1. Introduction
The field of medical imaging has experienced a transformative paradigm shift with the emergence of deep learning (DL), unlocking unprecedented opportunities for accurate disease classification and diagnosis as indicated in [
1,
2,
3,
4,
5]. In the area of brain tumor classification using Magnetic Resonance Imaging (MRI) scan images, these DL models have shown exceptional promise [
1,
2]. The significance of brain tumor diagnosis from MRI images lies in its pivotal role in modern healthcare, offering opportunities for early detection and timely treatment, as evident in [
2,
3,
6]. DL models have shown remarkable potential in automating this process, yet their effectiveness and reliability in real-world medical applications, particularly in brain tumor classification, remain under scrutiny. The primary challenge stems from the need to achieve both
high accuracy and
model interpretability, as emphasized in [
7].
Despite the impressive classification results produced by DL models, their “black-box” nature impedes a comprehensive understanding of the decision-making rationale. Also, their incorporation into clinical practice and decision-making needs adaptability, not just in terms of classification performance, but also in interpretability and explainability. This study investigates the junction of medical image analysis and DL interpretability. Brain tumors, characterized by their diversity and clinical significance as highlighted in [
2,
3], demand precise classification for tailored treatment plans and improved patient outcomes. While DL models have exhibited substantial prowess in this domain, their inherent opacity presents a challenge, as discussed in [
3,
6]. The central issue revolves around understanding how these models reach their conclusions, particularly crucial in the context of medical decision-making. The decision-making processes of DL models in medical imaging are currently under scrutiny through interpretability techniques, shedding light on how algorithms make predictions. The transparency and reliability of AI-driven medical diagnoses are augmented by these techniques, facilitating the identification of pertinent features and areas within images contributing to the model’s choices.
To address this challenge, we conduct a broad performance evaluation of various state-of-the-art DL frameworks with transfer learning for brain tumor MRI classification. This evaluation encompasses not only the assessment of classification accuracy but also a focus on evaluating the interpretability of the models. Adaptive path-based techniques, including Gradient-weighted Class Activation Mapping (Grad-Cam), Grad-Cam++, Integrated Gradient (IG), and Saliency Mapping, are leveraged to unravel the underlying controlling processes of these models. The ultimate objective is to enhance the clinical utility of DL based on brain tumor identification by providing more transparent and interpretable results.
Techniques for interpretability, such as Grad-Cam++ [
8], Grad-CAM [
9], Integrated Gradients [
10], and SHAP [
11], have distinguished themselves by highlighting the image regions most important to the model’s conclusions. For instance, Grad-CAM creates heatmaps that highlight significant image regions, aiding doctors in comprehending the model’s focal point. Although DL models for diagnosing brain tumors have attained amazing levels of accuracy [
12,
13], their intricate internal mechanisms raise questions about their dependability and generalizability. To solve this, academics have started looking into how interpretable these models are. To improve a DL model’s transparency, [
14] created a brand-new technique for viewing tumor localization. It is difficult to gauge how well DL models can be interpreted. According to [
15,
16], several criteria have been put out to evaluate consistency, localization accuracy, and sensitivity to perturbations. These measures are intended to offer a consistent framework for evaluating how closely the model’s explanations match clinical expectations and actual tumor sites. Medical practitioner’s ability to make decisions may be improved by combining interpretable DL models into clinical practice. Recent research [
17] has shown how radiologists and doctors can use interpretable model explanations to validate the model’s predictions and produce more accurate diagnoses. Radiologists participated in the study in [
18,
19,
20] to assess the interpretability of DL models in medical imaging. To get professional opinions on the value and comprehensibility of the model explanations, the study employed user studies. The research underlined how crucial interpretability is to fostering communication among medical specialists.
The exploration of multiple DL architectures holds considerable importance in evaluating their effectiveness within the specialized domain of brain tumor classification using MRI images. We conduct an ultra-careful analysis, each model, including AlexNet, DenseNet121, EfficientB0, GoogleNet, Inception V3, ResNet50, VGG16, VGG19, ViT Transformer, and Xception, is identified by a unique set of parameters and indicators, contributing to a degree of understanding of their merits. The evaluation criteria, spanning accuracy, precision, recall, and F1 score percentages, collectively offer a comprehensive and insightful depiction of the capabilities exhibited by these models. To further light up our findings, we implement interpretability techniques, such as Grad-CAM, Grad-CAM++, Integrated Gradient, and Saliency Mapping, unveiling the intricate decision-making processes and providing valuable insights into the regions and features significantly contributing to the models’ classifications. This integrated approach not only gauges the models’ performance but also enhances the transparency and interpretability of their decision-making procedure.
The rest of this paper is structured as follows.
Section 2 discusses the literature review on explainability in medical imaging, while
Section 3 covers the attribution methods used in this work and the transfer learning models.
Section 4 presents the dataset and our proposed method. In
Section 5, experimental results are presented, and conclusions are reported in
Section 6.
2. Literature on DL Models Explainability in Medical Imaging
The input image is passed through the algorithm via one forward as well as backward propagation. The resulting score is then computed forward, and the dependency gradient amongst the convolution layers is then determined to build an attribution map. This process is known as the Vanilla Gradient explainability method. It is an easy-to-understand attribution approach with little processing power requirements because of its simplicity. Vanilla Gradient and other attribution-based graphic aids for MRI imaging of brain tumors were evaluated using an attribution-based scheme named NeuroXAI [
21]. Both feature segmentation and classification were visualized using these techniques. Vanilla Gradient produced noisier attribution maps than the other attribution techniques and had gradient saturation, which means that a change in a neuron has no effect on the network’s output and so cannot be assessed. Similar results were observed utilizing Vanilla Gradient for feature visualization in [
22], where the dissimilitude intensification juncture from computed tomography (CT) images is anticipated. Furthermore, Vanilla Gradient is unable to distinguish between classes (such as healthy and diseased) [
23]. This demonstrates that Vanilla Gradient is unable to produce attribution maps that are distinct based on class. The deconvolution network (DeconvNet) is essentially comparable to Vanilla Gradient, with the primary distinction lying in the computation of gradients over a Rectified Linear Unit (ReLU) function [
24].
The human brain’s artery segmentation was studied using different attribution techniques for interlayer CNN visualization using TorchEsegeta, a structure for image-based DL algorithms that can be understood and explained [
25]. Since other techniques also indicated non-vessel activation, their primary focus was on the vessels, Vanilla Gradient, and DeconvNET produced results that were more comprehensible to humans than other attribution techniques like Deep Learning Important Features (DeepLIFT) and Gradient-weighted Class Activation Mapping (GradCAM++).
Guided back propagation (GBP) integrates both the deconvNET [
26] and the Vanilla Gradient. Comparing this approach to applying each technique separately yields less noisy attribution maps as there are fewer active voxels. As compared to Vanilla Gradient, GBP presented purpose-specific attribution maps in the NeuroXAI framework with a lot less noise [
21]. An extra refining mechanism for the GBP was suggested in [
26] to further reduce the quantity of noise and the influence of indiscriminate attributions on predicting brain disorders using MRI. GBP is likely to offer attribution maps with reduced noise, but it could furthermore produce attribution maps that are too sparse, which are unhelpful for comprehensive image characterization [
27]. Although they are not class discriminative, all three of the gradient-based techniques are quite sensitive to the way the neural network layers collect information. ReLU and pooling layers may also cause local gradients to saturate. As a result, significant characteristics may disappear as the network’s layers advance, which might lead to an inadequate model clarification or even a concentration on unimportant features. Layer-wise relevance propagation (LRP) is an Explainable Artificial Intelligence (XAI) technique that employs principles unique to LRP to propagate the class score backward through the neural layers to the input image [
28]. The core idea of LRP is to preserve inter-neuron interdependence, ensuring that information acquired by one layer of neurons is equally transferred to the next lower layer. LRP addresses the challenges posed by the saturation problem since the decomposition relies on propagating relevance scores between neurons rather than gradients. In a study focused on the detection of abdominal aortic aneurysms using CT images, LRP demonstrated a distinct class difference based on activation differences in the aortic lumen [
29].
Deep Learning Important Features (DeepLIFT) is an XAI technique that addresses the saturation problem by employing a neutral reference activation, such as the neuron activation in computed tomography (CT) scans without pathology or disease [
30]. The difference between a new neuron’s activation and the reference activation is described by this reference activation. An attribution map is generated by computing the contribution scores for each neuron based on these differences. To discern individuals with Multiple Sclerosis (MS) using MRI, DeepLIFT was compared to LRP and Vanilla Gradient [
31]. Based on the quantitative evaluation, DeepLIFT extracts target-specific characteristics much better than Vanilla Gradient and marginally better than LRP. Gradient saturation is something that both LRP and DeepLIFT can handle, which might be why it outperforms Vanilla Gradient in this classification challenge. Among the most popular model-specific attribution techniques is the class activation map (CAM) [
32,
33]. Rather than using numerous dense layers, it employs a Global Average Pooling (GAP) layer, which adds linearity before the last dense layer and after the last convolution layer. Low-dimension attribution maps are produced by CAM since they only utilize information from the last convolution layer. As a result, the low-dimension CAM can show if a model can generally focus on particular targets, but because of its poor specificity, it is unable to discriminatively define characteristics depending on class [
34,
35]. Additionally, it was revealed through perturbation analysis of several attribution methodologies that gradient-based approaches have a better rigor than CAM [
22]. However, CAM can be indicative when doing patch-based (more targeted) tumor analysis as opposed to whole image tumor analysis [
36,
37], or when the classes in a classification task exhibit obvious visual distinctions, such as between healthy and Alzheimer’s brains [
38].
