Submitted:
01 March 2024
Posted:
01 March 2024
You are already at the latest version
Abstract
Keywords:
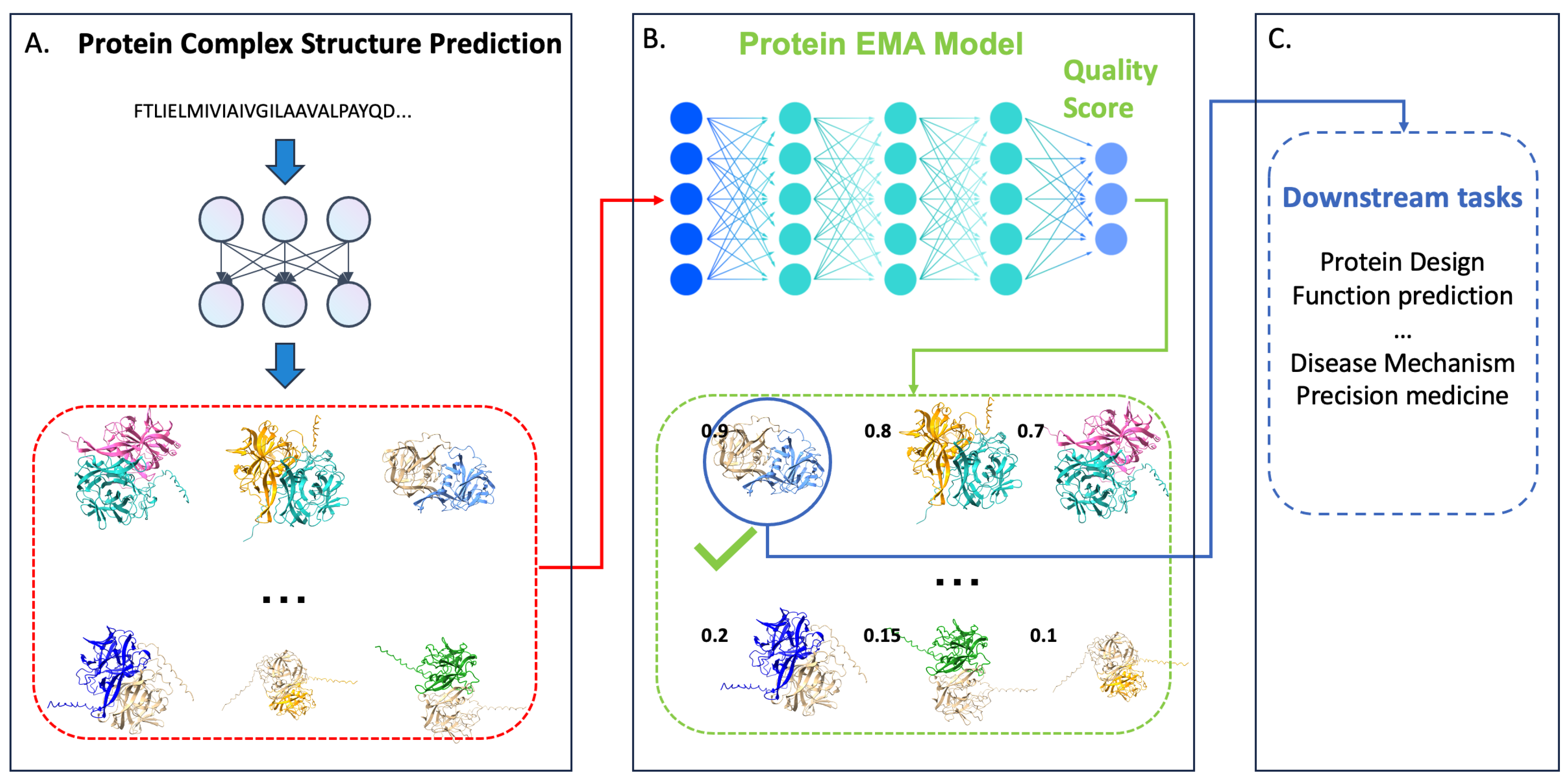
1. Introduction
2. Metrics for Evaluating the Quality of Protein Complex Structures and the Performance of EMA Methods
2.1. Global Structural Quality Evaluation of Protein Complex Structure
2.2. Interface Quality Evaluation of Protein Complex Structure
2.3. Local Structural Quality Evaluation of Protein Complex Structure
2.4. Metrics of Evaluating Protein Complex Structure EMA
3. Learning the Representation of Protein Complex Structure
3.1. Protein Complex Structure Representation
3.2. Graph Neural Network
3.2.1. Graph Convolutional Neural Network
3.2.2. Graph Attention Neural Network
3.2.3. Graph Transformer Neural Network
4. Datasets for Training and Test Protein Complex EMA Methods
| Data Sources | Number of Targets/Structures | Source |
|---|---|---|
| DockGround | 61/6100 | Link |
| Docking Benchmark | 230/* | Link |
| PPI4DOCK | 1417/54000 | Link |
| CARPI set | 15/19013 | Link |
| CASP15 | 38/9930 | Link |
| DBM55-AF2 | 15/450 | Link |
5. Deep Learning-based EMA Methods for Protein Complex Structure
| Name | Year* | Main Techniques |
Prediction | Representation Level |
Single-/ Multi-model |
|---|---|---|---|---|---|
| PAUL [44] | 2020 | Equivariant-GCN | iRMSD | Atom | Single |
| DOVE [17] | 2020 | 3D-CNN | The probability of an input decoy has a acceptable quality or not | Atom | Single |
| EGCN [12] | 2020 | GCN | iRMSD | Residue | Single |
| GNN_DOVE [18] | 2021 | GAT | The probability of an input decoy has a acceptable quality or not | Atom | Single |
| DGANN [66] | 2021 | GAT | The probability of an input decoy is near-native or not | Residue | Single |
| Trscore [41] | 2022 | 3D-CNN | The probability of an input decoy is near-native or not | Atom | Single |
| DeepRank_GNN [25] | 2022 | GNN | f-nat (fraction of native contacts) | Residue | Single |
| VoroIF-GNN [67] | 2023 | GAT | CAD score | Atom | Single |
| DeepUMQA3 [27,45] | 2023 | 2D-CNN | lDDT | Residue | Single |
| DProQA [24] | 2023 | GT | DockQ | Residue | Single |
| G-RANK [30] | 2023 | GVP | f-nat (fraction of native contacts) | Atom | Single |
| PIQLE [31] | 2023 | GAT | Interface score, Fold score, Residue score | Residue | Single |
| GraphGPSM [46] | 2023 | EGNN | TM-Score | Residue | Single |
| GraphCPLMQA [47] | 2023 | GT+EGNN+2DCNN | lDDT | Residue | Single |
| PointDE [42] | 2023 | Point cloud network | The probability of an input decoy is near-native or not | Atom | Single |
| ComplexQA [48] | 2023 | GCN | Interface residue score | Residue | Single |
| GCPNet-EMA [49] | 2024 | GCP | lDDT | Residue | Single |
| Name | Features |
|---|---|
| PAUL | Atomic positions and types |
| DOVE | Contact potentials, GOAP, ITScore |
| EGCN | Node features: side-chain pseudo atom’s charge, non-bonded radii, and distance-to-Ca, solvent accessible surface area(SASA). Edge features: Atom distance features |
| GNN_DOVE | Node features: atom physicochemical proprieties of atoms. Edge features: covalent bonds, atom distance. |
| DGANN | Node features: physical-chemical properties, PSSM, Information Content |
| Trscore | Atoms’ physicochemical features |
