Submitted:
26 February 2024
Posted:
26 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Review of Previous Work
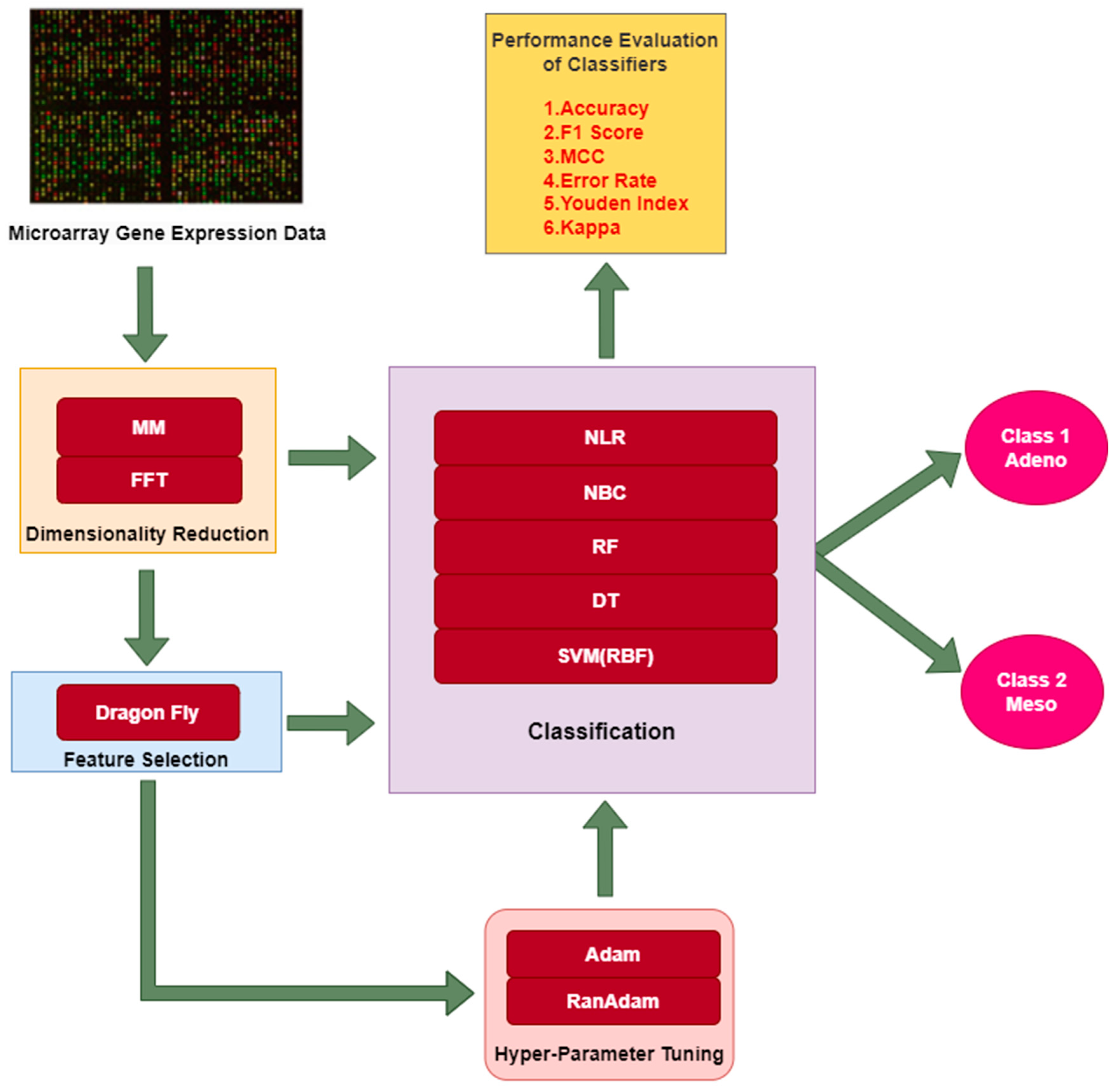
2. Materials and Methods
2.1. Details about the Dataset
2.2. Dimensionality Reduction (DimRe)
2.2.1. Mixture Model for DimRe
2.2.2. Fast Fourier Transform for DimRe
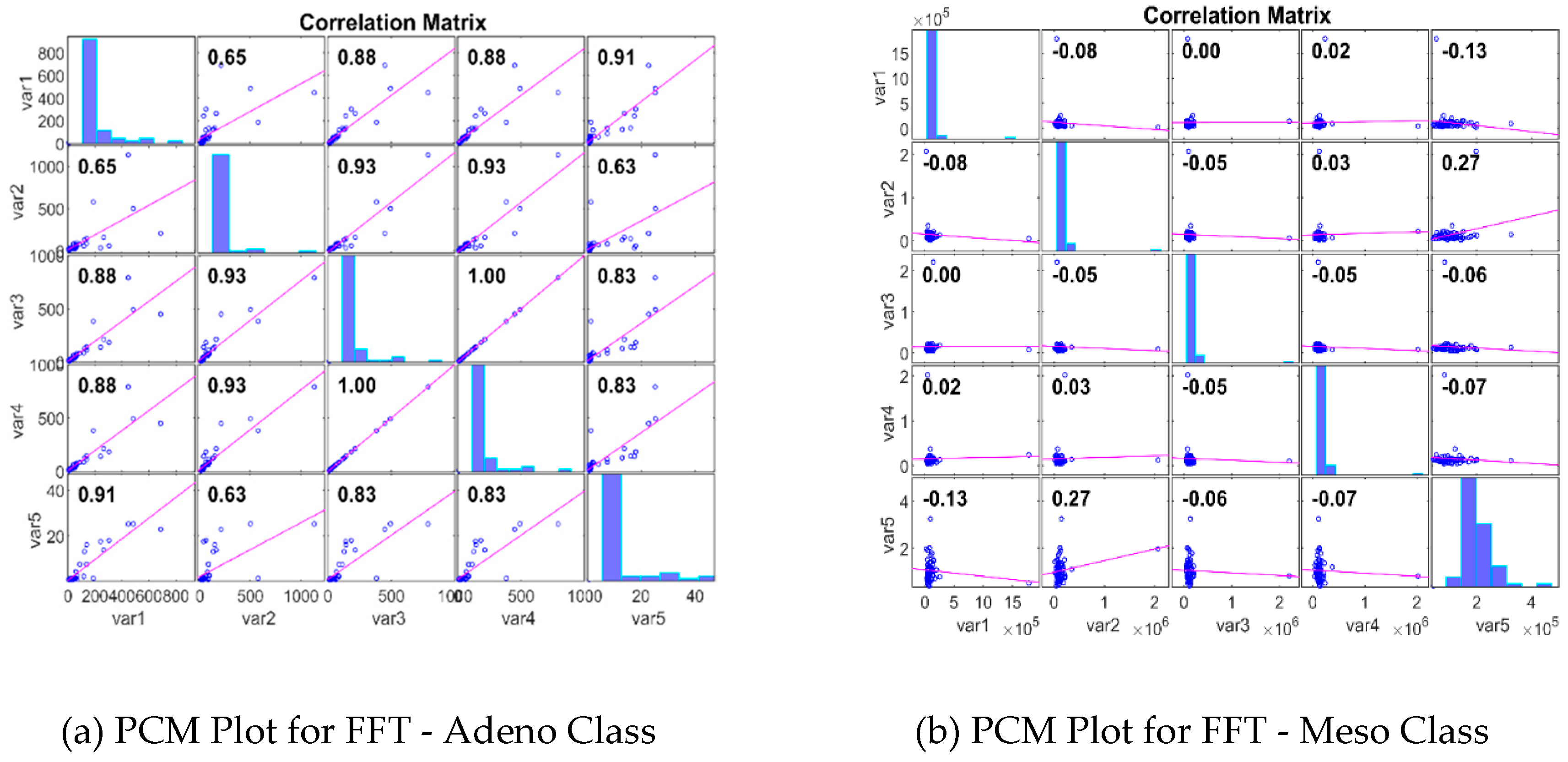
2.2.3. Impact Analysis of DimRe Methods through Statistics
2.3. Feature Selection (FS) Techniques
2.4. Classification
2.4.1. Nonlinear Regression
2.4.2. Naive Bayesian Classifier
2.4.3. Decision Tree Classifier
2.4.4. Random Forest
2.4.5. SVM (RBF)
2.5. Training and Testing
3. Results and Discussion
3.1. Hyper Parameter Tuning
3.1.1. Adam Hyper Parameter Tuning
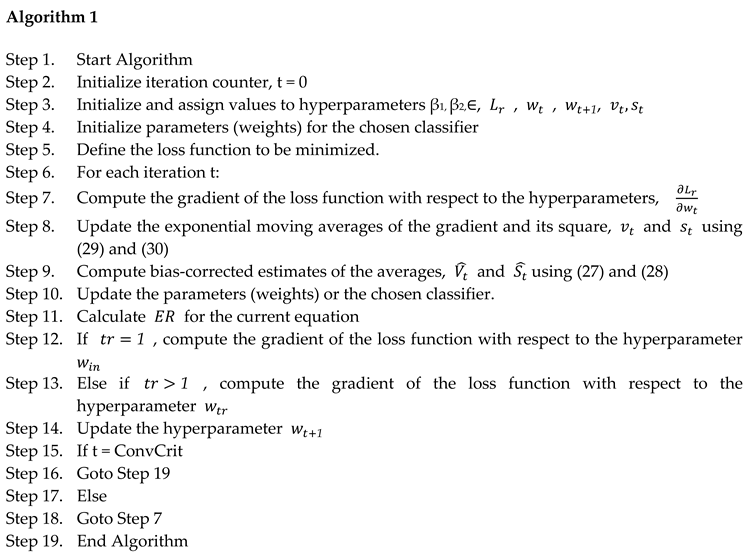
3.1.2. RanAdam Hyper Parameter Tuning

3.2. Computational Complexity (CC)
4. Limitations
5. Conclusion and Future Work
References
- Egeblad, Mikala, Elizabeth S. Nakasone, and Zena Werb. "Tumors as organs: complex tissues that interface with the entire organism." Developmental cell 18.6 (2010): 884-901. [CrossRef]
- Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med. 2011 Dec;32(4):605-44. [CrossRef] [PubMed] [PubMed Central]
- Schabath, Matthew B., and Michele L. Cote. "Cancer progress and priorities: lung cancer." Cancer epidemiology, biomarkers & prevention 28.10 (2019): 1563-1579. [CrossRef]