The use of XAI techniques has increased dramatically as a result of COVID-19 detection [
39]. Generally speaking, these techniques may be distinguished by either using the entire CT scan or only the lung segmentation for COVID-19 identification. In particular, there was a significant performance difference in attribution mapping for COVID-19 detection based on the entire picture. The most common attribution technique was Grad-CAM, an extension of CAM, which produced both very specific [
40,
41] and non-specific attributions [
31,
42,
43], but generally was able to approximately pinpoint the possible COVID-19 lesions to produce reliable predictions. A priori segmentation of the lungs was proposed to eliminate the impact of non-target specific characteristics [
43,
44,
45,
46,
47,
48,
49]. In this manner, only characteristics from the lungs may be extracted by both the DL algorithms and the XAI approaches. In this sense, the XAI techniques and the DL algorithms can only extract characteristics from the lungs. Compared to utilizing the whole CT image with Grad-CAM, this anatomically based XAI approach demonstrated greater specificity. This indicates the benefits of medical-based data reduction for DL and XAI algorithms, that is, lowering the number of trainable characteristics and/or getting rid of uninformative characteristics based on the input image. When the entire image was used, comparable non-target relevant attribution maps were observed as well (in the absence of data reduction) for cerebral hemorrhage detection [
50], and automated grading of expanded perivascular spaces in acute stroke [
51]. In a manner akin to the COVID-19 investigations, prior anatomical segmentation was employed to categorize and illustrate mortality risks based on cardiac PET [
52], Alzheimer’s disease [
53] and schizophrenia based on MRI [
54]. Grad-CAM’s low-dimensional attribution maps, however, continue to cause poor specificity even while data handling reduces the prevalence of non-target specific characteristics [
55,
56]. The authors proposed that the active characteristics surrounding the tumor correlate to areas harboring occult microscopic illness based on the Grad-CAM attribution maps, in research for the categorization of lung cancer histology based on CT images [
7]. It is more plausible, though, due to Grad-CAM’s poor dimensionality, as CT lacks the spatial resolution necessary to identify these tiny illnesses.
In classification tasks when there is a discernible radiological difference between the classes, Grad-CAM, like CAM, can be class discriminative [
10,
57,
58,
59,
60]. However, additional attribution methods like Vanilla Gradient and GBP should be utilized in cases of tasks with less clear radiological distinctions, such as predicting survival based on tumor feature, where Grad-CAM lacks fine-grained information [
22,
25]. In MRI imaging of brain tumors, research that paired GBP with Grad-CAM, a technique known as guided Grad-CAM (gGrad-CAM) showed improved localized attribution maps with greater resolution [
21]. These supports integrate the benefits of attribution techniques for accurate and comprehensible model visualization. Numerous further enhanced versions of Grad-CAM, including Grad-CAM++, have been developed. To improve target-specific feature localization over Grad-CAM, Grad-CAM++ was introduced [
6]. Grad-CAM may reduce the disparity in relevance between the various gradients since it averages the feature map gradients. Grad-CAM++ substitutes a weighted average, which quantifies each feature map unit’s significance, in its stead. In terms of knee osteoarthritis prediction using MRI, it demonstrated better target-specific attribution maps than Grad-CAM [
9].
4. Data and Proposed Method
4.1. Dataset
The dataset used in this study was curated by Navoneel Chakrabarty and Swati Kanchan [
73]. For easy access and reference, the dataset is currently available on Kaggle
1.
This brain tumor dataset comprises 3264 2D slices of T1-weighted contrast-enhanced images, encompassing three distinct types of brain tumors—glioma, meningioma, and pituitary tumors—along with images of a healthy brain. The dataset has been partitioned into 2937 images for training and 327 for testing purposes.
Figure 1 visually presents samples from each of the four classes within the dataset and
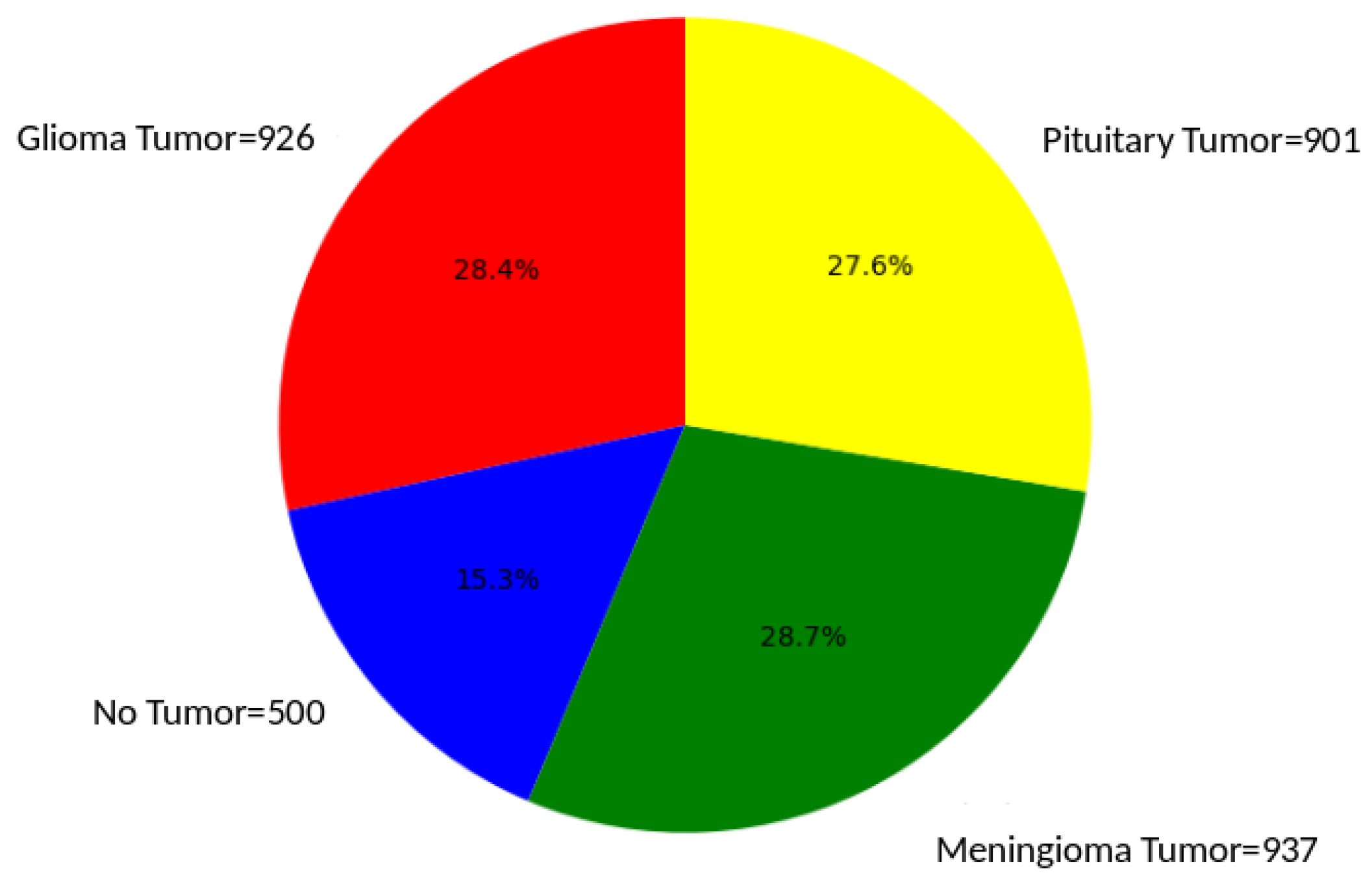
Figure 2 illustrates the proportion of each of the four classes in the dataset
Gliomas are tumors derived from glial cells and can manifest as either benign or malignant. Among them, glioblastoma multiforme stands out as a particularly aggressive variant, posing significant challenges in terms of therapeutic intervention [
3]. Pituitary Tumors which arise in the pituitary gland, these tumors can disrupt hormonal balance. They may present as growths that secrete hormones or as non-functioning growths. Common sub-types include prolactinomas and growth hormone-secreting tumors, each with its distinct clinical implications. Meningiomas are generally benign, slow-growing tumors originating from the meninges. The symptoms associated with meningiomas vary based on the size and location of the tumor, making their clinical presentation diverse and often dependent on individual cases [
3].
Understanding the unique features and characteristics of these brain tumors is essential for accurate diagnosis and effective treatment strategies. The use of this curated dataset allows for in-depth exploration and analysis of these distinct tumor types, contributing valuable insights to the field of medical imaging and diagnostics with deep learning and interpretability.