| DeepRank_GNN | Node features: residue type, residue charge, residue polarity, buried surface area, PSSM; conservation score, information content, residue depth, residue half sphere exposure. Edge feature: residue distance |
| VoroIF-GNN | Node features: contact surface areas, contact-solvent border length, sum of inter-contact border lengths; contact type-dependent descriptors. Edge feature: Inter-contact border length |
| DeepUMQA3 | Ultrafast Shape Recognition (USR), residue voxelization, inter-residue distance and orientations, amino acid properties; level of intra-monomer: sequence embedding, secondary structure, energy terms; inter-monomer level: attention map of the inter-monomer paired sequence, inter-monomer USR |
| DProQA | Node features: residue type, secondary structure type, relative accessible surface area, torsion angles, node positional encoding. Edge features: Three types of distance, edge positional encoding, contact indicator, permutation-invariant chain encoding |
| G-RANK | Node features: atom types; Edge features: edge direction, edge length |
| PIQLE | Node features: residue encoding, relative residue positioning, secondary structure, SASA, torsion angles, number of effective sequences (Neff). Edge features: multimeric interaction distance, multimeric interaction orientation |
| GraphGPSM | USR, residue voxelization, inter-residue distance and orientations, amino acid properties; level of intra-monomer: sequence embedding, secondary structure, energy terms; inter-monomer level: attention map of the inter-monomer, paired sequence, inter-monomer USR, Ca coordinates |
| GraphCPLMQA | MSA embedding, sequence embedding, structure embedding, triangular location and residue-level contact order, relative position encoding, dihedral and planar angles, voxelization and distance map, Meiler, Blosum62 and DSSP |
| PointDE | Atomic type, residue types and coordinate, chain identity |
| ComplexQA | Sequence features, three-dimensional structural and chemical features |
| GCPNet-EMA | Node features: residue type, positional encoding, virtual dihedral and bond Angles over the trace, residue backbone dihedral angles; Residue-wise ESM Embeddings, residue-wise AlphaFold 2 plDDT, residue-sequential forward and backward vectors; Edge features: Euclidean Distance between connected atoms, directional vector between connected atoms |
| Name | Training Data | Testing data | Source |
|---|---|---|---|
| PAUL | DBM4 | DBM5, PPI4DOCK | NA |
| DOVE | DBM4 | DockGround | https://kiharalab.org/dove/ |
| EGCN | DBM4 | CAPRI | https://github.com/Shen-Lab/EGCN |
| GNN_DOVE | Dockground, DBM4 | CAPRI | https://github.com/kiharalab/GNN_DOVE |
| DGANN | DBM4 | DBM5.5 | https://github.com/coffee19850519/PPDocking/tree/master |
| Trscore | DBM4 | DBM5 | https://github.com/BioinformaticsCSU/TRScore |
| DeepRank_GNN | DBM5 | CAPRI | https://github.com/DeepRank/Deeprank-GNN |
| VoroIF-GNN | Custom set | Custom set | https://www.voronota.com/expansion_js/ |
| DeepUMQA3 | Custom set | Custom set | http://zhanglab-bioinf.com/DeepUMQA/ |
| DProQA | Dockground, DBM5.5, Custom Dataset | Custom Dataset | https://github.com/jianlin-cheng/DProQA/tree/main |
| G-RANK | DBM5 | CAPRI | https://github.com/ha01994/grank |
| PIQLE | Dockground v2 | Dockground v1 | https://github.com/Bhattacharya-Lab/PIQLE |