- Lemjabbar-Alaoui, Hassan, et al. "Lung cancer: Biology and treatment options." Biochimica et Biophysica Acta (BBA)-Reviews on Cancer 1856.2 (2015): 189-210.
- Mustafa, Murtaza, et al. "Lung cancer: risk factors, management, and prognosis." IOSR Journal of Dental and Medical Sciences 15.10 (2016): 94-101.
- Causey, Jason L., et al. "Highly accurate model for prediction of lung nodule malignancy with CT scans." Scientific reports 8.1 (2018): 9286. [CrossRef]
- Mukae, Hiroshi, et al. "The Japanese respiratory society guidelines for the management of cough and sputum (digest edition)." Respiratory Investigation 59.3 (2021): 270-290. [CrossRef]
- Konstantina Kourou, Themis P. Exarchos, Konstantinos P. Exarchos, Michalis V. Karamouzis, Dimitrios I. Fotiadis, Machine learning applications in cancer prognosis and prediction, Computational and Structural Biotechnology Journal,Volume 13,2015,Pages 8-17,ISSN 2001-0370. [CrossRef]
- Leong, Steven, et al. "Diagnostic bronchoscopy--current and future perspectives." Journal of thoracic disease 5.Suppl 5 (2013): S498. [CrossRef]
- Visser, Eric P., et al. "Spatial resolution and sensitivity of the Inveon small-animal PET scanner." Journal of Nuclear Medicine 50.1 (2009): 139-147. [CrossRef]
- Rivera, M. Patricia, Atul C. Mehta, and Momen M. Wahidi. "Establishing the diagnosis of lung cancer: Diagnosis and management of lung cancer: American College of Chest Physicians evidence-based clinical practice guidelines." Chest 143.5 (2013): e142S-e165S.
