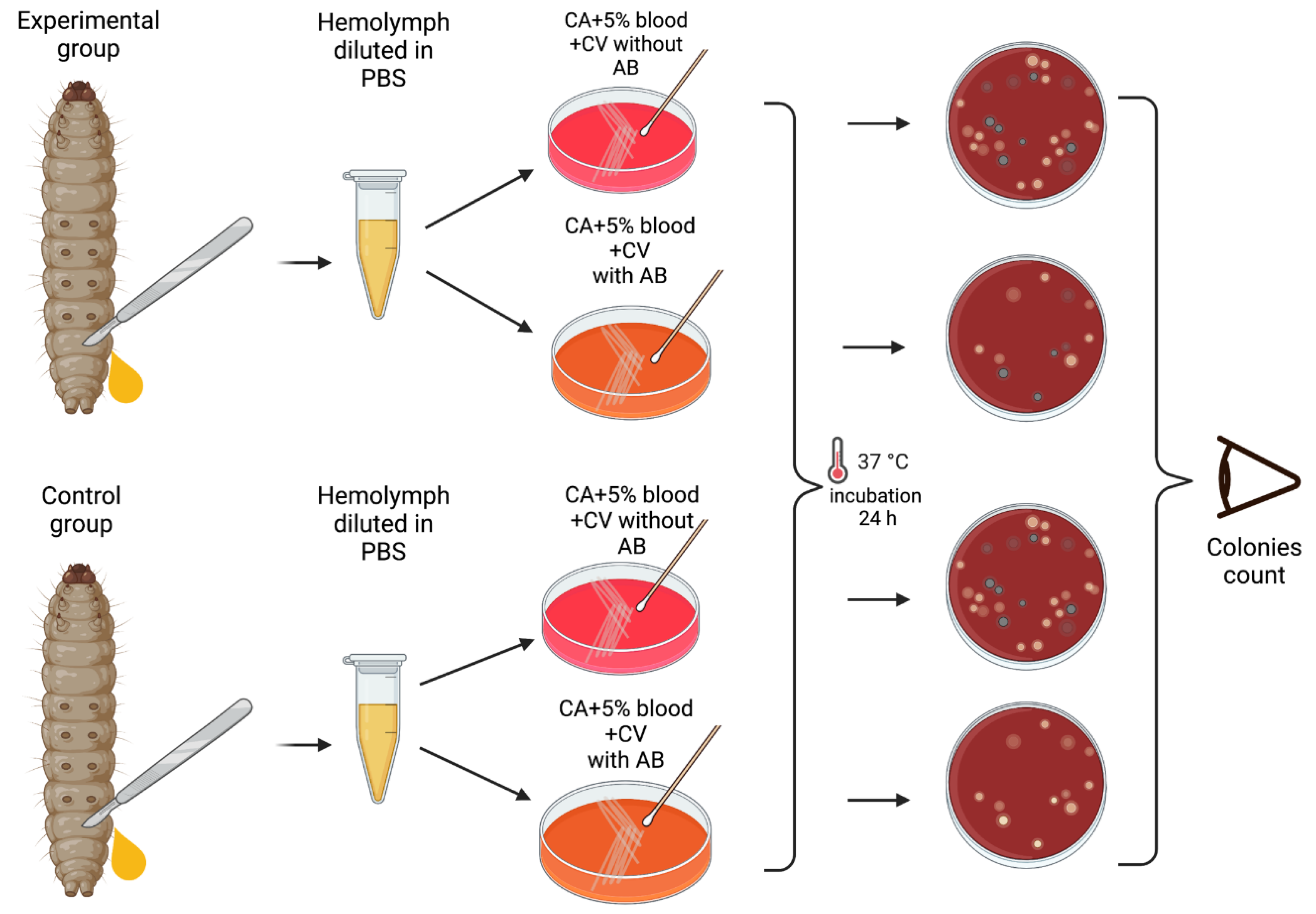
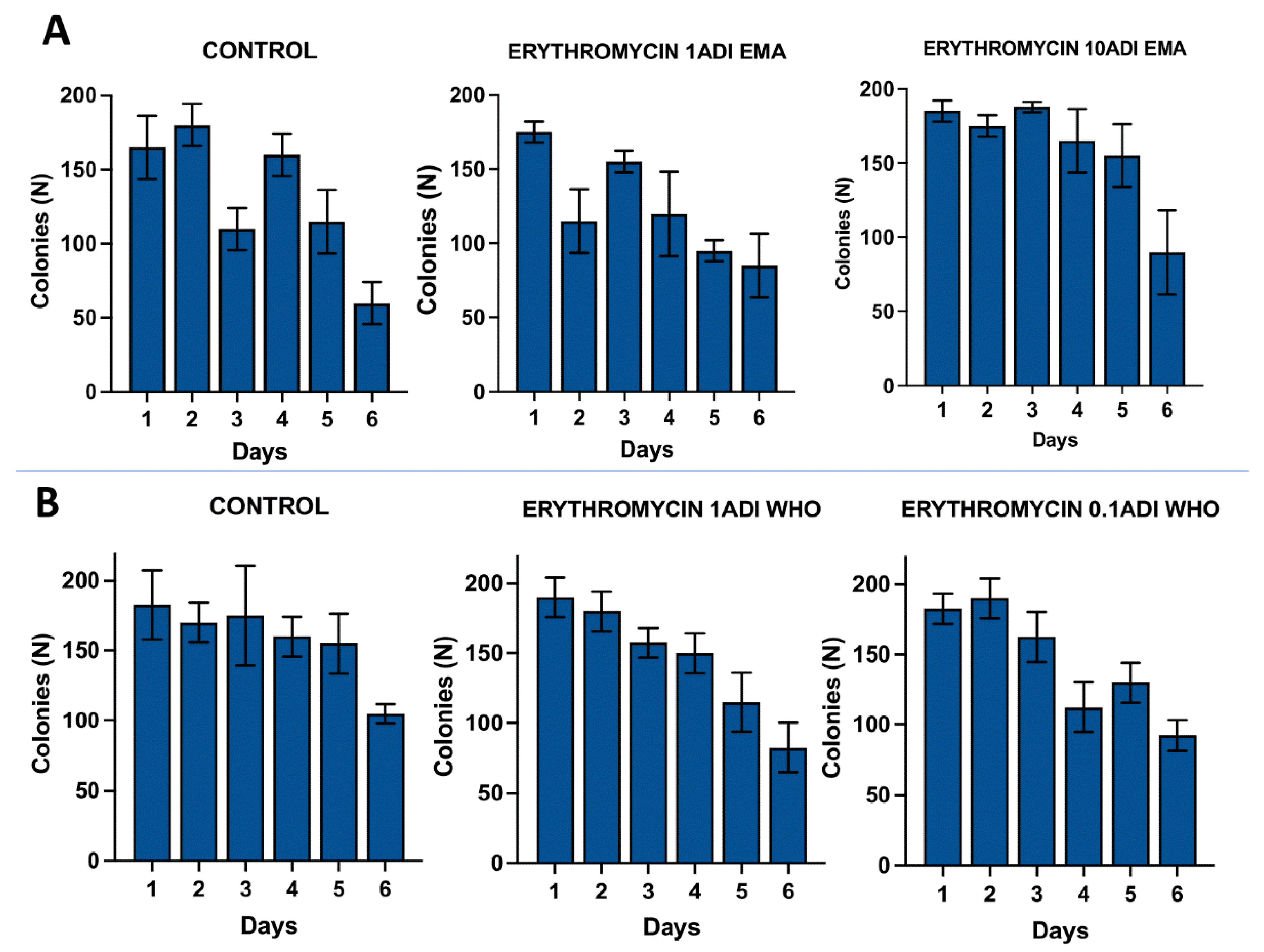
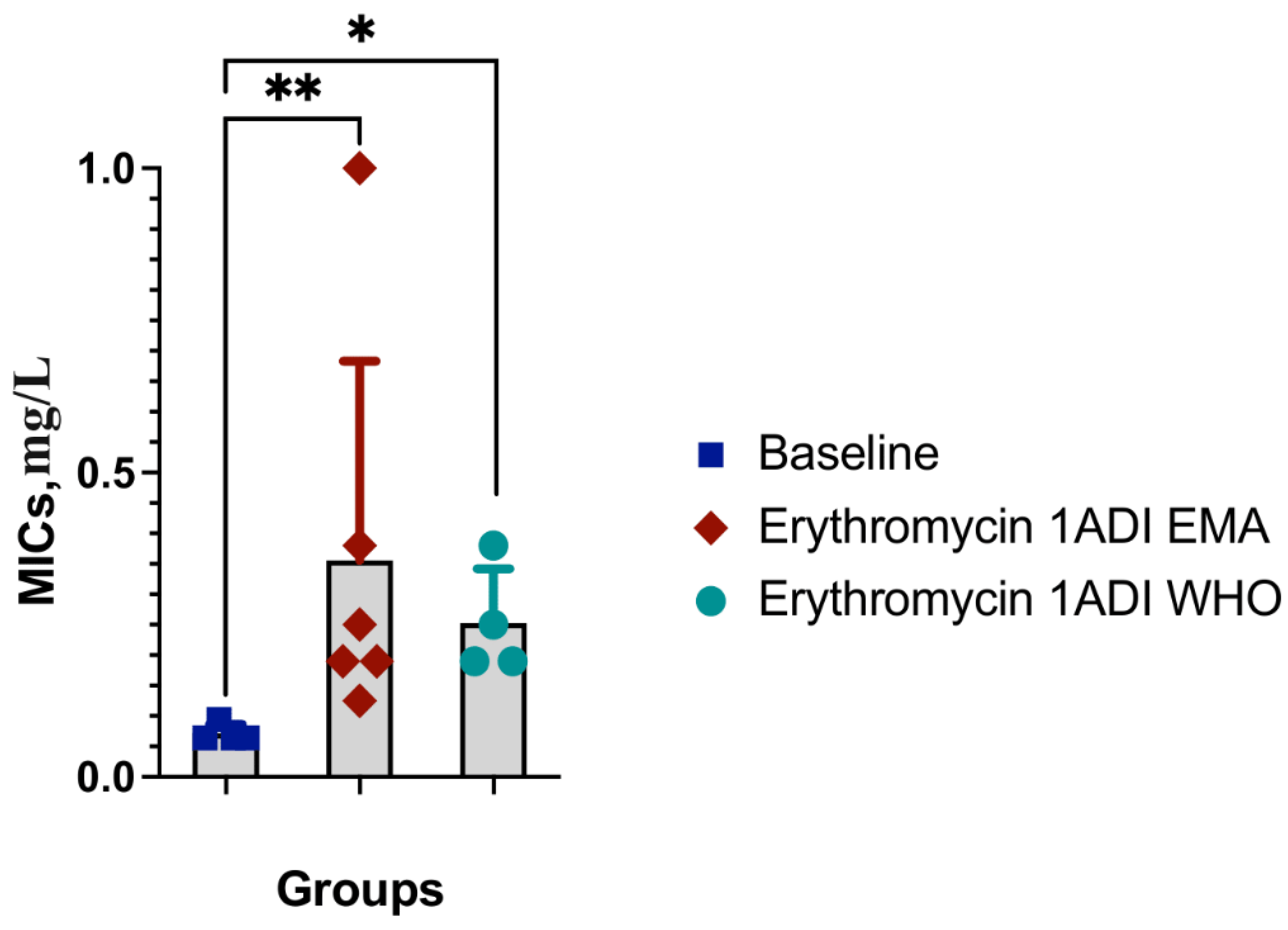
Results
| Gene Product | CDS/Gene | Strain | ||||||
|---|---|---|---|---|---|---|---|---|
| 2802b | 1403b | 1403c | 0303c | Ctrl_B2 | Ctrl_B4 | ATCC | ||
| Hypothetical Protein | OPMNIGBM_00355 | - | - | - | c.888A>C | - | - | - |
| Hypothetical Protein | OPMNIGBM_00536 | - | - | - | p.Arg77Trp | - | - | - |
| Tyrosine recombinase | xerS | - | - | - | p.Val324Leu | - | - | - |
| Hypothetical Protein | OPMNIGBM_00800 | - | - | - | p.Ser121Gly | - | - | - |
| Putative TrmH family tRNA/rRNA methyltransferase | OPMNIGBM_00823 | - | - | - | p.Ser203Arg | - | - | - |
| Hypothetical Protein | OPMNIGBM_00925 | - | - | - | p.Trp47Leu | - | - | - |
| Hypothetical Protein | OPMNIGBM_01263 | - | - | - | p.Leu8Ser | - | - | - |
| Hypothetical Protein | OPMNIGBM_00216 | - | p.Glu159* | - | NA | - | - | - |
| Hypothetical Protein | OPMNIGBM_00913 | - | p.His44Asn | - | NA | - | - | - |
| Arylsulfatase | OPMNIGBM_00351 | c.750T>C | - | c.750T>C | NA | - | - | - |
| Heat-inducible transcription repressor | hrcA | c.954C>A | - | c.954C>A | NA | - | - | - |