| GraphGPSM | Custom set | CASP15 | http://zhanglab-bioinf.com/GraphGPSM/ |
| GraphCPLMQA | Custom set | CASP15 | http://zhanglab-bioinf.com/GraphCPLMQA/ |
| PointDE | DOCKGROUND | CAPRI, Custom Dataset | https://github.com/AI-ProteinGroup/PointDE |
| ComplexQA | DockGround, DBM5, Custom Dataset | Custom set | https://github.com/Cao-Labs/ComplexQA/tree/main |
| CGPNet-EMA | Custom set | CASP15, Custom set | https://github.com/BioinfoMachineLearning/GCPNet-EMA |
6. Future Work
Author Contributions
Funding
Conflicts of Interest
References
- Scott, D.E.; Bayly, A.R.; Abell, C.; Skidmore, J. Small molecules, big targets: drug discovery faces the protein–protein interaction challenge. Nature Reviews Drug Discovery 2016, 15, 533–550. [Google Scholar] [CrossRef] [PubMed]
- Athanasios, A.; Charalampos, V.; Vasileios, T.; Md Ashraf, G. Protein-protein interaction (PPI) network: recent advances in drug discovery. Current drug metabolism 2017, 18, 5–10. [Google Scholar] [CrossRef]
- Macalino, S.J.Y.; Basith, S.; Clavio, N.A.B.; Chang, H.; Kang, S.; Choi, S. Evolution of in silico strategies for protein-protein interaction drug discovery. Molecules 2018, 23, 1963. [Google Scholar] [CrossRef] [PubMed]
- Baker, D. Prediction and design of macromolecular structures and interactions. Philosophical Transactions of the Royal Society B: Biological Sciences 2006, 361, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Lippow, S.M.; Tidor, B. Progress in computational protein design. Current opinion in biotechnology 2007, 18, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J. Protein complex prediction with AlphaFold-Multimer. biorxiv 2021, 2021-10. [Google Scholar]
- Skolnick, J.; Kolinski, A.; Ortiz, A. Derivation of protein-specific pair potentials based on weak sequence fragment similarity. Proteins: structure, function, and bioinformatics 2000, 38, 3–16. [Google Scholar] [CrossRef]
- Lu, H.; Skolnick, J. A distance-dependent atomic knowledge-based potential for improved protein structure selection. Proteins: Structure, Function, and Bioinformatics 2001, 44, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.Y.; Zou, X. An iterative knowledge-based scoring function for protein–protein recognition. Proteins: Structure, Function, and Bioinformatics 2008, 72, 557–579. [Google Scholar] [CrossRef]
- Vreven, T.; Hwang, H.; Weng, Z. Integrating atom-based and residue-based scoring functions for protein–protein docking. Protein Science 2011, 20, 1576–1586. [Google Scholar] [CrossRef]
- Basu, S.; Wallner, B. Finding correct protein–protein docking models using ProQDock. Bioinformatics 2016, 32, i262–i270. [Google Scholar] [CrossRef]
- Cao, Y.; Shen, Y. Energy-based graph convolutional networks for scoring protein docking models. Proteins: Structure, Function, and Bioinformatics 2020, 88, 1091–1099. [Google Scholar] [CrossRef]
- Geng, C.; Jung, Y.; Renaud, N.; Honavar, V.; Bonvin, A.M.; Xue, L.C. iScore: a novel graph kernel-based function for scoring protein–protein docking models. Bioinformatics 2020, 36, 112–121. [Google Scholar] [CrossRef]
- Lyskov, S.; Gray, J.J. The RosettaDock server for local protein–protein docking. Nucleic acids research 2008, 36, W233–W238. [Google Scholar] [CrossRef] [PubMed]