- Lubitz, Carrie C., et al. "Microarray analysis of thyroid nodule fine-needle aspirates accurately classifies benign and malignant lesions." The Journal of Molecular Diagnostics 8.4 (2006): 490-498. [CrossRef]
- Dhaun, Neeraj, et al. "Utility of renal biopsy in the clinical management of renal disease." Kidney international 85.5 (2014): 1039-1048. [CrossRef]
- Nguyen, Danh V., and David M. Rocke. "Tumor classification by partial least squares using microarray gene expression data." Bioinformatics 18.1 (2002): 39-50. [CrossRef]
- Saheed, Yakub Kayode. "Effective dimensionality reduction model with machine learning classification for microarray gene expression data." Data Science for Genomics. Academic Press, 2023. 153-164.
- Jäger, Jochen, Rimli Sengupta, and Walter L. Ruzzo. "Improved gene selection for classification of microarrays." Biocomputing 2003. 2002. 53-64.
- De Souza, Jovani Taveira, Antonio Carlos De Francisco, and Dayana Carla De Macedo. "Dimensionality reduction in gene expression data sets." IEEE Access 7 (2019): 61136-61144. [CrossRef]
- Rafique, Omar, and Ajaz Hussain Mir. "Weighted dimensionality reduction and robust Gaussian mixture model based cancer patient subtyping from gene expression data." Journal of Biomedical Informatics 112 (2020): 103620. [CrossRef]
- Inamura, Kentaro, et al. "Two subclasses of lung squamous cell carcinoma with different gene expression profiles and prognosis identified by hierarchical clustering and non-negative matrix factorization." Oncogene 24.47 (2005): 7105-7113. [CrossRef]
- Hsu, Ying-Lin, Po-Yu Huang, and Dung-Tsa Chen. "Sparse principal component analysis in cancer research." Translational cancer research 3.3 (2014): 182.
- Mollaee, Maryam, and Mohammad Hossein Moattar. "A novel feature extraction approach based on ensemble feature selection and modified discriminant independent component analysis for microarray data classification." Biocybernetics and Biomedical Engineering 36.3 (2016): 521-529. [CrossRef]
- Chen, Joe W., and Joseph Dhahbi. "Lung adenocarcinoma and lung squamous cell carcinoma cancer classification, biomarker identification, and gene expression analysis using overlapping feature selection methods." Scientific reports 11.1 (2021): 13323.
- Wang, Zixuan, et al. "Genetic algorithm-based feature selection with manifold learning for cancer classification using microarray data." BMC bioinformatics 24.1 (2023): 139. [CrossRef]
- Lee, George, Carlos Rodriguez, and Anant Madabhushi. "Investigating the efficacy of nonlinear dimensionality reduction schemes in classifying gene and protein expression studies." IEEE/ACM Transactions on Computational Biology and Bioinformatics 5.3 (2008): 368-384. [CrossRef]
- A. A. Raweh, M. Nassef and A. Badr, "A Hybridized Feature Selection and Extraction Approach for Enhancing Cancer Prediction Based on DNA Methylation," in IEEE Access, vol. 6, pp. 15212-15223, 2018. [CrossRef]
- Otoom, Ahmed Fawzi, Emad E. Abdallah, and Maen Hammad. "Breast cancer classification: comparative performance analysis of image shape-based features and microarray gene expression data." International Journal of Bio-Science & Bio-Technology 7.2 (2015): 37-46. [CrossRef]
- Orsenigo, Carlotta, and Carlo Vercellis. "A comparative study of nonlinear manifold learning methods for cancer microarray data classification." Expert systems with Applications 40.6 (2013): 2189-2197. [CrossRef]
- Fan, Liwei, Kim-Leng Poh, and Peng Zhou. "A sequential feature extraction approach for naïve bayes classification of microarray data." Expert Systems with Applications 36.6 (2009): 9919-9923. [CrossRef]
- Chen, Kun-Huang, et al. "Applying particle swarm optimization-based decision tree classifier for cancer classification on gene expression data." Applied Soft Computing 24 (2014): 773-780. [CrossRef]
- Díaz-Uriarte, Ramón, and Sara Alvarez de Andrés. "Gene selection and classification of microarray data using random forest." BMC bioinformatics 7 (2006): 1-13.
- Azzawi, Hasseeb, et al. "Lung cancer prediction from microarray data by gene expression programming." IET systems biology 10.5 (2016): 168-178. [CrossRef]
- Kotsiantis, Sotiris B., Ioannis D. Zaharakis, and Panayiotis E. Pintelas. "Machine learning: a review of classification and combining techniques." Artificial Intelligence Review 26 (2006): 159-190. [CrossRef]
- Ioannou, G., Tagaris, T. & Stafylopatis, A. AdaLip: An Adaptive Learning Rate Method per Layer for Stochastic Optimization. Neural Process Lett 55, 6311–6338 (2023). [CrossRef]
- Alrefai, Nashat, and Othman Ibrahim. "Optimized feature selection method using particle swarm intelligence with ensemble learning for cancer classification based on microarray datasets." Neural Computing and Applications 34.16 (2022): 13513-13528. [CrossRef]
- Quitadadmo, Andrew, James Johnson, and Xinghua Shi. "Bayesian hyperparameter optimization for machine learning based eQTL analysis." Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics. 2017.
- Wisesty, Untari Novia, Febryanti Sthevanie, and Rita Rismala. "Momentum Backpropagation Optimization for Cancer Detection Based on DNA Microarray Data." International Journal of Artificial Intelligence Research 4.2 (2020): 127-134. [CrossRef]
- Rakshitha, Kiran P., and N. C. Naveen. "Op-RMSprop (Optimized-Root Mean Square Propagation) Classification for Prediction of Polycystic Ovary Syndrome (PCOS) using Hybrid Machine Learning Technique." International Journal of Advanced Computer Science and Applications 13.6 (2022).
- Sena Yağmur, and Nalan ÖZKURT. "Convolutional neural network hyperparameter tuning with Adam optimizer for ECG classification." 2020 innovations in intelligent systems and applications conference (ASYU). IEEE, 2020.
- Gavin J. Gordon, Roderick V. Jensen, Li-Li Hsiao, Steven R. Gullans, Joshua E. Blumenstock, Sridhar Ramaswamy, William G. Richards, David J. Sugarbaker, Raphael Bueno; Translation of Microarray Data into Clinically Relevant Cancer Diagnostic Tests Using Gene Expression Ratios in Lung Cancer and Mesothelioma1. Cancer Res 1 September 2002; 62 (17): 4963–4967.