| 50S ribosomal protein L1 | rplA | p.Asn34Lys | - | p.Asn34Lys | NA | - | - | - |
| Hyothetical protein | OPMNIGBM_00258 | p.Cys102Arg | - | - | NA | - | - | - |
| ATP dependant DNA helicase | recG | c.837A>G | - | - | NA | - | - | - |
| Hypothetical Protein | OPMNIGBM_00430 | p.Asp27Asn | - | - | NA | - | - | - |
| Autolysin | lytA_2 | c.108C>T | - | - | NA | - | - | - |
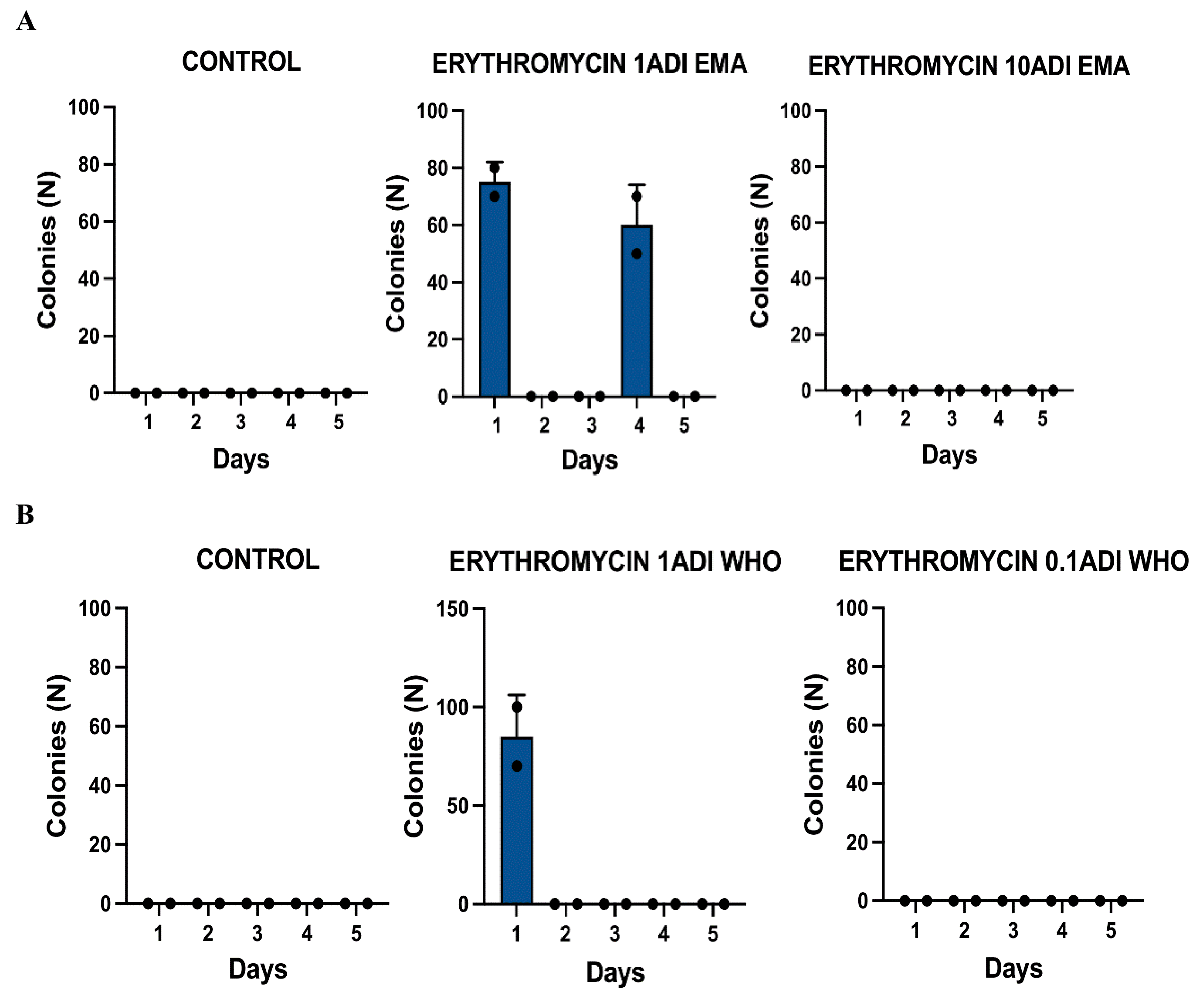
| Dose | Strain ID | Groups | Erythromycin injected | MIC | Day of experiment | Ribosomal protein mutations |
|---|---|---|---|---|---|---|
| ng | mg/L | rplA | ||||
| 1EMA ADI | 2802b | Test | 1.75 | 0.25 | 1 | Asn34Lys |
| 1EMA ADI | 0303c | Test | 1.75 | 0.190 | 4 | |
| 1WHO ADI | 1403b | Test | 0.25 | 0.25 | 1 | |
| 1WHO ADI | 1403c | Test | 0.25 | 0.38 | 1 | Asn34Lys |
| No dose | ATCC 49619 | Reference | 0.064 | |||
| No dose | Ctrl-B2 | Control | 0.125 | |||
| No dose | Ctrl-B4 | Control | 0.094 |
Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgements
Competing interests
Consent for publication
References