4.2. Proposed Method
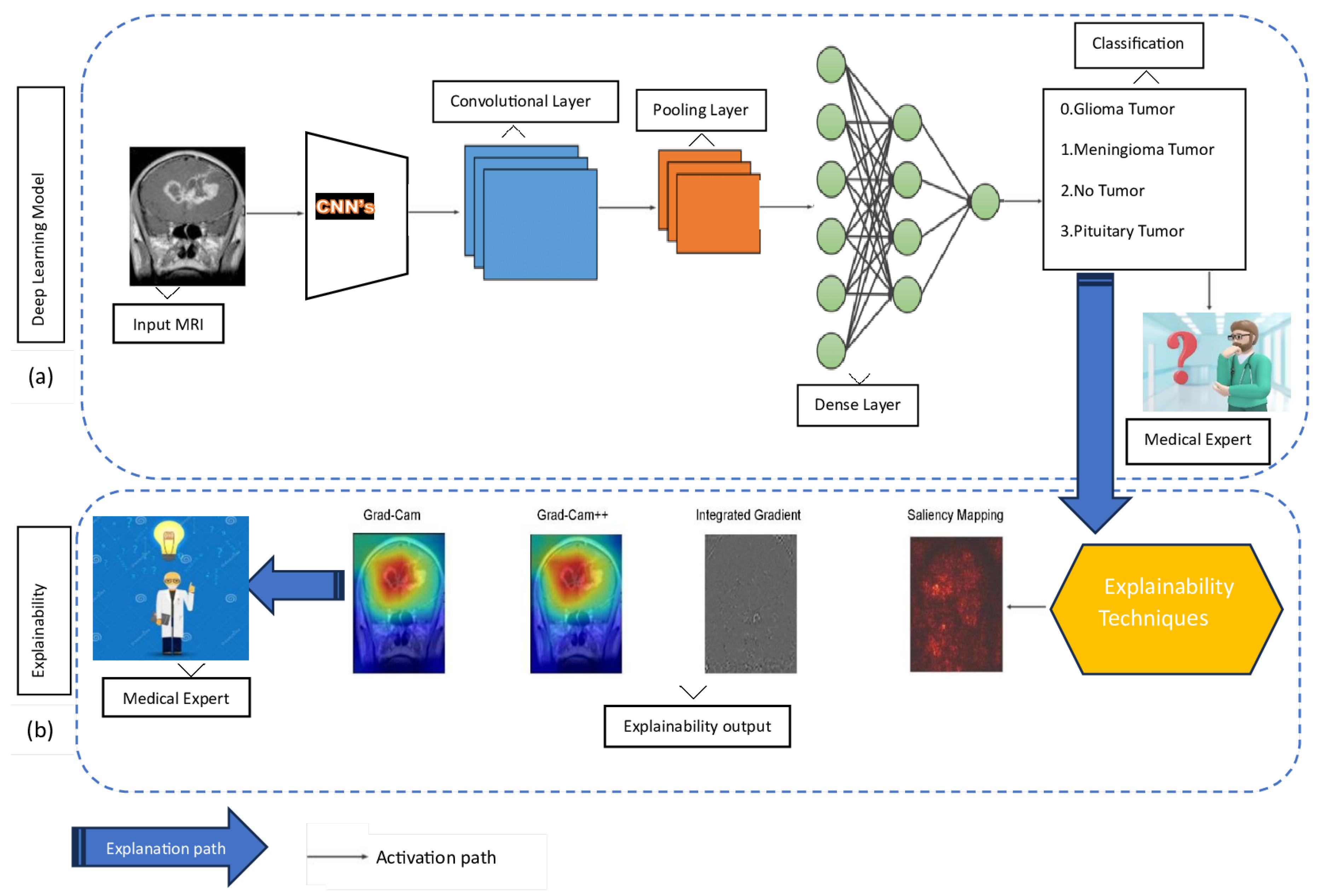
This paper proposes a comprehensive framework for brain tumor classification while integrating the model’s explainability. This approach comprises two phases: phase (a) for classification and phase (b) for explainability, as illustrated in
Figure 3.
Phase (a): This phase integrates a CNN model with TL for the classification of brain tumor types based on MRI data. In this approach, the acquired features of a pre-trained CNN model serve as an initial foundation, proving particularly advantageous in scenarios involving sparsely labeled data. The data is fed into a CNN, which subsequently processes the data through a convolutional layer to capture intricate patterns and spatial hierarchies within the MRI images. Following this, the pooling layer is employed to down-sample and reduce the feature space, optimizing computational efficiency. Progressing along the activation path, the dense layer plays a pivotal role in transforming high-level features for effective classification. Finally, the model makes decisions about tumor types based on the combination of these learned features. When the decision is made, a medical expert becomes curious and seeks to understand how the model makes decisions based on their expertise.
Phase (b): This phase uses explainability techniques, including Grad-Cam, Grad-Cam++, Integrated Gradient, and Saliency Mapping. The explanation aims to shed light on how the CNN model arrives at its classifications, providing valuable insights to the medical expert. Grad-Cam and Grad-Cam++ offer visualization of crucial regions in the MRI images that contribute to the model’s decisions. Integrated Gradient provides a comprehensive understanding of feature importance by perturbing input features, while Saliency Mapping highlights salient features influencing the classification. Together, these explainability techniques bridge the gap between the model’s predictions and human interpretability, fostering trust and comprehension in the application of DL models to medical imaging. After the model is explained, the focus shifts smoothly to the part where medical experts take over and make sense of it. The explained visualizations and insights provided by techniques like Grad-Cam, Grad-Cam++, Integrated Gradient, and Saliency Mapping serve as a bridge between the complex world of DL classifications and the expertise of medical experts. These outputs are presented to medical experts, allowing them to interpret and comprehend the rationale behind the model’s decisions.
4.3. Training, Regularization, and Testing
This section elaborates on the model training process, hyperparameter tuning, and model testing, with a specific emphasis on the computational infrastructure used.
The models were trained using a DL approach, leveraging a CNN architecture with TL. Dataset was subjected to shuffling, ensuring a random order. Afterward, the dataset was divided, allocating 90 % for training purposes and reserving 10 % for validation. This approach aims to minimize bias and foster robustness in model training and evaluation. The dataset was split into training and validation sets, with the training set containing 2937 images and validation 327 images. The CNNs were implemented using Tensorflow version 2.12.0. The training process involved feeding the network with batches of images, where the model learned to identify patterns and features associated with each class of brain tumor. The choice of the loss function, optimizer (e.g., Adam, SGD), and learning rate played crucial roles in guiding the training process. These choices were fine-tuned based on the performance of the model on the validation set.
Hyperparameters, such as the number of layers, filter sizes, and dropout rates, were optimized to enhance the model’s predictive capabilities. This involved a systematic exploration of different hyperparameter combinations using techniques like random search. The goal was to find the configuration that resulted in the best model performance as shown in
Table 1. Additionally, TL was employed, using pre-trained models such as ResNet or VGG and fine-tuning them for the specific task of brain tumor classification.
The testing phase aimed to evaluate the trained model’s generalization to new, unseen data. A separate test set, consisting of 327 images, was used for this purpose. Metrics such as accuracy, precision, recall, and the so-called F1 score were computed to assess the model’s performance on each brain tumor class. The models were rigorously evaluated to ensure robustness and reliability in real-world scenarios. Careful attention was paid to handling imbalances in class distribution to avoid biased performance metrics.
The model training and hyperparameter tuning of our pre-trained models demanded substantial computational resources, and the efficiency of the process hinged on the specifications of our Dell computer, featuring an Intel Core i7 processor. The use of a powerful GPU, specifically the P100 model, played a pivotal role in accelerating the training speed. With 32 GB of system RAM, our system efficiently handled the computational load, facilitated by the Kaggle kernel environment. Implementation was carried out in Python, using Keras and TensorFlow version 2.12.0.
5. Results
In this section, we present the outcomes of our study, offering a comprehensive overview of the key findings and their interpretation. Through this detailed analysis, we aim to provide a thorough understanding of the performance, interpretability, and decision-making processes inherent in the deep learning models employed in this work. Our contributions extend to the broader field of model interpretability, enhancing the collective knowledge within the research community.