- Liu, TC., Kalugin, P.N., Wilding, J.L. et al. GMMchi: gene expression clustering using Gaussian mixture modeling. BMC Bioinformatics 23, 457 (2022). [CrossRef]
- Cheong Hee Park, Haesun Park,Fingerprint classification using fast Fourier transform and nonlinear discriminant analysis,Pattern Recognition,Volume 38, Issue 4,2005,Pages 495-503,ISSN 0031-3203. [CrossRef]
- Kim, Philip M., and Bruce Tidor. "Subsystem identification through dimensionality reduction of large-scale gene expression data." Genome research 13.7 (2003): 1706-1718. [CrossRef]
- Almugren, Nada, and Hala Alshamlan. "A survey on hybrid feature selection methods in microarray gene expression data for cancer classification." IEEE access 7 (2019): 78533-78548. [CrossRef]
- Cai, Zhihua, et al. "Classification of lung cancer using ensemble-based feature selection and machine learning methods." Molecular BioSystems 11.3 (2015): 791-800. [CrossRef]
- Alhenawi, Esra'A., et al. "Feature selection methods on gene expression microarray data for cancer classification: A systematic review." Computers in Biology and Medicine 140 (2022): 105051. [CrossRef]
- Kang, Chuanze, et al. "Feature selection and tumor classification for microarray data using relaxed Lasso and generalized multi-class support vector machine." Journal of theoretical biology 463 (2019): 77-91. [CrossRef]
- Dagnew, Guesh, and B. H. Shekar. "Ensemble learning-based classification of microarray cancer data on tree-based features." Cognitive Computation and Systems 3.1 (2021): 48-60.
- Cui, Xueting, et al. "A hybrid improved dragonfly algorithm for feature selection." IEEE Access 8 (2020): 155619-155629. [CrossRef]
- 49. Majdi Mafarja, Ibrahim Aljarah, Ali Asghar Heidari, Hossam Faris, Philippe Fournier-Viger, Xiaodong Li, Seyedali Mirjalili, Binary dragonfly optimization for feature selection using time varying transfer functions, Knowledge-Based Systems, Volume 161, 2018,Pages 185-204, ISSN 0950-7051. [CrossRef]
- Chnoor M. Rahman, Tarik A. Rashid, "Dragonfly Algorithm and Its Applications in Applied Science Survey", Computational Intelligence and Neuroscience, vol. 2019, Article ID 9293617, 21 pages, 2019. [CrossRef]
- Peng, Yanxiong, Wenyuan Li, and Ying Liu. "A hybrid approach for biomarker discovery from microarray gene expression data for cancer classification." Cancer informatics 2 (2006): 117693510600200024. [CrossRef]
- Mohapatra, P., Sreejit Chakravarty, and P. K. Dash. "Microarray medical data classification using kernel ridge regression and modified cat swarm optimization based gene selection system." Swarm and Evolutionary Computation 28 (2016): 144-160. [CrossRef]
- Huynh, Phuoc-Hai, Van-Hoa Nguyen, and Thanh-Nghi Do. "A coupling support vector machines with the feature learning of deep convolutional neural networks for classifying microarray gene expression data." Modern approaches for intelligent information and database systems (2018): 233-243.
- Dai, Wenseng, Yang-Yu Chuang, and Chi-Jie Lu. "A clustering-based sales forecasting scheme using support vector regression for computer server." Procedia Manufacturing 2 (2015): 82-86. [CrossRef]