- Gullberg E, Albrecht LM, Karlsson C, et al. Selection of a Multidrug Resistance Plasmid by Sublethal Levels of Antibiotics and Heavy Metals Baquero F, ed. mBio 2014; 5. [CrossRef]
- Stanton IC, Murray AK, Zhang L, et al. Evolution of antibiotic resistance at low antibiotic concentrations including selection below the minimal selective concentration. Commun Biol 2020; 3. [CrossRef]
- Gullberg E, Cao S, Berg OG, et al. Selection of Resistant Bacteria at Very Low Antibiotic Concentrations Lipsitch M, ed. PLoS Pathog 2011; 7: e1002158. [CrossRef]
- Kraupner N, Ebmeyer S, Hutinel M, et al. Selective concentrations for trimethoprim resistance in aquatic environments. Environ Int 2020; 144: 106083. [CrossRef]
- Bengtsson-Palme J, Larsson DGJ. Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environ Int 2016; 86: 140–9. [CrossRef]
- EFSA Panel on Biological Hazards (BIOHAZ), Koutsoumanis K, Allende A, et al. Maximum levels of cross-contamination for 24 antimicrobial active substances in non-target feed. Part 1: Methodology, general data gaps and uncertainties. EFS2 2021; 19.
- Mitchell JM, Griffiths MW, Mcewen SA, et al. Antimicrobial Drug Residues in Milk and Meat: Causes, Concerns, Prevalence, Regulations, Tests, and Test Performance. J Food Prot 1998; 61: 742–56. [CrossRef]