5.1. Classification Results
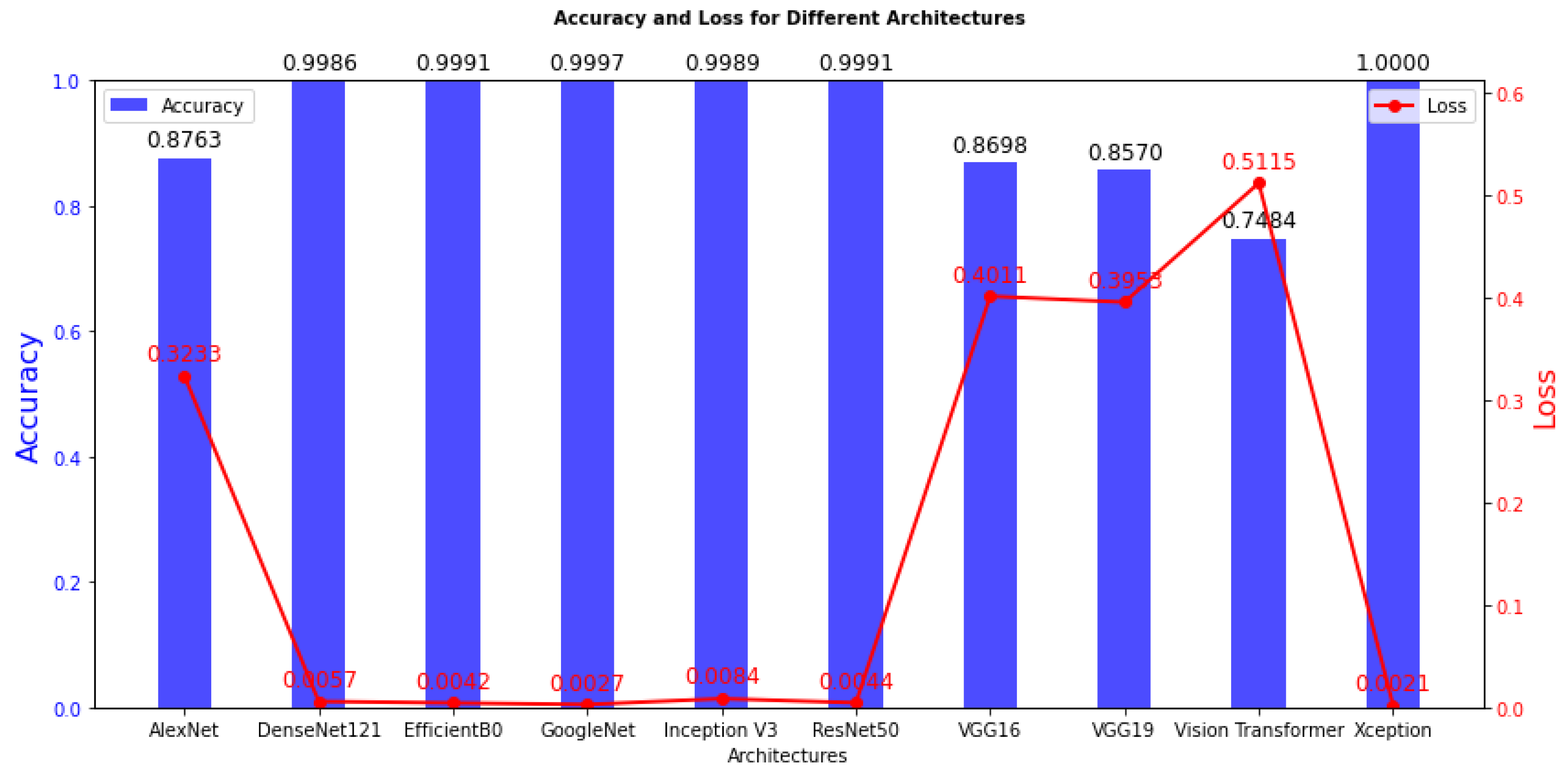
We begin by discussing the training results, and offering insights into the model training process. The detailed training outcomes are presented in
Table 2 and visually represented in
Figure 4. Moving on to the test results, a summary is provided in
Table 3, and
Figure 5 visually highlights the superior performance of the best model.
This section also undertakes a thorough analysis and interpretation of the confusion matrix derived from the classification. A detailed examination follows, shedding light on the interpretability results. Specifically, results for adaptive path-based techniques, such as Grad-Cam, Grad-Cam++, Integrated Gradient, and Saliency mapping, are discussed in-depth. These techniques have enhanced our understanding of how deep learning models make classification decisions.
The training outcomes, as detailed in
Table 2, offer insights into the model’s ability to learn from the training data. High training accuracy and low training loss often signify successful training, yet these metrics may not necessarily ensure performance on the test data. Among the models investigated in this study, namely DenseNet121, EfficientB0, GoogleNet, Inception V3, ResNet50, and Xception, stand out with an acceptable training accuracy, ranging from 99.86% to 100%. These high accuracy scores indicate that these models have effectively learned the statistical regularities present in the training data. Furthermore, the associated training loss values are remarkably low, showcasing the models’ efficiency in minimizing errors during the training phase. It is imperative to note that achieving high training accuracy does not necessarily guarantee superior performance on unseen datasets, emphasizing the importance of comprehensive evaluation of the test data for a more robust assessment of model generalization.
In contrast, models with notably lower training accuracy, spanning from 74.84% to 87.63%, such as AlexNet, VGG16, VGG19, and Vision Transformer, while still demonstrating commendable performance on the training data, exhibit comparatively higher training loss values. This suggests a slightly higher level of modeling error during the learning process for these models. The inferior performance of VGG16, VGG19, ViT Transformer, and AlexNet may be due to a combination of huge parameter counts and excessive model complexity, which may not be adequately aligned with the task’s features. To increase the generalization capacity of these models, regularization strategies like dropout or batch normalization may need to be further refined or optimized.
The classification results, as detailed in