- A. Kelemen, Hong Zhou, P. Lawhead and Yulan Liang, "Naive Bayesian classifier for microarray data," Proceedings of the International Joint Conference on Neural Networks, 2003., Portland, OR, 2003, pp. 1769-1773 vol.3. [CrossRef]
- Hastie, Trevor, et al. The elements of statistical learning: data mining, inference, and prediction. Vol. 2. New York: springer, 2009.
- James, Gareth, et al. An introduction to statistical learning. Vol. 112. New York: springer, 2013.
- El Kafrawy, Passent, et al. "An efficient SVM-based feature selection model for cancer classification using high-dimensional microarray data." IEEE Access 9 (2021): 155353-155369.
- Vapnik, Vladimir. The nature of statistical learning theory. Springer science & business media, 2013.
- Xiong, Zheng, et al. "Evaluating explorative prediction power of machine learning algorithms for materials discovery using k-fold forward cross-validation." Computational Materials Science 171 (2020): 109203.
- 61. Daud Muhajir, Muhammad Akbar, Affindi Bagaskara, Retno Vinarti,Improving classification algorithm on education dataset using hyperparameter tuning, Procedia Computer Science, Volume 197,2022, Pages 538-544, ISSN 1877-0509. [CrossRef]
- Elgeldawi, Enas, et al. "Hyperparameter tuning for machine learning algorithms used for arabic sentiment analysis." Informatics. Vol. 8. No. 4. MDPI, 2021. [CrossRef]
- Kaur, S., Aggarwal, H. & Rani, R. Hyper-parameter optimization of deep learning model for prediction of Parkinson’s disease. Machine Vision and Applications 31, 32 (2020). [CrossRef]
- Masud, Mehedi, et al. "Pre-trained convolutional neural networks for breast cancer detection using ultrasound images." ACM Transactions on Internet Technology (TOIT) 21.4 (2021): 1-17. [CrossRef]
- Fathi, Hanaa, et al. "An efficient cancer classification model using microarray and high-dimensional data." Computational Intelligence and Neuroscience 2021 (2021). [CrossRef]
- Guan, Peng, et al. "Lung cancer gene expression database analysis incorporating prior knowledge with support vector machine-based classification method." Journal of experimental & clinical cancer research 28.1 (2009): 1-7. [CrossRef]
- Gupta, Surbhi, et al. "Deep learning techniques for cancer classification using microarray gene expression data." Frontiers in Physiology (2022): 2002. [CrossRef]
- Mramor, Minca, et al. "Visualization-based cancer microarray data classification analysis." Bioinformatics 23.16 (2007): 2147-2154. [CrossRef]
- Ke, Lin, et al. "Improved swarm-optimization-based filter-wrapper gene selection from microarray data for gene expression tumor classification." Pattern Analysis and Applications (2022): 1-18. [CrossRef]
- Xia, Daniel, et al. "Minimalist approaches to cancer tissue-of-origin classification by DNA methylation." Modern Pathology 33.10 (2020): 1874-1888. [CrossRef]
- Morani, Federica, et al. "Identification of overexpressed genes in malignant pleural mesothelioma." International journal of molecular sciences 22.5 (2021): 2738. [CrossRef]
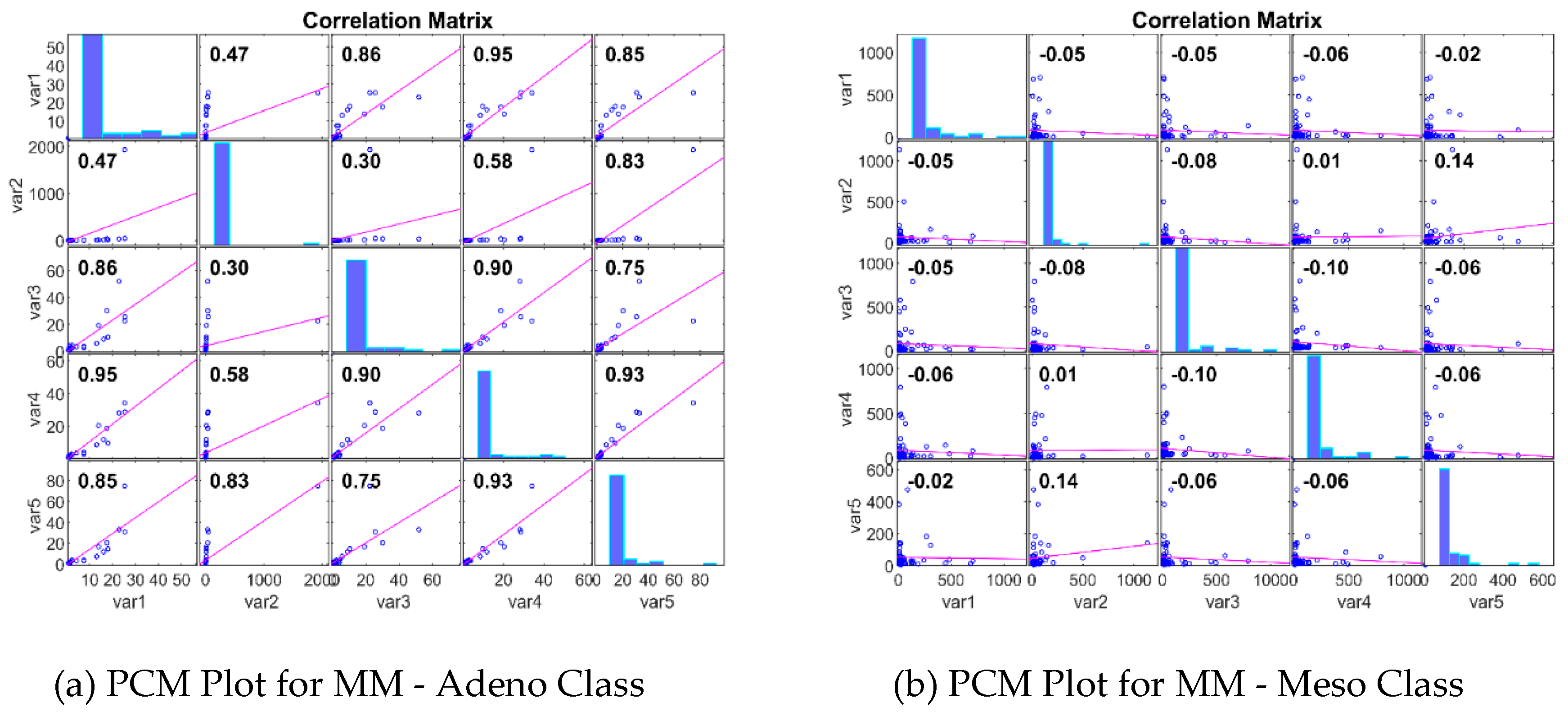