- González N, Abdellati S, De Baetselier I, et al. Ciprofloxacin Concentrations 1/1000th the MIC Can Select for Antimicrobial Resistance in N. gonorrhoeae—Important Implications for Maximum Residue Limits in Food. Antibiotics 2022; 11: 1430. [CrossRef]
- Kenyon, C. Positive association between the use of macrolides in food-producing animals and pneumococcal macrolide resistance: a global ecological analysis. Int J Infect Dis 2022; 116: 344–7. [CrossRef]
- European Centre for Disease Prevention and Control (ECDC), European Food Safety Authority (EFSA), European Medicines Agency (EMA). ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals. EFS2 2017; 15.
- Allel K, Day L, Hamilton A, et al. Global antimicrobial-resistance drivers: an ecological country-level study at the human–animal interface. Lancet Planet Health 2023; 7: e291–303. [CrossRef]
- Tang KL, Caffrey NP, Nóbrega DB, et al. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planet Health 2017; 1: e316–27. [CrossRef]
- FAO/WHO. Codex Alimentarius: Guidelines for the simple evaluation of dietary exposure to food additives. CAC/GL 3-1989 Adopted 1989. Revision 2014. https://www.fao.org/input/download/standards/6/cxg_003e.pdf.
- Murray AK, Stanton I, Gaze WH, Snape J. Dawning of a new ERA: Environmental Risk Assessment of antibiotics and their potential to select for antimicrobial resistance. Water Res 2021; 200: 117233. [CrossRef]
- Subirats J, Domingues A, Topp E. Does Dietary Consumption of Antibiotics by Humans Promote Antibiotic Resistance in the Gut Microbiome? J Food Prot 2019; 82: 1636–42. [CrossRef]
- EMEA. The European Agency for the Evaluation of Medicinal Products. Committee for veterinary medicinal products: enrofloxacin summary report (2).1998. https://www.ema.europa.eu/en/documents/mrl-report/enrofloxacin-modification-bovine-porcine-poultry-summary-report-2-committee-veterinary-medicinal_en.pdf.
- EMEA. The European Agency for the Evaluation of Medicinal Products. Committee for veterinary medicinal products: erythromycin summary report (1).2000. https://www.ema.europa.eu/en/documents/mrl-report/erythromycin-erythromycin-thiocyante-erythromycin-stearate-summary-report-1-committee-veterinary_en.pdf.