Table 3, provide valuable insights into the model’s capability to classify the test data, it becomes evident that EfficientB0 emerges as the superior model among all those considered in this study. Upon closer examination of the results, it is apparent that some of the models may not be suited for this specific task, with some requiring more computational resources due to higher parameter counts, as observed in VGG16 and VGG19.
Figure 5 compares the 10 deep learning models used in this study, highlighting each model’s performance across key criteria such as accuracy, precision, recall, and F1 score percentages. The models include AlexNet, DenseNet121, EfficientB0, GoogleNet, Inception V3, ResNet50, VGG16, VGG19, ViT Transformer, and Xception. This graphical representation offers a concise summary of these models’ effectiveness in classifying brain tumors from MRI images, enabling a quick and informative comparison.
EfficientB0, exhibiting the best performance, shows great promise and warrants further consideration, particularly in medical imaging applications. The insights gained from this comprehensive evaluation contribute to the selection of an optimal deep learning model for the task of classifying brain tumors, emphasizing the potential impact of EfficientB0 in medical imaging.
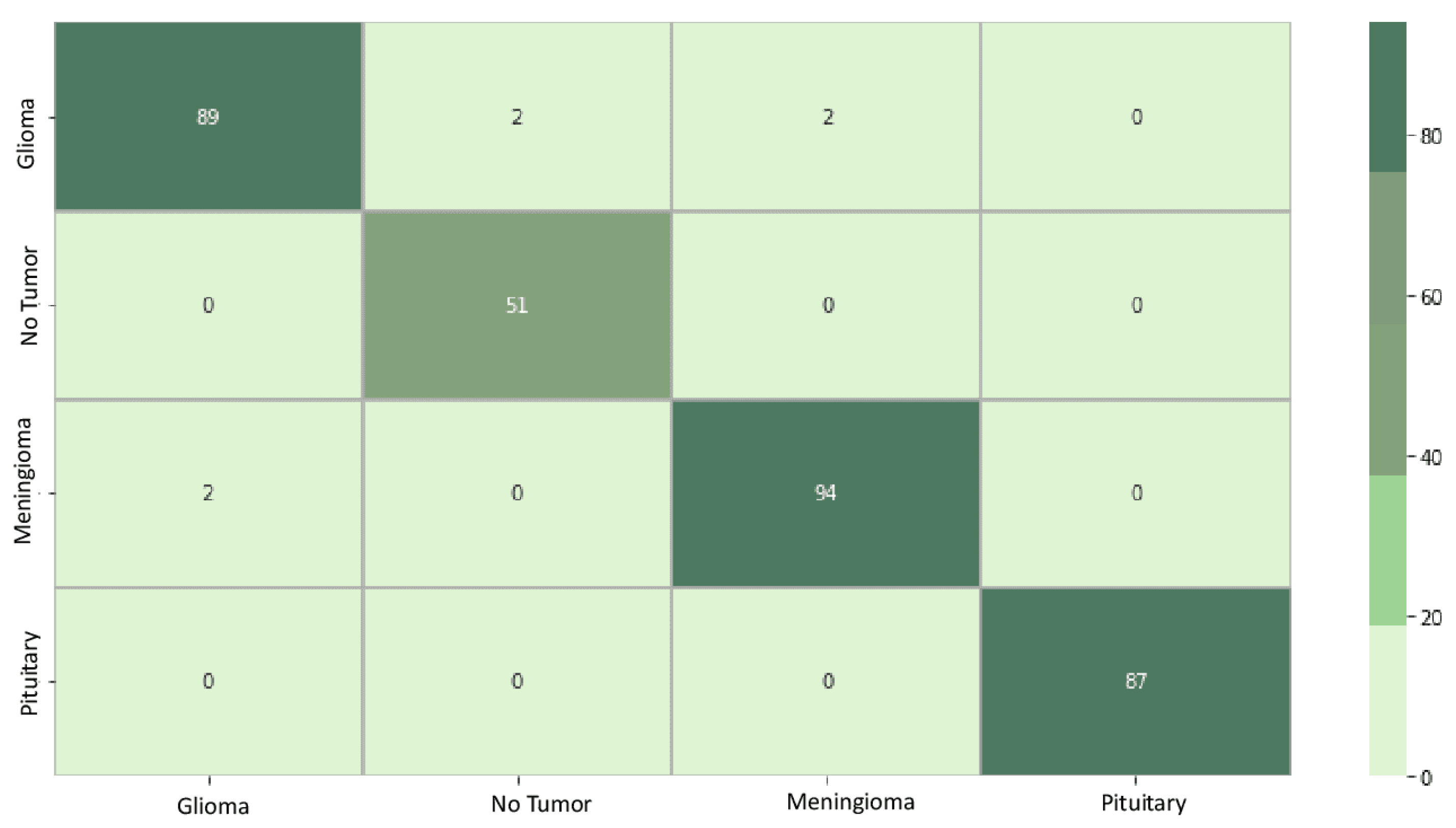
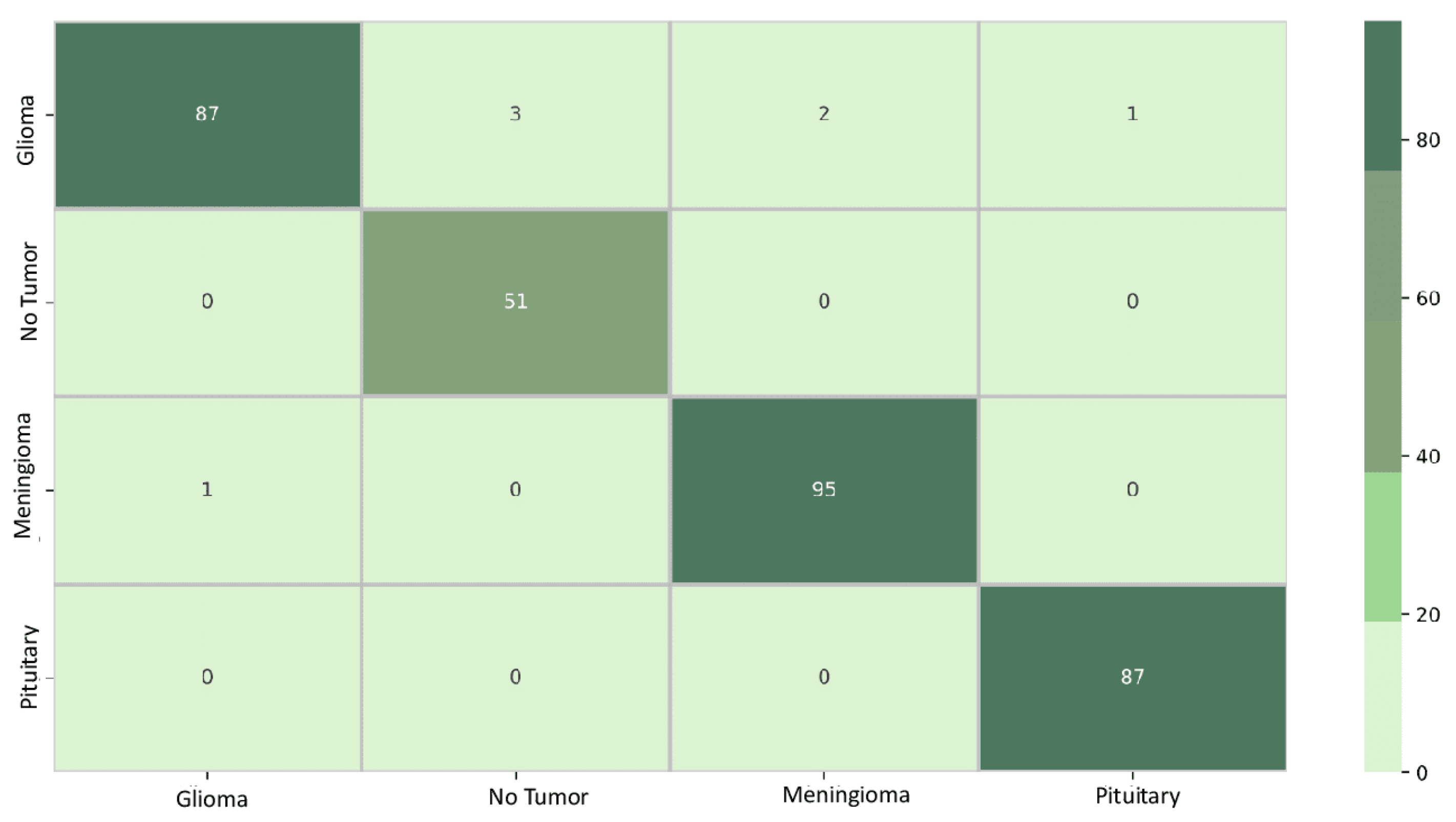
Figure 6 displays the confusion matrix for the EfficientNetB0 model. The diagonal elements represent samples that were accurately predicted. Out of the total 327 samples, 321 were predicted correctly, resulting in an overall accuracy of 98%. The element at row 1 and column 2, a value of 2, indicates that EfficientNetB0 incorrectly learned the classification boundary between classes 1 and 2. This implies that the model confused data initially belonging to class 2 with class 1. Conversely, the element at row 4 and column 1, a value of 0, signifies that the classification boundary between classes 1 and 4 was correctly learned by EfficientNetB0, and the model did not confuse data initially belonging to class 4 with class 1.
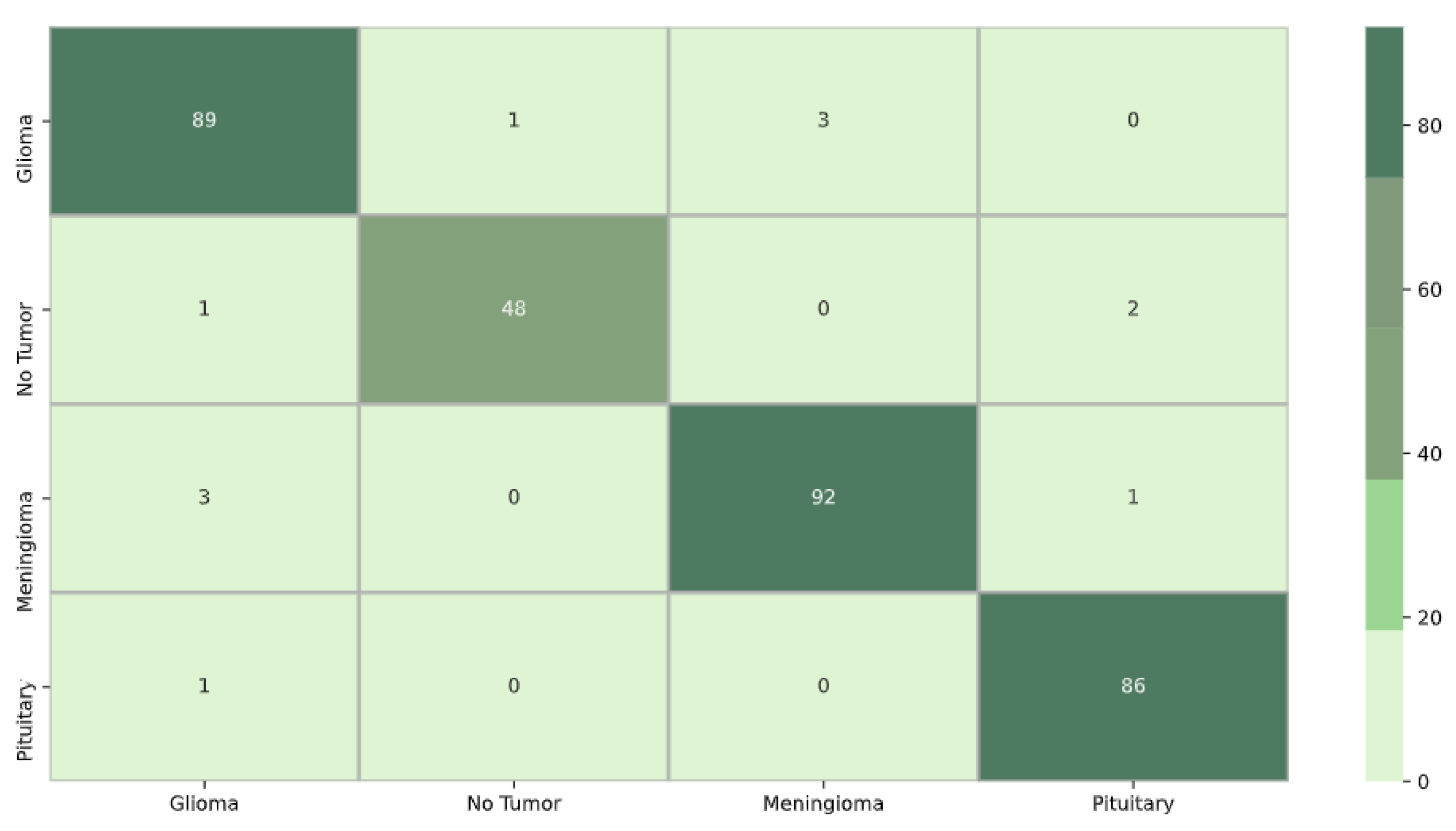
For model DenseNet121 in
Figure 7, the confusion matrix shows that out of the total 327 samples, 320 were accurately predicted, resulting in an overall accuracy of 97%. The value 3 in row 1 and column 2 indicates that the classification boundary between classes 1 and 2 was incorrectly learned by DenseNet121, suggesting that the model confused data initially belonging to class 2 with class 1. Similarly, the presence of 0 in row 2 and column 1 implies that the classification boundary between classes 2 and 1 was correctly learned by DenseNet121, and the model did not confuse data initially belonging to class 1 with class 2.