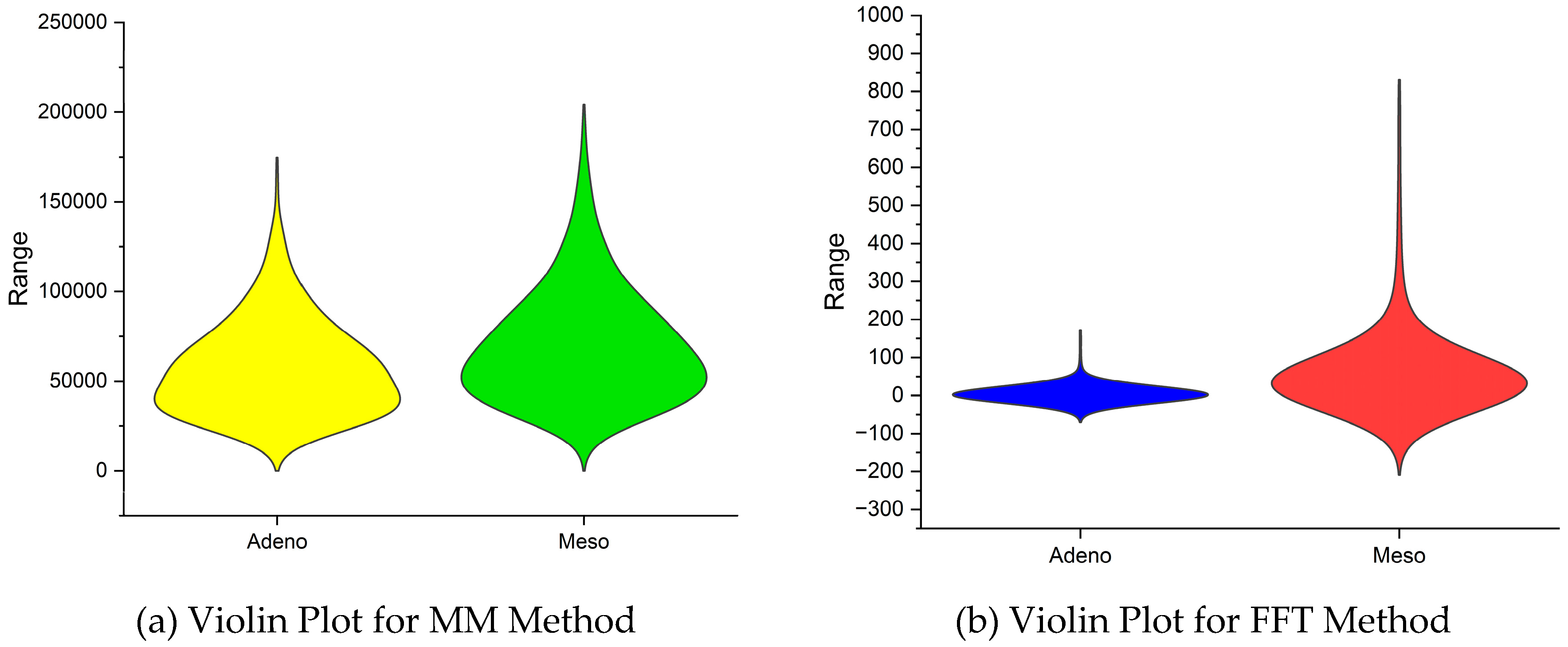
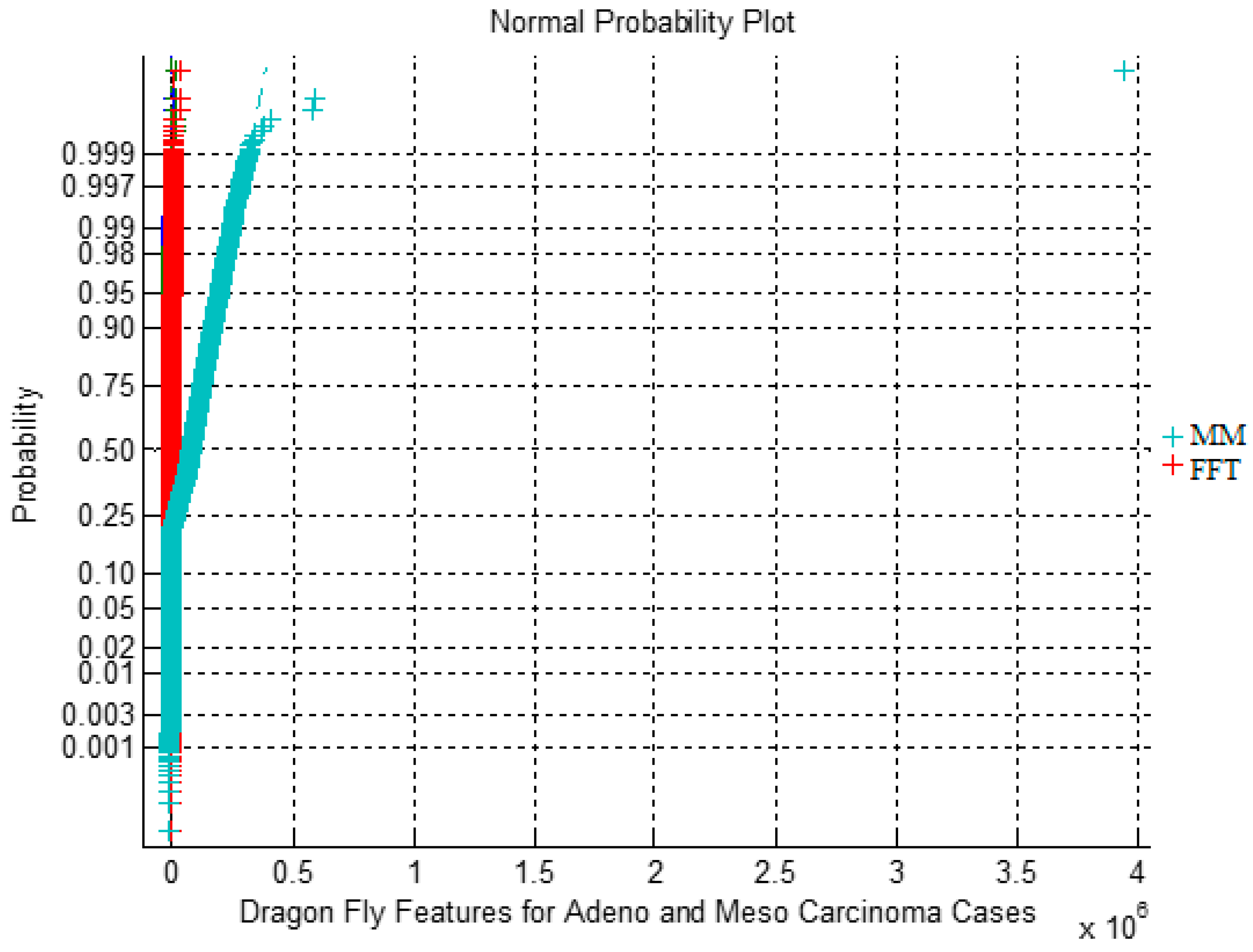

|
Sl.No |
Statistical Features |
Mixture Model | FFT | ||
|---|---|---|---|---|---|
| Adeno carcinoma | Meso cancer | Adeno carcinoma | Meso cancer | ||
| 1 | Mean | 12.77239 | 84.4254 | 50051.74 | 64399.1406 |
| 2 | Variance | 28701.74 | 72406.87 | 8.14E+08 | 1207801420 |
| 3 | Skewness | 25.62594 | 11.83928 | 22.08858 | 17.9010876 |
| 4 | Kurtosis | 1008.477 | 211.3989 | 1392.65 | 1072.04601 |
| 5 | PCC | 0.84004 | 0.926835 | 0.944664 | 0.94001594 |
| 6 | T-test | 0.017655 | 3.14E-18 | 2.06E-24 | 1.096E-021 |
| 7 | p-value <0.01 | 0.493103 |
0.5 |
0.5 | 0.5 |
| 8 | Canonical Correlation Analysis (CCA) | 0.3852 |
0.3371 |
||
| Classifiers | Mixture Model DimRe Method and Without FS | FFT DimRe Method and Without FS | Mixture Model DimRe Method and With DF FS | FFT DimRe Method and With DF FS | ||||
| Training MSE | Testing MSE | Training MSE | Testing MSE | Training MSE | Testing MSE | Training MSE | Testing MSE | |
| Nonlinear Regression | 3.84E-07 | 5.63E-05 | 3.11E-06 | 0.000016 | 1.44E-06 | 3.6E-06 | 2.54E-09 | 6.24E-08 |
| Naïve Bayesian | 1.56E-09 | 2.93E-07 | 5.61E-09 | 3.24E-08 | 3.48E-06 | 4.2E-05 | 3.03E-07 | 5.04E-05 |
| Random Forest | 1.23E-08 | 1.94E-05 | 1.44E-07 | 6.89E-05 | 3.06E-07 | 5.76E-06 | 6.4E-06 | 2.92E-05 |
| Decision Tree | 3.25E-06 | 5.48E-05 | 2.56E-06 | 4.49E-05 | 2.89E-07 | 2.6E-05 | 8.1E-07 | 4.76E-05 |
| SVM(RBF) | 2.6E-08 | 1.69E-06 | 8.1E-08 | 2.5E-07 | 1.96E-09 | 5.18E-07 | 1.02E-08 | 1.56E-07 |
| Classifier | Parameter Value |
|---|---|
| NR | T1= 0.85, T2=0.65, n1, n2, and n3 is retrieved from (15), b0 = 0.01, Convergence Criteria (ConvCrit) = MSE |
| NB | Smoothing parameter, α = 0.06, Prior Probability = 0.15, ConvCrit = MSE |
| RF | Number of trees NT = 100, Depth D = 10, ConvCrit = MSE |
| DT | Depth D = 10, ConvCrit = MSE |
| SVM (RBF) | Width of the radial basis function, =1, , ConvCrit = MSE |
|
Truth of Clinical Situation |
Observed | ||
| Adeno | Meso | ||
|
Actual |
Adeno | TP | FN |
| Meso | FP | TN | |
| Performance Metrics | Derived from Confusion matrix |
|---|---|
| Accuracy | |
| F1 Score | |
| Mathews Correlation Coefficient | |
| Error Rate | |
| Youden Index | |
| Kappa | )/(1-Eacc) |
| DimRe Method | Mixture Model | FFT | ||||||||
| Classifiers | NR | NB | RF | DT | SVM (RBF) |
NR | NB | RF | DT | SVM (RBF) |
| Parameters | ||||||||||
| Accuracy | 67.403 | 76.243 | 75.691 | 65.746 | 59.669 | 72.928 | 88.950 | 62.983 | 54.144 | 60.221 |
| F1 Score | 78.067 | 84.912 | 84.397 | 76.692 | 70.445 | 81.369 | 93.464 | 74.131 | 66.122 | 69.492 |
| MCC | 0.197 | 0.307 | 0.317 | 0.179 | 0.194 | 0.404 | 0.583 | 0.170 | 0.067 | 0.315 |
| Error Rate | 32.597 | 23.757 | 24.309 | 34.254 | 40.331 | 27.072 | 11.050 | 37.017 | 45.856 | 39.779 |
| Youden Index | 24.839 | 35.505 | 37.398 | 22.839 | 25.742 | 51.978 | 53.398 | 22.065 | 8.839 | 41.763 |
| Kappa | 0.178 | 0.298 | 0.304 | 0.159 | 0.153 | 0.353 | 0.578 | 0.145 | 0.052 | 0.230 |
| DimRe Method | Mixture Model | FFT | ||||||||
| Classifiers | Nonlinear Regression | Naïve Bayesian | Random Forest | Decision Tree | SVM (RBF) |