- EMEA. The European Agency for the Evaluation of Medicinal Products. Committee for medicinal products for veterinary use. European public MRL assessment report (EPMAR).Tylvalosin.2007. https://www.ema.europa.eu/en/documents/mrl-report/tylvalosin-acetylisovaleryltylosin-modification-acceptable-daily-intake-adi-european-public-mrl_en.pdf.
- EMA. European Medicines Agency. Committee for Medicinal Products for Veterinary Use: VICH GL36(R2): Studies to evaluate the safety of residues of veterinary drugs in human food: general approach to establish a microbiological ADI.2019. https://www.ema.europa.eu/en/documents/scientific-guideline/vich-gl36r2-studies-evaluate-safety-residues-veterinary-drugs-human-food-general-approach-establish_en.pdf.
- FAO/WHO. Codex Alimentarius Commission (CAC): Maximum Residue Limits.2021. http://www.fao.org/fao-who-codexalimentarius/codex-texts/maximum-residue-limits/en/.
- FAO/WHO. Codex Alimentarius Commission Procedural Manual. Twenty-eighth edition, revised, 2023. Rome. [CrossRef]
- WHO. Evaluations of the Joint FAO/WHO Expert Committee on Food Additives (JECFA). Erythromycin. 2006. https://apps.who.int/food-additives-contaminants-jecfa-database/Home/Chemical/3938. 3938.
- EMEA. The European Agency for the Evaluation of Medicinal Products. Committee for veterinary medicinal products: erythromycin summary report (3).2002. https://www.ema.europa.eu/en/documents/mrl-report/erythromycin-extension-all-food-producing-species-summary-report-3-committee-veterinary-medicinal_en.pdf.
- Andrea A, Krogfelt K, Jenssen H. Methods and Challenges of Using the Greater Wax Moth (Galleria mellonella) as a Model Organism in Antimicrobial Compound Discovery. Microorganisms 2019; 7: 85. [CrossRef]
- Cutuli MA, Petronio Petronio G, Vergalito F, et al. Galleria mellonella as a consolidated in vivo model hosts: New developments in antibacterial strategies and novel drug testing. Virulence 2019; 10: 527–41. [CrossRef]
- Cools F, Torfs E, Aizawa J, et al. Optimization and Characterization of a Galleria mellonella Larval Infection Model for Virulence Studies and the Evaluation of Therapeutics Against Streptococcus pneumoniae. Front Microbiol 2019; 10.
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 13.0., 2023. http://www.eucast.org.
- Laumen JGE, Van Dijck C, Abdellati S, et al. Antimicrobial susceptibility of commensal Neisseria in a general population and men who have sex with men in Belgium. Sci Rep 2022; 12. [CrossRef]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–20. [CrossRef]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014; 30: 2068–9. [CrossRef]
- Lee JT, Li X, Hyde C, et al. PfaSTer: a machine learning-powered serotype caller for Streptococcus pneumoniae genomes. Microb Genom 2023; 9. [CrossRef]
- Chen L, Zheng D, Liu B, et al. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res 2015; 44: D694–7. [CrossRef]
- ia B, Raphenya AR, Alcock B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2016; 45: D566–73. [CrossRef]
- Felmingham D, Cantón R, Jenkins SG. Regional trends in β-lactam, macrolide, fluoroquinolone and telithromycin resistance among Streptococcus pneumoniae isolates 2001–2004. J Infect 2007; 55: 111–8. [CrossRef]
- Huang L-D, Yang M-J, Huang Y-Y, et al. Molecular Characterization of Predominant Serotypes, Drug Resistance, and Virulence Genes of Streptococcus pneumoniae Isolates From East China. Front Microbiol 2022; 13. [CrossRef]
- 36. Holmes AR, McNab R, Millsap KW, et al. The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol Microbiol 2001; 41: 1395–408. [CrossRef]