- Torchala, M.; Moal, I.H.; Chaleil, R.A.; Fernandez-Recio, J.; Bates, P.A. SwarmDock: a server for flexible protein–protein docking. Bioinformatics 2013, 29, 807–809. [Google Scholar] [CrossRef] [PubMed]
- Vangone, A.; Rodrigues, J.; Xue, L.; van Zundert, G.; Geng, C.; Kurkcuoglu, Z.; Nellen, M.; Narasimhan, S.; Karaca, E.; van Dijk, M.; et al. Sense and simplicity in HADDOCK scoring: Lessons from CASP-CAPRI round 1. Proteins: Structure, Function, and Bioinformatics 2017, 85, 417–423. [Google Scholar] [CrossRef]
- Wang, X.; Terashi, G.; Christoffer, C.W.; Zhu, M.; Kihara, D. Protein docking model evaluation by 3D deep convolutional neural networks. Bioinformatics 2020, 36, 2113–2118. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Flannery, S.T.; Kihara, D. Protein docking model evaluation by graph neural networks. Frontiers in Molecular Biosciences 2021, 8, 647915. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.S.; Liu, J.; Zhou, X.G.; Zhang, G.J. DeepUMQA: ultrafast shape recognition-based protein model quality assessment using deep learning. Bioinformatics 2022, 38, 1895–1903. [Google Scholar] [CrossRef]
- Morehead, A.; Chen, X.; Wu, T.; Liu, J.; Cheng, J. EGR: Equivariant Graph Refinement and Assessment of 3D Protein Complex Structures. arXiv, 2022; arXiv:2205.10390. [Google Scholar]
- Olechnovic, K.; Venclovas, C. VoroIF-GNN: Voronoi tessellation-derived protein-protein interface assessment using a graph neural network. bioRxiv 2023, 2023–04. [Google Scholar] [CrossRef]
- Edmunds, N.S.; Alharbi, S.M.; Genc, A.G.; Adiyaman, R.; McGuffin, L.J. Estimation of model accuracy in CASP15 using the M odFOLDdock server. Proteins: Structure, Function, and Bioinformatics, 2023. [Google Scholar]
- Roy, R.S.; Liu, J.; Giri, N.; Guo, Z.; Cheng, J. Combining pairwise structural similarity and deep learning interface contact prediction to estimate protein complex model accuracy in CASP15. Proteins: Structure, Function, and Bioinformatics, 2023. [Google Scholar]
- Chen, X.; Morehead, A.; Liu, J.; Cheng, J. A gated graph transformer for protein complex structure quality assessment and its performance in CASP15. Bioinformatics 2023, 39, i308–i317. [Google Scholar] [CrossRef] [PubMed]
- Réau, M.; Renaud, N.; Xue, L.C.; Bonvin, A.M. DeepRank-GNN: a graph neural network framework to learn patterns in protein–protein interfaces. Bioinformatics 2023, 39, btac759. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, K.; Zhang, G. Improved model quality assessment using sequence and structural information by enhanced deep neural networks. Briefings in bioinformatics 2023, 24, bbac507. [Google Scholar] [CrossRef]
- Liu, J.; Liu, D.; Zhang, G.J. DeepUMQA3: a web server for accurate assessment of interface residue accuracy in protein complexes. Bioinformatics, 2023; btad591. [Google Scholar]
- Liu, D.; Zhang, B.; Liu, J.; Li, H.; Song, L.; Zhang, G. GraphCPLMQA: Assessing protein model quality based on deep graph coupled networks using protein language model. bioRxiv, 2023; 2023–05. [Google Scholar]
- Han, Y.; Zhang, S.; He, F. A Point Cloud-Based Deep Learning Model for Protein Docking Decoys Evaluation. Mathematics 2023, 11, 1817. [Google Scholar] [CrossRef]
- Kim, H.Y.; Kim, S.; Park, W.Y.; Kim, D. G-RANK: an equivariant graph neural network for the scoring of protein–protein docking models. Bioinformatics Advances 2023, 3, vbad011. [Google Scholar] [CrossRef]