Nonlinear Regression | Naïve Bayesian | Random Forest | Decision Tree | SVM (RBF) |
| Parameters | ||||||||||
| Accuracy | 67.956 | 68.508 | 53.039 | 60.221 | 91.160 | 85.083 | 58.011 | 53.591 | 67.956 | 82.873 |
| F1 Score | 77.863 | 78.967 | 65.021 | 71.875 | 94.558 | 90.970 | 68.333 | 65.854 | 78.519 | 88.889 |
| MCC | 0.277 | 0.209 | 0.057 | 0.124 | 0.715 | 0.481 | 0.217 | 0.042 | 0.203 | 0.554 |
| Error Rate | 32.044 | 31.492 | 46.961 | 39.779 | 8.840 | 14.917 | 41.989 | 46.409 | 32.044 | 17.127 |
| Youden Index | 35.742 | 26.172 | 7.505 | 16.172 | 76.538 | 48.731 | 28.860 | 5.613 | 25.505 | 66.538 |
| Kappa | 0.240 | 0.191 | 0.043 | 0.103 | 0.711 | 0.481 | 0.163 | 0.033 | 0.184 | 0.524 |
| Classifiers | Optimal Values | Initial Values | |||||
| β1 | β2 | ||||||
| NR | 0.5 | 0.5 | 0.2 | 0.28 | 0.42 | 0.1 | 0.15 |
| NB | 0.6 | 0.4 | 0.26 | 0.32 | 0.5 | 0.1 | 0.2 |
| RF | 0.45 | 0.55 | 0.38 | 0.4 | 0.38 | 0.1 | 0.25 |
| DT | 0.55 | 0.45 | 0.33 | 0.41 | 0.6 | 0.15 | 0.2 |
| SVM(RBF) | 0.35 | 0.65 | 0.32 | 0.45 | 0.5 | 0.1 | 0.2 |
| Classifiers With Adam Hyper parameter Tuning |
Mixture Model DimRe Method and With DF FS | FFT DimRe Method and With DF FS | ||
| Training Accuracy | Testing Accuracy |
Training Accuracy | Testing Accuracy |
|
| Nonlinear Regression | 90.31 | 88.23 | 91.34 | 89.84 |
| Naïve Bayesian | 91.23 | 89.29 | 92.56 | 90.39 |
| Random Forest | 92.97 | 91.84 | 93.47 | 91.95 |
| Decision Tree | 86.31 | 82.87 | 92.54 | 90.39 |
| SVM(RBF) | 98.66 | 96.47 | 93.79 | 90.84 |
| Classifiers With RanAdam Hyper parameter Tuning |
Mixture Model DimRe Method and With DF FS | FFT DimRe Method and With DF FS | ||
| Training Accuracy | Testing Accuracy |
Training Accuracy | Testing Accuracy |
|
| Nonlinear Regression | 92.62 | 89.74 | 92.44 | 90.64 |
| Naïve Bayesian | 95.87 | 93.22 | 93.52 | 90.51 |
| Random Forest | 94.25 | 92.86 | 94.62 | 92.19 |
| Decision Tree | 92.37 | 90.219 | 95.61 | 93.53 |
| SVM(RBF) | 93.66 | 90.72 | 99.41 | 98.86 |
| DimRe Method | Mixture Model | FFT Method | ||||||||
| Classifiers | Nonlinear Regression | Naïve Bayesian | Random Forest | Decision Tree | SVM (RBF) |
Nonlinear Regression | Naïve Bayesian | Random Forest | Decision Tree | SVM (RBF) |
| Parameters | ||||||||||
| Accuracy | 80.110 | 87.293 | 87.845 | 82.873 | 94.475 | 87.845 | 88.398 | 88.950 | 88.398 | 87.845 |
| F1 Score | 87.413 | 92.256 | 92.667 | 89.199 | 96.667 | 92.466 | 92.929 | 93.243 | 93.023 | 92.414 |
| MCC | 0.417 | 0.570 | 0.572 | 0.494 | 0.805 | 0.618 | 0.607 | 0.631 | 0.586 | 0.630 |
| Error Rate | 19.890 | 12.707 | 12.155 | 17.127 | 5.525 | 12.155 | 11.602 | 11.050 | 11.602 | 12.155 |
| Youden Index | 47.849 | 59.075 | 57.183 | 56.301 | 80.538 | 67.419 | 62.968 | 66.194 | 57.849 | 69.978 |
| Kappa | 0.406 | 0.569 | 0.572 | 0.483 | 0.805 | 0.612 | 0.606 | 0.630 | 0.586 | 0.620 |
| DimRe Method | Mixture Model | FFT Method | ||||||||
| Classifiers | Nonlinear Regression | Naïve Bayesian | Random Forest | Decision Tree | SVM (RBF) |
Nonlinear Regression | Naïve Bayesian | Random Forest | Decision Tree | SVM (RBF) |