- Romero-Steiner S, Pilishvili T, Sampson JS, et al. Inhibition of Pneumococcal Adherence to Human Nasopharyngeal Epithelial Cells by Anti-PsaA Antibodies. Clin Vaccine Immunol 2003; 10: 246–51. [CrossRef]
- Marion C, Stewart JM, Tazi MF, et al. Streptococcus pneumoniae Can Utilize Multiple Sources of Hyaluronic Acid for Growth Camilli A, ed. Infect Immun 2012; 80: 1390–8. [CrossRef]
- Croney CM, Nahm MH, Juhn SK, et al. Invasive and Noninvasive Streptococcus pneumoniae Capsule and Surface Protein Diversity following the Use of a Conjugate Vaccine. Clin Vaccine Immunol 2013; 20: 1711–8. [CrossRef]
- Mellroth P, Sandalova T, Kikhney A, et al. Structural and Functional Insights into Peptidoglycan Access for the Lytic Amidase LytA of Streptococcus pneumoniae Rappuoli R, ed. mBio 2014; 5. [CrossRef]
- Rai P, He F, Kwang J, et al. Pneumococcal Pneumolysin Induces DNA Damage and Cell Cycle Arrest. Sci Rep 2016; 6. [CrossRef]
- Wren JT, Blevins LK, Pang B, et al. Pneumococcal Neuraminidase A (NanA) Promotes Biofilm Formation and Synergizes with Influenza A Virus in Nasal Colonization and Middle Ear Infection Pirofski L, ed. Infect Immun 2017; 85. [CrossRef]
- Subramanian, K.; Henriques-Normark, B.; Normark, S. Emerging concepts in the pathogenesis of theStreptococcus pneumoniae: From nasopharyngeal colonizer to intracellular pathogen. Cell Microbiol 2019; 21. [CrossRef]
- Zhao W, Pan F, Wang B, et al. Epidemiology Characteristics of Streptococcus pneumoniae From Children With Pneumonia in Shanghai: A Retrospective Study. Front Cell Infect Microbiol 2019; 9. [CrossRef]
- El Moujaber G, Osman M, Rafei R, et al. Molecular mechanisms and epidemiology of resistance in Streptococcus pneumoniae in the Middle East region. J Med Microbiol 2017; 66: 847–58. [CrossRef]
- Gill, M.J.; Brenwald, N.P.; Wise, R. Identification of an Efflux Pump Gene, pmrA, Associated with Fluoroquinolone Resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother 1999; 43: 187–9. [CrossRef]
- Piddock, L.J.V.; Johnson, M.M.; Simjee, S.; Pumbwe, L. Expression of Efflux Pump Gene pmrA in Fluoroquinolone-Resistant and -Susceptible Clinical Isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 2002; 46: 808–12. [CrossRef]
- Lee, H.; Hsu, F.-F.; Turk, J.; Groisman, E.A. The PmrA-Regulated pmrC Gene Mediates Phosphoethanolamine Modification of Lipid A and Polymyxin Resistance in Salmonella enterica. J Bacteriol 2004; 186: 4124–33. [CrossRef]
- El Garch F, Lismond A, Piddock LJV, et al. Fluoroquinolones induce the expression of patA and patB, which encode ABC efflux pumps in Streptococcus pneumoniae. J Antimicrob Chemother 2010; 65: 2076–82. [CrossRef]
- Garvey, M.I.; Baylay, A.J.; Wong, R.L.; Piddock, L.J.V. Overexpression of patA and patB, Which Encode ABC Transporters, Is Associated with Fluoroquinolone Resistance in Clinical Isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 2011; 55: 190–6. [CrossRef]
- Arthur, M.; Courvalin, P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother 1993; 37: 1563–71. [CrossRef]
- Arthur, M.; Molinas, C.; Depardieu, F.; Courvalin, P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol 1993; 175: 117–27. [CrossRef]
- Monteiro da Silva BN, Faria AR, Souza S da SR, et al. Expression of VanA-type vancomycin resistance in a clinical isolate of Enterococcus faecium showing insertion of IS19 in the vanS gene. Int J Antimicrob Agents 2020; 55: 105897. [CrossRef]
- Clewell DB, Weaver KE, Dunny GM, et al. Extrachromosomal and Mobile Elements in Enterococci: Transmission, Maintenance, and Epidemiology. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston: Massachusetts Eye and Ear Infirmary, 2014.