- Shuvo, M.H.; Karim, M.; Roche, R.; Bhattacharya, D. PIQLE: protein-protein interface quality estimation by deep graph learning of multimeric interaction geometries. Bioinformatics Advances.
- Moal, I.H.; Barradas-Bautista, D.; Jiménez-García, B.; Torchala, M.; van der Velde, A.; Vreven, T.; Weng, Z.; Bates, P.A.; Fernández-Recio, J. IRaPPA: information retrieval based integration of biophysical models for protein assembly selection. Bioinformatics 2017, 33, 1806–1813. [Google Scholar] [CrossRef]
- Lensink, M.F.; Brysbaert, G.; Raouraoua, N.; Bates, P.A.; Giulini, M.; Honorato, R.V.; van Noort, C.; Teixeira, J.M.; Bonvin, A.M.; Kong, R. Impact of AlphaFold on structure prediction of protein complexes: The CASP15-CAPRI experiment. Proteins: Structure, Function, and Bioinformatics, 2023. [Google Scholar]
- Zemla, A. LGA: a method for finding 3D similarities in protein structures. Nucleic acids research 2003, 31, 3370–3374. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins: Structure, Function, and Bioinformatics 2004, 57, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Wallner, B. DockQ: a quality measure for protein-protein docking models. PloS one 2016, 11, e0161879. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo-and hetero-oligomers beyond binary interactions by homology. Scientific reports 2017, 7, 10480. [Google Scholar] [CrossRef]
- Olechnovič, K.; Kulberkytė, E.; Venclovas, Č. CAD-score: a new contact area difference-based function for evaluation of protein structural models. Proteins: Structure, Function, and Bioinformatics 2013, 81, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Mariani, V.; Biasini, M.; Barbato, A.; Schwede, T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 2013, 29, 2722–2728. [Google Scholar] [CrossRef] [PubMed]
- Barradas-Bautista, D.; Cao, Z.; Vangone, A.; Oliva, R.; Cavallo, L. A random forest classifier for protein–protein docking models. Bioinformatics Advances 2022, 2, vbab042. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; He, J.; Lin, P.; Huang, S.Y.; Wang, J. TRScore: a 3D RepVGG-based scoring method for ranking protein docking models. Bioinformatics 2022, 38, 2444–2451. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liu, N.; Huang, Y.; Min, X.; Zeng, X.; Ge, S.; Zhang, J.; Xia, N. PointDE: Protein Docking Evaluation Using 3D Point Cloud Neural Network. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2023. [Google Scholar]
- Chen, C.; Chen, X.; Morehead, A.; Wu, T.; Cheng, J. 3D-equivariant graph neural networks for protein model quality assessment. Bioinformatics 2023, 39, btad030. [Google Scholar] [CrossRef] [PubMed]
- Eismann, S.; Townshend, R.J.; Thomas, N.; Jagota, M.; Jing, B.; Dror, R.O. Hierarchical, rotation-equivariant neural networks to select structural models of protein complexes. Proteins: Structure, Function, and Bioinformatics 2021, 89, 493–501. [Google Scholar] [CrossRef]
- Liu, J.; Liu, D.; He, G.; Zhang, G. Estimating protein complex model accuracy based on ultrafast shape recognition and deep learning in CASP15. Proteins: Structure, Function, and Bioinformatics, 2023. [Google Scholar]