Figure 8 depicts the confusion matrix for model Xception. Out of the total of 327 samples, 315 were accurately predicted, resulting in an overall accuracy of 96%. The value 1 in row 1 and column 2 indicates that the classification boundary between classes 1 and 2 was incorrectly learned by Xception, suggesting that the model confused data initially belonging to class 2 with class 1. Additionally, the presence of 0 in row 1 and column 4 signifies that the classification boundary between classes 1 and 4 was correctly learned by Xception, and the model did not confuse data initially belonging to class 4 with class 1.
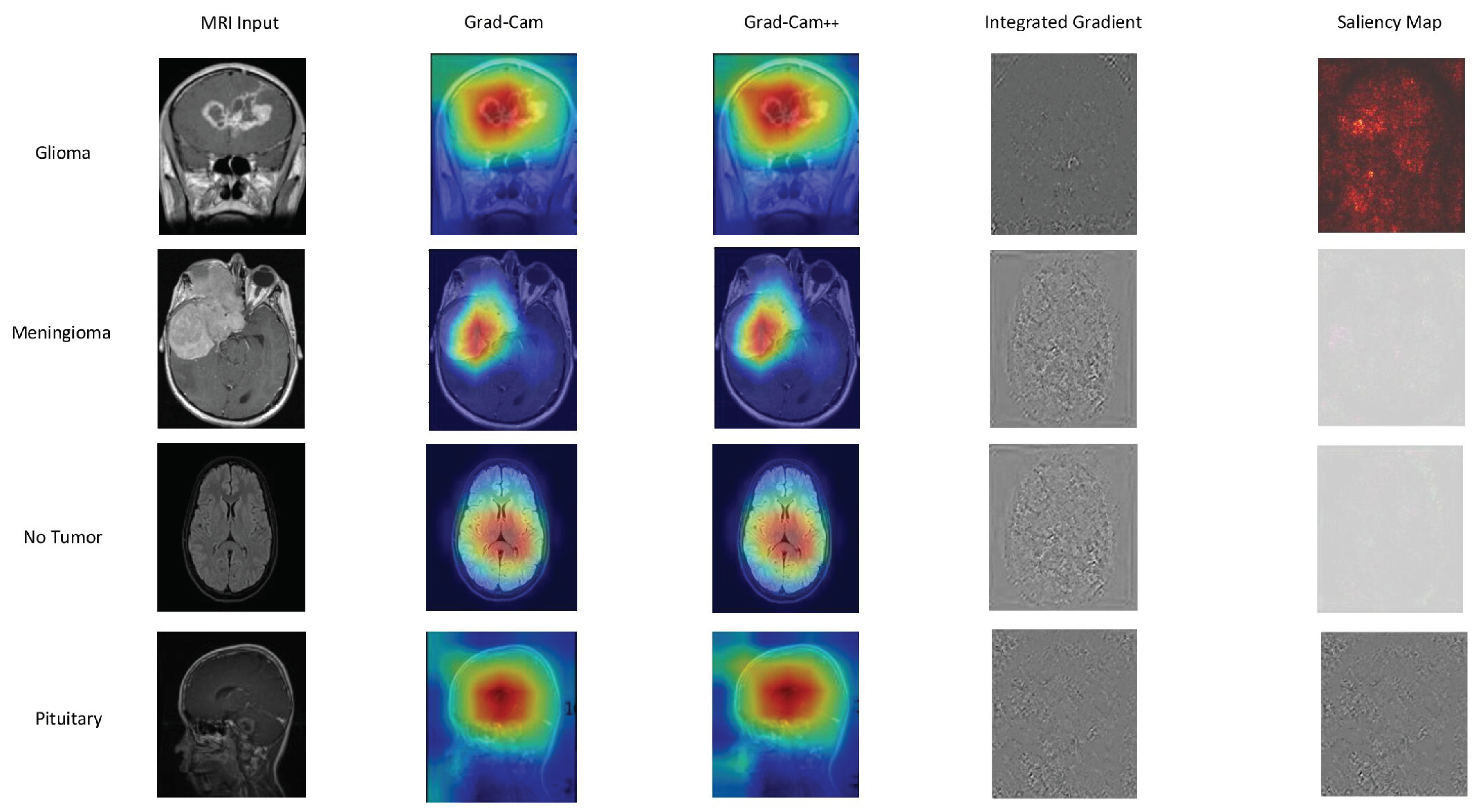
5.2. Interpretability Results
In
Figure 9,
Figure 10, and
Figure 11, we present a detailed look into the explainability results of our top-performing models: EfficientNetB0, DenseNet121, and Xception.
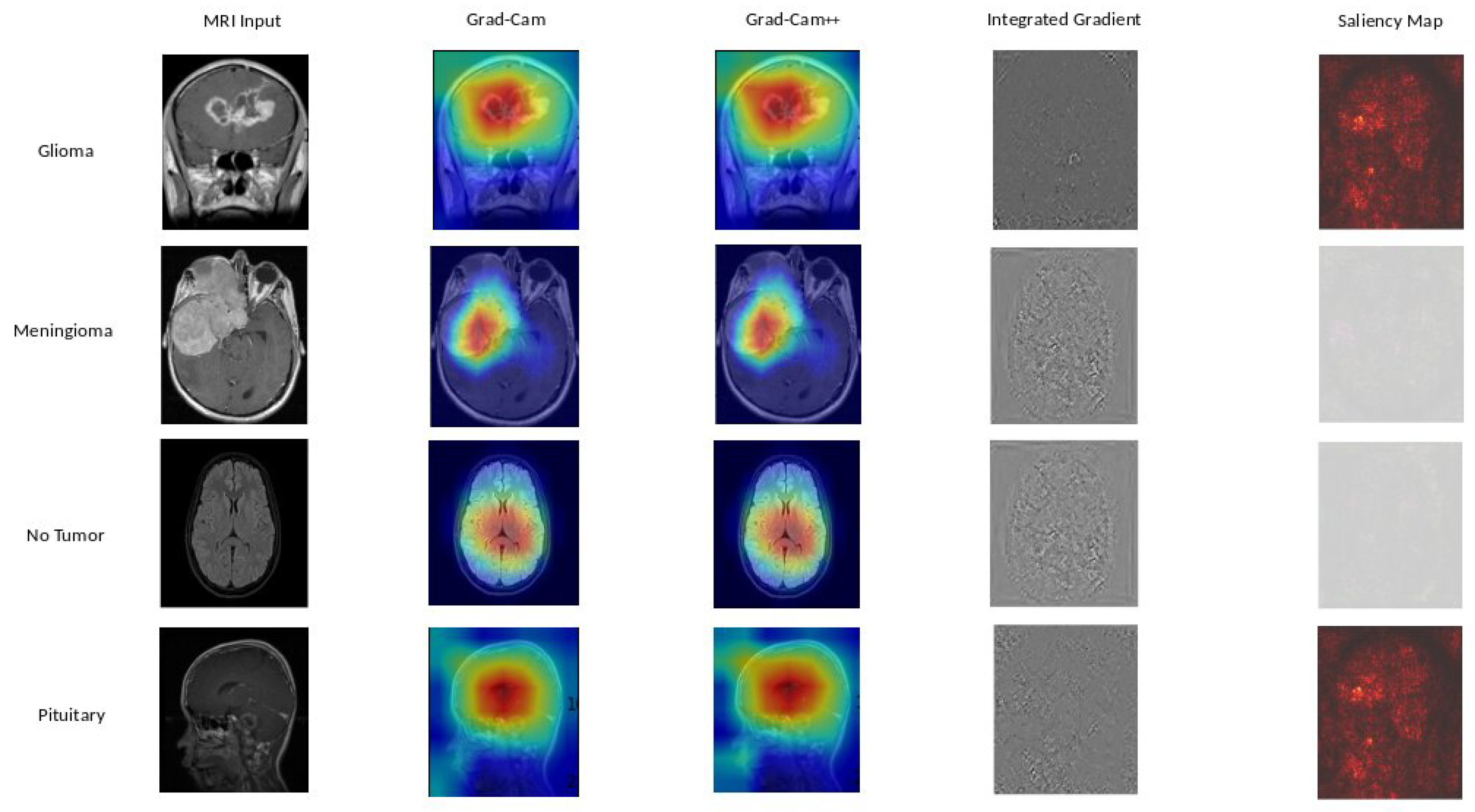
Figure 9 provide insights into the interpretability of EfficientNetB0, highlighting crucial image regions using techniques such as GradCAM, GradCAM++, Integrated Gradients, and Saliency Mapping. Moving to
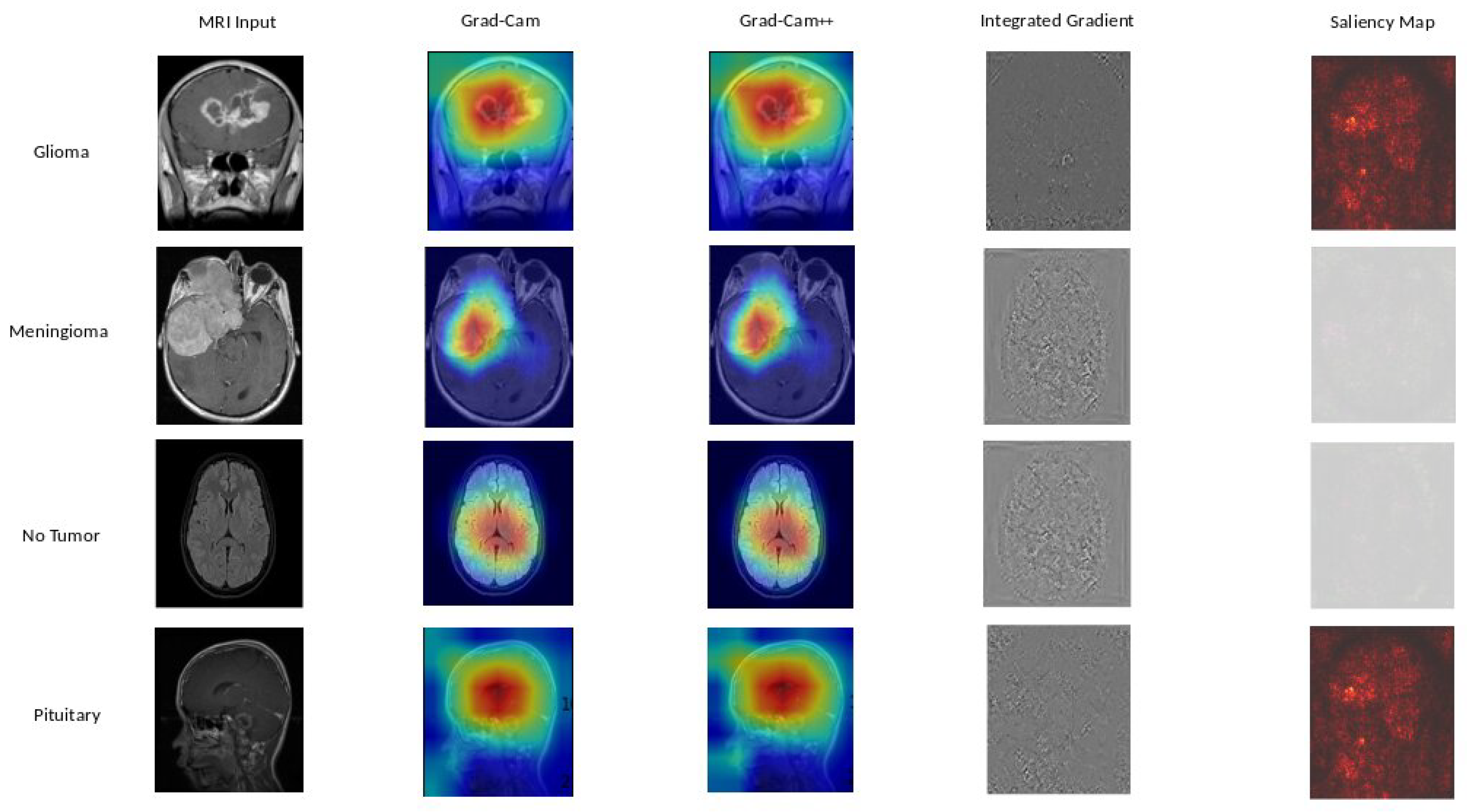
Figure 10, we explore the explainability of DenseNet121, our 2nd best model, uncovering the significant features influencing its predictions.
Figure 11 reveals the interpretability degrees of Xception, our 3rd best model, showcasing the impact of various image regions on classification decisions. These visualizations offer a transparent view into the decision-making processes of our models, facilitating understanding and trust.
Based on our obtained results, both the Grad-Cam and Grad-Cam++ methods demonstrate similar outcomes, offering visual explanations for the decision-making processes of EfficientNetB0, DenseNet121, and Xception in predicting brain tumors. These methods accurately pinpoint the exact location of the tumor, and the visualizations generated by both approaches closely align, suggesting a consistent portrayal of the crucial regions or features influencing the model’s predictions. For a more in-depth examination, refer to
Figure 9 for insights into EfficientNetB0,
Figure 10 for additional perspectives on DenseNet121, and for Xception, consult
Figure 11. Our evaluation of visual explanations centers on their capacity to accurately describe the tumor’s location within the brain, to foster confidence in human interpreters.
However, it is essential to note that Integrated Gradient faced challenges in precisely pinpointing the tumor’s location, and Saliency Mapping exhibited some noise. Despite these challenges, both Grad-Cam and Grad-Cam++ consistently provided excellent visual explanations for the decision processes of EfficientNetB0, DenseNet121, and Xception, significantly enhancing our understanding of their predictive mechanisms.
5.3. Discussion
The findings of evaluating the performance of different DL models provide intriguing new information on the relationship between total accuracy and model complexity, which is reflected by the number of parameters shown in
Table 3. DenseNet121 and EfficientB0 show impressive accuracy values of 97-98% with a lot fewer parameters, indicating how well they use information for the tasks at hand. On the other hand, accuracy is about 85% for VGG16 and VGG19, which have importantly greater parameter counts of more than 138 million. There is a clear exchange between model accuracy and complexity, highlighting the necessity of striking a balance. Interestingly, models with a reasonable amount of parameters, such as AlexNet, ResNet50, GoogleNet, and Xception, achieve competitive accuracy in the 80-96% range, striking a medium ground. Even with a low number of parameters, the ViT Transformer shows greater accuracy variability, highlighting the impact of architectural design on model performance. This study emphasizes how crucial it is to carefully weigh model complexity about the particular task at hand. Overly complicated models may not necessarily result in appreciably higher accuracy and may potentially be more prone to overfitting.