- Hong, H.-J.; Hutchings, M.I.; Buttner, M.J. Vancomycin Resistance VanS/VanR Two-Component Systems. Adv Exp Med Biol 2008; 631: 200–13. [CrossRef]
- Gooch HCC, Kiu R, Rudder S, et al. Enterococcus innesii sp. nov., isolated from the wax moth Galleria mellonella. Int J Syst Evol Microbiol 2021; 71. [CrossRef]
- Allonsius CN, Van Beeck W, De Boeck I, et al. The microbiome of the invertebrate model host Galleria mellonella is dominated by Enterococcus. Anim microbiome 2019; 1. [CrossRef]
- Ambrose, K.D.; Nisbet, R.; Stephens, D.S. Macrolide Efflux in Streptococcus pneumoniae Is Mediated by a Dual Efflux Pump (mel and mef) and Is Erythromycin Inducible. Antimicrob Agents Chemother 2005; 49: 4203–9. [CrossRef]
- Schroeder, M.R.; Stephens, D.S. Macrolide Resistance in Streptococcus pneumoniae. Front Cell Infect Microbiol 2016; 6. [CrossRef]
- Tait-Kamradt A, Davies T, Appelbaum PC, et al. Two New Mechanisms of Macrolide Resistance in Clinical Strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob Agents Chemother 2000; 44: 3395–401. [CrossRef]
- Tait-Kamradt A, Davies T, Cronan M, et al. Mutations in 23S rRNA and Ribosomal Protein L4 Account for Resistance in Pneumococcal Strains Selected In Vitro by Macrolide Passage. Antimicrob Agents Chemother 2000; 44: 2118–25. [CrossRef]
- Canu A, Malbruny B, Coquemont M, et al. Diversity of Ribosomal Mutations Conferring Resistance to Macrolides, Clindamycin, Streptogramin, and Telithromycin in Streptococcus pneumoniae. Antimicrob Agents Chemother 2002; 46: 125–31. [CrossRef]
- Nevskaya, N. Ribosomal protein L1 recognizes the same specific structural motif in its target sites on the autoregulatory mRNA and 23S rRNA. Nucleic Acids Res 2005; 33: 478–85. [CrossRef]
- Calvopiña, K.; Dulyayangkul, P.; Avison, M.B. Mutations in Ribosomal Protein RplA or Treatment with Ribosomal Acting Antibiotics Activates Production of Aminoglycoside Efflux Pump SmeYZ in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 2020; 64. [CrossRef]
- Waldner M, Kinnear A, Yacoub E, et al. Genome-Wide Association Study of Nucleotide Variants Associated with Resistance to Nine Antimicrobials in Mycoplasma bovis. Microorganisms 2022; 10: 1366. [CrossRef]
- Schlünzen F, Zarivach R, Harms J, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 2001; 413: 814–21. [CrossRef]
- Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK Antibiotics Bound to Mutated Large Ribosomal Subunits Provide a Structural Explanation for Resistance. Cell 2005; 121: 257–70. [CrossRef]
- Wilson, D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 2013; 12: 35–48. [CrossRef]
- Ching, C.; Zaman, M.H. Development and selection of low-level multi-drug resistance over an extended range of sub-inhibitory ciprofloxacin concentrations in Escherichia coli. Sci Rep 2020; 10. [CrossRef]
- Ching, C.; Zaman, M.H. Identification of Multiple Low-Level Resistance Determinants and Coselection of Motility Impairment upon Sub-MIC Ceftriaxone Exposure in Escherichia coli Gales AC, ed. mSphere 2021; 6. [CrossRef]
- Gomez JE, Kaufmann-Malaga BB, Wivagg CN, et al. Ribosomal mutations promote the evolution of antibiotic resistance in a multidrug environment. eLife 2017; 6. [CrossRef]
- Laumen JGE, Manoharan-Basil SS, Verhoeven E, et al. Molecular pathways to high-level azithromycin resistance in Neisseria gonorrhoeae. J Antimicrob Chemother 2021; 76: 1752–8. [CrossRef]
- Fyfe, C.; Grossman, T.H.; Kerstein, K.; Sutcliffe, J. Resistance to Macrolide Antibiotics in Public Health Pathogens. Cold Spring Harb Perspect Med 2016; 6: a025395. [CrossRef]
- Seiler, C.; Berendonk, T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbio 2012; 3. [CrossRef]
- Jin M, Lu J, Chen Z, et al. Antidepressant fluoxetine induces multiple antibiotics resistance in Escherichia coli via ROS-mediated mutagenesis. Environ Int 2018; 120: 421–30. [CrossRef]
- Hao H, Gokulan K, Piñeiro SA, et al. Effects of Acute and Chronic Exposure to Residual Level Erythromycin on Human Intestinal Epithelium Cell Permeability and Cytotoxicity. Microorganisms 2019; 7: 325. [CrossRef]
- Van Boeckel TP, Glennon EE, Chen D, et al. Reducing antimicrobial use in food animals. Science 2017; 357: 1350–2. [CrossRef]
- FDA, Center For Veterinary Medicine. 2021 Summary Report On Antimicrobials Sold or Distributed for Use in Food-Producing animals. 2022. https://www.fda.gov/media/163739/download.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