- He, G.; Liu, J.; Liu, D.; Zhang, G. GraphGPSM: a global scoring model for protein structure using graph neural networks. Briefings in Bioinformatics 2023, 24, bbad219. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, B.; Liu, J.; Li, H.; Song, L.; Zhang, G. Assessing protein model quality based on deep graph coupled networks using protein language model. Briefings in Bioinformatics 2024, 25, bbad420. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, S.; Hou, J.; Si, D.; Zhu, J.; Cao, R. ComplexQA: a deep graph learning approach for protein complex structure assessment. Briefings in Bioinformatics 2023, 24, bbad287. [Google Scholar] [CrossRef]
- Morehead, A.; Liu, J.; Cheng, J. Protein Structure Accuracy Estimation using Geometry-Complete Perceptron Networks. Protein Science 2024. [Google Scholar] [CrossRef]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. arXiv, 2016; arXiv:1609.02907. [Google Scholar]
- Velickovic, P.; Cucurull, G.; Casanova, A.; Romero, A.; Lio, P.; Bengio, Y.; et al. Graph attention networks. stat 2017, 1050, 10–48550. [Google Scholar]
- Dwivedi, V.P.; Bresson, X. A generalization of transformer networks to graphs. arXiv, 2020; arXiv:2012.09699. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.u.; Polosukhin, I. Attention is All you Need. In Proceedings of the Advances in Neural Information Processing Systems; Guyon, I.; Luxburg, U.V.; Bengio, S.; Wallach, H.; Fergus, R.; Vishwanathan, S.; Garnett, R., Eds. Curran Associates, Inc., Vol. 30. 2017. [Google Scholar]
- Liu, S.; Gao, Y.; Vakser, I.A. Dockground protein–protein docking decoy set. Bioinformatics 2008, 24, 2634–2635. [Google Scholar] [CrossRef] [PubMed]
- Tovchigrechko, A.; Vakser, I.A. Development and testing of an automated approach to protein docking. Proteins: Structure, Function, and Bioinformatics 2005, 60, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Tovchigrechko, A.; Vakser, I.A. GRAMM-X public web server for protein–protein docking. Nucleic acids research 2006, 34, W310–W314. [Google Scholar] [CrossRef]
- Chen, R.; Mintseris, J.; Janin, J.; Weng, Z. A protein–protein docking benchmark. Proteins: Structure, Function, and Bioinformatics 2003, 52, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Vreven, T.; Moal, I.H.; Vangone, A.; Pierce, B.G.; Kastritis, P.L.; Torchala, M.; Chaleil, R.; Jiménez-García, B.; Bates, P.A.; Fernandez-Recio, J.; et al. Updates to the integrated protein–protein interaction benchmarks: docking benchmark version 5 and affinity benchmark version 2. Journal of molecular biology 2015, 427, 3031–3041. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Guerois, R. PPI4DOCK: large scale assessment of the use of homology models in free docking over more than 1000 realistic targets. Bioinformatics 2016, 32, 3760–3767. [Google Scholar] [CrossRef]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.H.; Vreven, T.; Weng, Z. ZDOCK server: interactive docking prediction of protein–protein complexes and symmetric multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef]
- Lensink, M.F.; Wodak, S.J. Score_set: a CAPRI benchmark for scoring protein complexes. Proteins: Structure, Function, and Bioinformatics 2014, 82, 3163–3169. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Bryant, P.; Pozzati, G.; Elofsson, A. Improved prediction of protein-protein interactions using AlphaFold2. Nat Commun 13: 1265, 2022.