| Parameters | ||||||||||
| Accuracy | 86.740 | 91.160 | 91.160 | 88.398 | 87.293 | 88.950 | 85.635 | 88.398 | 90.608 | 98.343 |
| F1 Score | 91.892 | 94.667 | 94.702 | 93.023 | 92.256 | 93.289 | 91.216 | 93.069 | 94.352 | 98.997 |
| MCC | 0.557 | 0.689 | 0.681 | 0.586 | 0.570 | 0.621 | 0.520 | 0.576 | 0.665 | 0.943 |
| Error Rate | 13.260 | 8.840 | 8.840 | 11.602 | 12.707 | 11.050 | 14.365 | 11.602 | 9.392 | 1.657 |
| Youden Index | 58.409 | 68.860 | 66.301 | 57.849 | 59.075 | 63.634 | 54.516 | 55.290 | 65.634 | 95.441 |
| Kappa | 0.556 | 0.689 | 0.680 | 0.586 | 0.569 | 0.620 | 0.519 | 0.575 | 0.665 | 0.942 |
| Classifiers |
Mixture Model DimRe Method and With DF FS | FFT DimRe Method and With DF FS | ||
| Accuracy Improvement by Adam Method (%) |
Accuracy Improvement by RanAdam Method (%) |
Accuracy Improvement by Adam Method (%) |
Accuracy Improvement by RanAdam Method (%) |
|
| Nonlinear Regression | 15.172 | 21.65 | 3.145 | 4.347 |
| Naïve Bayesian | 21.519 | 24.84 | 34.375 | 32.258 |
| Random Forest | 39.623 | 41.81 | 39.752 | 39.375 |
| Decision Tree | 27.333 | 31.875 | 23.125 | 25 |
| SVM(RBF) | 3.509 | 4.43 | 5.66 | 15.73 |
| Classifiers | Without FS | With DF FS | With DF FS and Adam Tuning | With DF FS and RanAdam Tuning |
|---|---|---|---|---|
| Nonlinear Regression | O(2n3 log2n) | O(2n6 log 2n) | O(2n6 log 2n) | O(2n4 log2n) |
| Naïve Bayesian | O(2n4 log2n) | O(2n7 log 2n) | O(2n7 log 2n) | O(2n5log2n) |
| Random Forest | O(2n3 log2n) | O(2n6 log 2n) | O(2n6 log 2n) | O(2n4 log2n) |
| Decision Tree | O(2n3 log2n) | O(2n6 log 2n) | O(2n6 log 2n) | O(2n4 log2n) |
| SVM(RBF) | O(2n2 log4n) | O(2n5 log 4n) | O(2n5 log 4n) | O(2n3 log4n) |
| S.No | Author (with year) | Database | Classifier | Classes | Performance Accuracy in% |
|---|---|---|---|---|---|
| 1 | Azzawi (2015) [31] | National Library of Medicine and Kent Ridge Bio-medical Dataset | SVM, MLP, RBFN | Adenocarcinoma, Meso |
91.39 91.72 89.82 |
| 2 | Gordon (2002) [39] | Gordon MAGE Data | MAGE ratios | Adenocarcinoma, Meso |
90 |
| 3 | Fathi et al. (2021) [65] | Gordon MAGE Data | Decision Tree with feature fusion | Adenocarcinoma, Meso |
85 |
| 4 | Guan et al.(2009) [66] | Affymetrix Human GeneAtlas U95Av2 microarray dataset |
SVM (RBF) with gene based feature | Adenocarcinoma, Meso |
94 |
| 5 | Gupta et al. (2022) [67] | TCGA dataset | Deep CNN | Adenocarcinoma, Meso |
92 |
| 6 | Mramor et al. (2007) [68] | Gordon MAGE Data | SVM, Naïve Bayes, KNN, Decision Tree | Adenocarcinoma, Meso |
94.67 90.35 75.28 91.21 |
| 7 | Lin Ke (2022) [69] | Gordon MAGE Data | DT - C4.5 | Adenocarcinoma, Meso |
93 |
| 8 | Daniel Xia et al. (2020) [70] | Gordon MAGE Data | Minimalist Cancer Classifier | Adenocarcinoma, Meso |
90.6 |
| 9 | Morani et al.(2021) [71] | TCGA and GEO Dataset | Multivariate cox regression analysis | Adenocarcinoma, Meso |
90 |
| 10 | This Research | Gordon MAGE Data | RanAdam Hyper parameter tuning for FFT DimRe Techniques with DF FS and SVM (RBF) Classification | Adenocarcinoma, Meso |
98.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