Grad-CAM, Grad-CAM++, Integrated Gradient, and Saliency Mapping show important areas that match with glioma, meningioma, and pituitary classifications. What stands out is the remarkable similarity in outcomes between Grad-CAM and Grad-CAM++ methods, precisely pinpointing tumor locations. This consistency paints a clear picture of the essential regions influencing predictions across all three models, see in
Figure 9,
Figure 10, and
Figure 11 . These visualizations serve a crucial role, not just in making our models transparent, but also in fostering a deeper understanding and instilling trust in how decisions are made.
6. Conclusions
In essence, the objective of this investigation was to assess the proficiency of diverse DL models in categorizing brain tumors. Following a series of comprehensive experiments and detailed analysis, it becomes apparent that these models exhibit varying degrees of competence for the assigned task. Models such as DenseNet121, EfficientNetB0, ResNet50 GoogleNet, and Inception V3 distinguished themselves as top performers with almost flawless levels of accuracy, precision, recall, and F1 scores. For detailed classification results, refer to
Table 3. Conversely, AlexNet and the innovative ViT Transformer a recent contender in the field, displayed potential but fell behind in terms of accuracy and achieving an optimal equilibrium between precision and recall. This research accentuates the significance of carefully selecting the most suitable DL model that aligns with the specific requirements of the application. It further underscores how advancements in neural network architectures, exemplified by the ViT Transformer, persist in shaping the field of DL and computer vision, presenting captivating prospects for future advancements.
In summary, both Grad-Cam and Grad-CAM++ consistently provide a more acceptable insight into model interpretability compared to other methods tested in our study. Put simply, these methods precisely reveal the location of tumors, significantly enhancing our understanding of how DL models make decisions in classifying brain tumors. Therefore, it can be concluded that Grad-Cam and Grad-CAM++ heatmaps have improved our interpretative accuracy, playing a pivotal role in refining our understanding of DL model decision-making processes. These methodologies have been instrumental in enhancing the precision of our interpretations. This study contributes to selecting the correct DL model for brain tumor classification tasks while shedding light on ongoing challenges in making these models transparent and interpretable. Our contributions are outlined as follows:
Model Evaluation: The study comprehensively assesses various DL architectures, providing valuable insights into which models are most effective for brain tumor classification. This evaluation is crucial for guiding the selection of appropriate models in real-world medical imaging applications.
Brain Tumor Diagnosis: Diagnosing a brain tumor is a challenging process that requires the correct and rapid examination of MRI scan images. The study’s findings directly contribute to enhancing the accuracy and reliability of DL models for identifying brain tumors, focusing on this specific medical area. This is critical for early diagnosis and treatment planning for patients.
Model Interpretability: The incorporation of explainability approaches, such as Grad-Cam, Grad-Cam++, Integrated Gradient, and Saliency Mapping, represents a significant scientific contribution. By using these methods, the study increases the interpretability of DL models, shedding light on the decision-making processes and providing valuable intuition into how these models arrive at their classifications, particularly in the context of brain tumor diagnosis.
Figure 1.
Sample of each image in the dataset. A Glioma Tumor image typically exhibits abnormal growth in the brain, indicating potential malignancy. No Tumor images represent a healthy state without any abnormal growth or lesions. Meningioma Tumor images showcase tumors arising from the meninges, the protective layers around the brain, and the spinal cord. Pituitary Tumor images depict tumors in the pituitary gland, influencing hormone regulation and potentially affecting various bodily functions.
Figure 1.