- Gabb, H.A.; Jackson, R.M.; Sternberg, M.J. Modelling protein docking using shape complementarity, electrostatics and biochemical information. Journal of molecular biology 1997, 272, 106–120. [Google Scholar] [CrossRef]
- Huang, X.; Pearce, R.; Zhang, Y. FASPR: an open-source tool for fast and accurate protein side-chain packing. Bioinformatics 2020, 36, 3758–3765. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; He, F.; Chen, Y.; Qin, W.; Yu, H.; Xu, D. Quality assessment of protein docking models based on graph neural network. Frontiers in Bioinformatics 2021, 1, 693211. [Google Scholar] [CrossRef]
- Olechnovic, K.; VoroIF-GNN, V.C. Voronoi tessellation-derived protein-protein interface assessment using a graph neural network. Proteins Struct Funct Bioinformatics 2023. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Zhang, X.; Ma, N.; Han, J.; Ding, G.; Sun, J. Repvgg: Making vgg-style convnets great again. In Proceedings of the Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 2021, pp. 13733–13742.
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp.; pp. 770–778.
- Pierce, B.; Weng, Z. ZRANK: reranking protein docking predictions with an optimized energy function. Proteins: Structure, Function, and Bioinformatics 2007, 67, 1078–1086. [Google Scholar] [CrossRef]
- Pierce, B.; Weng, Z. A combination of rescoring and refinement significantly improves protein docking performance. Proteins: Structure, Function, and Bioinformatics 2008, 72, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Skolnick, J. GOAP: a generalized orientation-dependent, all-atom statistical potential for protein structure prediction. Biophysical journal 2011, 101, 2043–2052. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.; Vreven, T.; Janin, J.; Weng, Z. Protein–protein docking benchmark version 4.0. Proteins: Structure, Function, and Bioinformatics 2010, 78, 3111–3114. [Google Scholar] [CrossRef]
- Dominguez, C.; Boelens, R.; Bonvin, A.M. HADDOCK: a protein- protein docking approach based on biochemical or biophysical information. Journal of the American Chemical Society 2003, 125, 1731–1737. [Google Scholar] [CrossRef]
- Renaud, N.; Geng, C.; Georgievska, S.; Ambrosetti, F.; Ridder, L.; Marzella, D.F.; Réau, M.F.; Bonvin, A.M.; Xue, L.C. DeepRank: a deep learning framework for data mining 3D protein-protein interfaces. Nature communications 2021, 12, 7068. [Google Scholar] [CrossRef]
- Jing, B.; Eismann, S.; Suriana, P.; Townshend, R.J.; Dror, R. Learning from protein structure with geometric vector perceptrons. arXiv, 2020; arXiv:2009.01411. [Google Scholar]
- Morehead, A.; Cheng, J. Geometry-complete perceptron networks for 3d molecular graphs. arXiv, 2022; arXiv:2211.02504. [Google Scholar]
- Ma, X.; Qin, C.; You, H.; Ran, H.; Fu, Y. Rethinking network design and local geometry in point cloud: A simple residual MLP framework. arXiv, 2022; arXiv:2202.07123. [Google Scholar]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023, 379, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Leaver-Fay, A.; Tyka, M.; Lewis, S.M.; Lange, O.F.; Thompson, J.; Jacak, R.; Kaufman, K.W.; Renfrew, P.D.; Smith, C.A.; Sheffler, W.; et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. In Methods in enzymology; Elsevier, 2011; Vol. 487, pp. 545–574.
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic acids research 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