Sample of each image in the dataset. A Glioma Tumor image typically exhibits abnormal growth in the brain, indicating potential malignancy. No Tumor images represent a healthy state without any abnormal growth or lesions. Meningioma Tumor images showcase tumors arising from the meninges, the protective layers around the brain, and the spinal cord. Pituitary Tumor images depict tumors in the pituitary gland, influencing hormone regulation and potentially affecting various bodily functions.
Figure 2.
Brain tumor distribution. This pie chart effectively visualizes the relative proportions of different brain tumor types, offering a clear and concise representation of the distribution within the studied sample. The dataset comprises 926 MRI images of Glioma Tumors, 500 images with No Tumors, 901 images featuring Pituitary Tumors, and 937 images showing Meningioma Tumors.
Figure 2.
Brain tumor distribution. This pie chart effectively visualizes the relative proportions of different brain tumor types, offering a clear and concise representation of the distribution within the studied sample. The dataset comprises 926 MRI images of Glioma Tumors, 500 images with No Tumors, 901 images featuring Pituitary Tumors, and 937 images showing Meningioma Tumors.
Figure 3.
Comprehensive Framework for Brain Tumor Classification: Integrating Classification Accuracy and Model Explainability. (a) In the initial phase of our study, we focused on classifying different types of tumors. However, despite achieving classification results, we found ourselves in the dark about why the models made specific decisions. (b) To address this gap, we transitioned to the next stage of our investigation, delving into explainability methods. In this phase (b), we aim to shed light on the decision-making processes of our models. By employing explainability tools, we seek to unravel the factors and features that influenced the model’s classifications. This shift allows us to not only categorize tumors accurately but also understand the underlying rationale behind the model’s predictions, contributing to a more informed and interpretable outcome.
Figure 3.
Comprehensive Framework for Brain Tumor Classification: Integrating Classification Accuracy and Model Explainability. (a) In the initial phase of our study, we focused on classifying different types of tumors. However, despite achieving classification results, we found ourselves in the dark about why the models made specific decisions. (b) To address this gap, we transitioned to the next stage of our investigation, delving into explainability methods. In this phase (b), we aim to shed light on the decision-making processes of our models. By employing explainability tools, we seek to unravel the factors and features that influenced the model’s classifications. This shift allows us to not only categorize tumors accurately but also understand the underlying rationale behind the model’s predictions, contributing to a more informed and interpretable outcome.
Figure 4.
Training Accuracy and loss are the two main keys that are shown in this figure, which shows the training outcomes of several models. A distinct DL model is represented by each bar in the plot, and the height of the bar indicates the model’s training accuracy. Additionally, the loss values for each model are displayed as a line plot superimposed on the same graph.
Figure 4.
Training Accuracy and loss are the two main keys that are shown in this figure, which shows the training outcomes of several models. A distinct DL model is represented by each bar in the plot, and the height of the bar indicates the model’s training accuracy. Additionally, the loss values for each model are displayed as a line plot superimposed on the same graph.
Figure 5.
Visualizing the comparison of different DL models to provide a clear overview of their performance. The main performance indicators used to assess the effectiveness of various models are highlighted in the legend. The top model is named as EfficientNetB0 due to its excellent results in accuracy, precision, recall, and F1 score, which demonstrate its ability to provide well-balanced and accurate predictions on the provided test dataset.
Figure 5.
Visualizing the comparison of different DL models to provide a clear overview of their performance. The main performance indicators used to assess the effectiveness of various models are highlighted in the legend. The top model is named as EfficientNetB0 due to its excellent results in accuracy, precision, recall, and F1 score, which demonstrate its ability to provide well-balanced and accurate predictions on the provided test dataset.
Figure 6.
EfficientNetB0 confusion matrix. The matrix systematically breaks down the model’s predictions, highlighting instances of true positives (correctly identified cases), true negatives (correctly rejected cases), false positives (incorrectly identified cases), and false negatives (missed cases) for each tumor class.
Figure 6.
EfficientNetB0 confusion matrix. The matrix systematically breaks down the model’s predictions, highlighting instances of true positives (correctly identified cases), true negatives (correctly rejected cases), false positives (incorrectly identified cases), and false negatives (missed cases) for each tumor class.
Figure 7.
DenseNet121 confusion matrix.
Figure 7.
DenseNet121 confusion matrix.
Figure 8.
Xception confusion matrix.
Figure 8.
Xception confusion matrix.
Figure 9.
EfficientNetB0 explainability: We used a variety of explainability approaches, such as Grad-CAM, Grad-CAM++, Integrated Gradient, and Saliency Mapping, in our evaluation of EfficientNetB0 for brain tumor classification. These techniques played an important role in assisting in identifying the specific regions in MRI scan images that corresponded to the tumor types such as glioma, meningioma, no tumor, and pituitary. By using these techniques, we were able to determine the critical locations for the categorization of each tumor type and obtain important insights into EfficientNetB0’s decision-making process.
Figure 9.
EfficientNetB0 explainability: We used a variety of explainability approaches, such as Grad-CAM, Grad-CAM++, Integrated Gradient, and Saliency Mapping, in our evaluation of EfficientNetB0 for brain tumor classification. These techniques played an important role in assisting in identifying the specific regions in MRI scan images that corresponded to the tumor types such as glioma, meningioma, no tumor, and pituitary. By using these techniques, we were able to determine the critical locations for the categorization of each tumor type and obtain important insights into EfficientNetB0’s decision-making process.
Figure 10.
DenseNet121 explainability: We used a variety of explainability approaches, such as Grad-CAM, Grad-CAM++, Integrated Gradient, and Saliency Mapping, in our evaluation of DenseNet121 for brain tumor classification. These techniques played an important role in assisting in identifying the specific regions in MRI scan images that corresponded to the tumor types such as glioma, meningioma, no tumor, and pituitary. By using these techniques, we were able to determine the critical locations for the categorization of each tumor type and obtain important insights into DenseNet121’s decision-making process.
Figure 10.
DenseNet121 explainability: We used a variety of explainability approaches, such as Grad-CAM, Grad-CAM++, Integrated Gradient, and Saliency Mapping, in our evaluation of DenseNet121 for brain tumor classification. These techniques played an important role in assisting in identifying the specific regions in MRI scan images that corresponded to the tumor types such as glioma, meningioma, no tumor, and pituitary. By using these techniques, we were able to determine the critical locations for the categorization of each tumor type and obtain important insights into DenseNet121’s decision-making process.
Figure 11.
Xception explainability: We used a variety of explainability approaches, such as Grad-CAM, Grad-CAM++, Integrated Gradient, and Saliency Mapping, in our evaluation of Xception for brain tumor classification. These techniques played an important role in assisting in identifying the specific regions in MRI scan images that corresponded to the tumor types such as glioma, meningioma, no tumor, and pituitary. By using these techniques, we were able to determine the critical locations for the categorization of each tumor type and obtain important insights into Xception’s decision-making process.
Figure 11.
Xception explainability: We used a variety of explainability approaches, such as Grad-CAM, Grad-CAM++, Integrated Gradient, and Saliency Mapping, in our evaluation of Xception for brain tumor classification. These techniques played an important role in assisting in identifying the specific regions in MRI scan images that corresponded to the tumor types such as glioma, meningioma, no tumor, and pituitary. By using these techniques, we were able to determine the critical locations for the categorization of each tumor type and obtain important insights into Xception’s decision-making process.
Table 1.
Training Hyperparameters
Table 1.
Training Hyperparameters
| Hyperparameter |
Setting |
| Batch size |
32 |
| Learning rate |
|
| Epochs |
10 |
| Training split |
90% |
| Test split |
10% |
| Optimizer |
Adam |
| Input size |
pixels |
| Loss function |
Categorical cross-entropy |
Table 2.
Training Results
Table 2.
Training Results
| Model Name |
Training Accuracy |
Loss |
| AlexNet |
0.8763 |
0.3233 |
| DenseNet121 |
0.9986 |
0.0057 |
| EfficientB0 |
0.9991 |
0.0042 |
| GoogleNet |
0.9997 |
0.0.0027 |
| Inception V3 |
0.9989 |
0.0084 |
| ResNet50 |
0.9991 |
0.0044 |
| VGG16 |
0.8698 |
0.4011 |
| VGG19 |
0.8570 |
0.3953 |
| Vision Transformer |
0.7484 |
0.5115 |
| Xception |
1.0000 |
0.0021 |
Table 3.
Test dataset classification results where various metrics are considered for each model.
Table 3.
Test dataset classification results where various metrics are considered for each model.
| Model Name |
Parameters |
Accuracy % |
Precision % |
Recall % |
F1 Score % |
| AlexNet |
61.9M |
78 |
80 |
77 |
77 |
| DenseNet121 |
8.1M |
97 |
97 |
97 |
97 |
| EfficientB0 |
5.3M |
98 |
98 |
98 |
98 |
| GoogleNet |
11.2M |
91 |
93 |
92 |
92 |
| Inception V3 |
23.9M |
96 |
97 |
96 |
96 |
| ResNet50 |
25.6M |
95 |
96 |
96 |
96 |
| VGG16 |
138.4M |
85 |
85 |
86 |
85 |
| VGG19 |
143.7M |
85 |
85 |
85 |
85 |
| ViT Transformer |
86M |
70 |
72 |
72 |
70 |
| Xception |
22.9M |
96 |
97 |
96 |
96 |