1. Introduction
Through the clinical experience of the wise ancestors and continuous exploration, the science of traditional Chinese medicine has developed more mature, with a complete theoretical basis and clinical diagnosis and treatment system. Syndrome differentiation and treatment is the main method of traditional Chinese medicine (TCM) diagnosis and treatment, of which the key link is symptom description, and this part of information mostly exists in the form of text in TCM literature resources. In order to carry out knowledge reasoning from these text information, we first need to apply information extraction technology. The development of entity recognition and extraction technology has roughly gone through three stages
[1]: entity extraction based on pattern matching, entity extraction based on machine learning and entity extraction. Accurate identification and extraction of the elements of TCM syndrome differentiation is the basis for realizing intelligent differentiation of TCM. On this basis, the use of knowledge graph technology methods can more deeply mine TCM knowledge, and provide more powerful support for clinical auxiliary diagnosis and treatment applications. At present, there have been many related studies on the use of entity recognition models
[2,3,4,5] for TCM text entity recognition. In recent years, there has been progress in entity recognition in the field of traditional Chinese medicine. However, the diversity, complexity and irregularity of data require more accurate annotation methods. Different models need to be compared to select more accurate and efficient entity recognition to support research and clinical application. In this study, TCM symptom information is annotated and processed, and entity recognition technology is selected for automatic recognition to improve the accuracy and provide support for TCM information extraction and mining.
2. Data and Methods
2.1. Data Sources
The data sources of this study were mainly from three aspects. The data of teaching cases were from the seventh edition of Internal Medicine of Traditional Chinese Medicine textbooks and related teaching reference materials, the data of clinical cases were mainly from the Essence of Modern Chinese famous medical Cases, and the data of literature were mainly from the medical cases in relevant papers published on CNKI. The cough was selected in this study, and all data were collected by manual entry and review. The required text was input into excel sheet for storage, and 100 cases of each data were collected.
2.2. Model Methods
2.2.1. Named entity recognition
Named entity recognition (NER) refers to the extraction of words with specific meanings from the specified free text. In general texts, entities usually refer to organization names, place names, and personal names. However, in the field of traditional Chinese medicine, entities mainly refer to symptoms, tongue images, pulse conditions, disease names, drugs, parts, times, and diagnosis and treatment sites. Named entity recognition (NER) technology has gone through three stages. In the early research, it is generally based on manually built rules and dictionaries
[6]. However, it can only be applied to specific scenarios, and cannot be promoted and applied in a large area. Later, traditional machine learning methods represented by CRF
[7] and the combination of rules and dictionary methods have reduced the dependence on manual work in entity recognition, but there are some problems such as low efficiency of feature extraction and long training time of model. In recent years, deep learning has made great progress. More text feature information can be captured, which greatly improves the training efficiency of the model, and the automatic extraction function greatly reduces the dependence on manual work.
2.2.2. BiLSTM-CRF
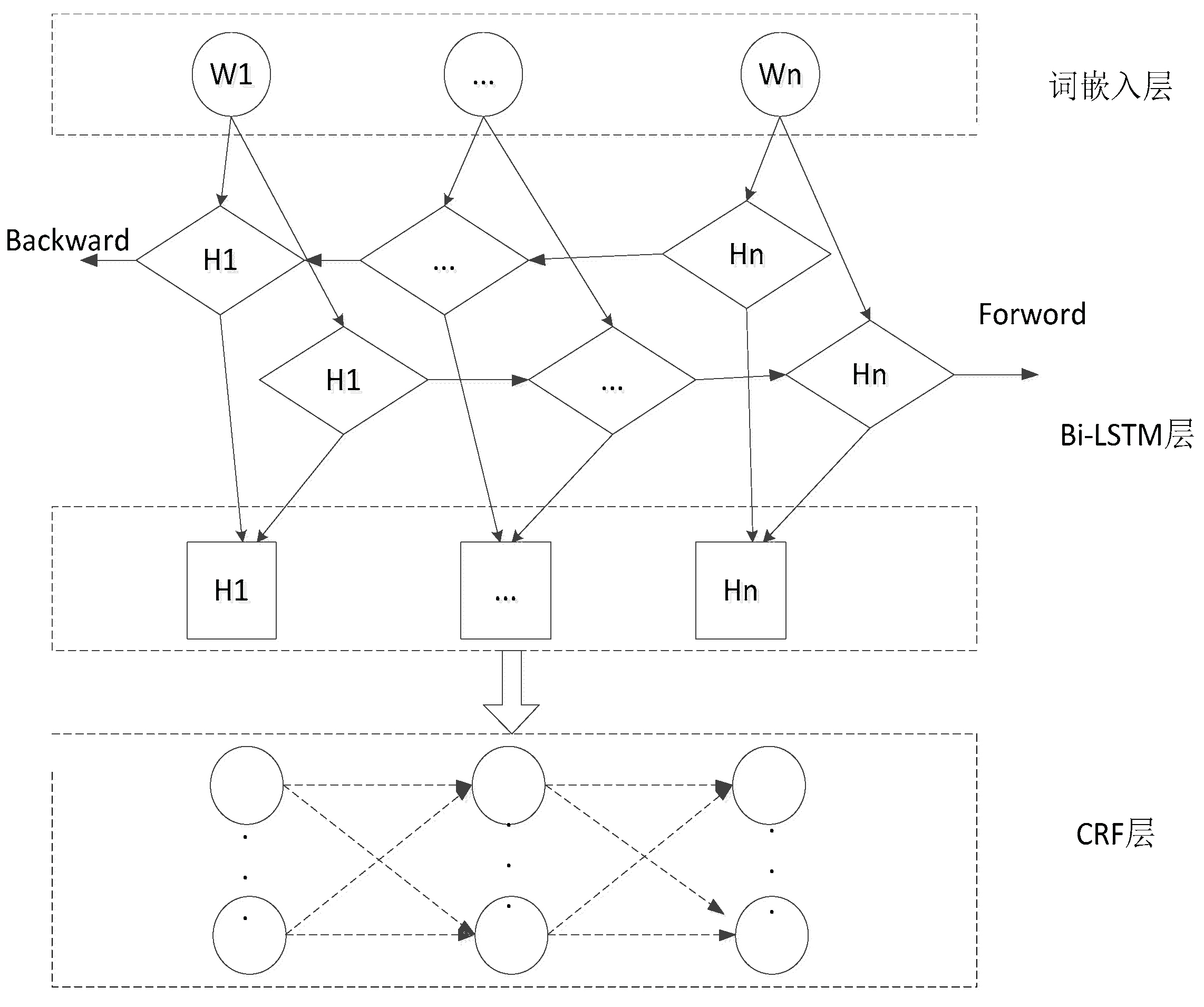
The bilSTM-CRF model mainly includes a four-layer structure of word embedding layer, BiLSTM layer, CRF layer and output layer, as shown in
Figure 1. (1) Word embedding layer: The external text input sequence is converted into the word vector sequence of the corresponding word, and the word vector is concatenated according to the window size. (2) BiLSTM layer: it models the semantic information of the word vector sequence, extracts the text feature expression, and outputs to the hidden layer before and after splicing. (3) CRF layer: The transition probability matrix is obtained by multiplying the output sequence of the hidden layer with the parameter matrix, and then the maximum likelihood estimation method is used to solve the model parameters. In the prediction process, the dimension bit algorithm is used to dynamically decode the label sequence that maximizes the objective function. (4) Output layer: directly output the entity prediction label.
2.2.3. BERT-BiLSTM-CRF
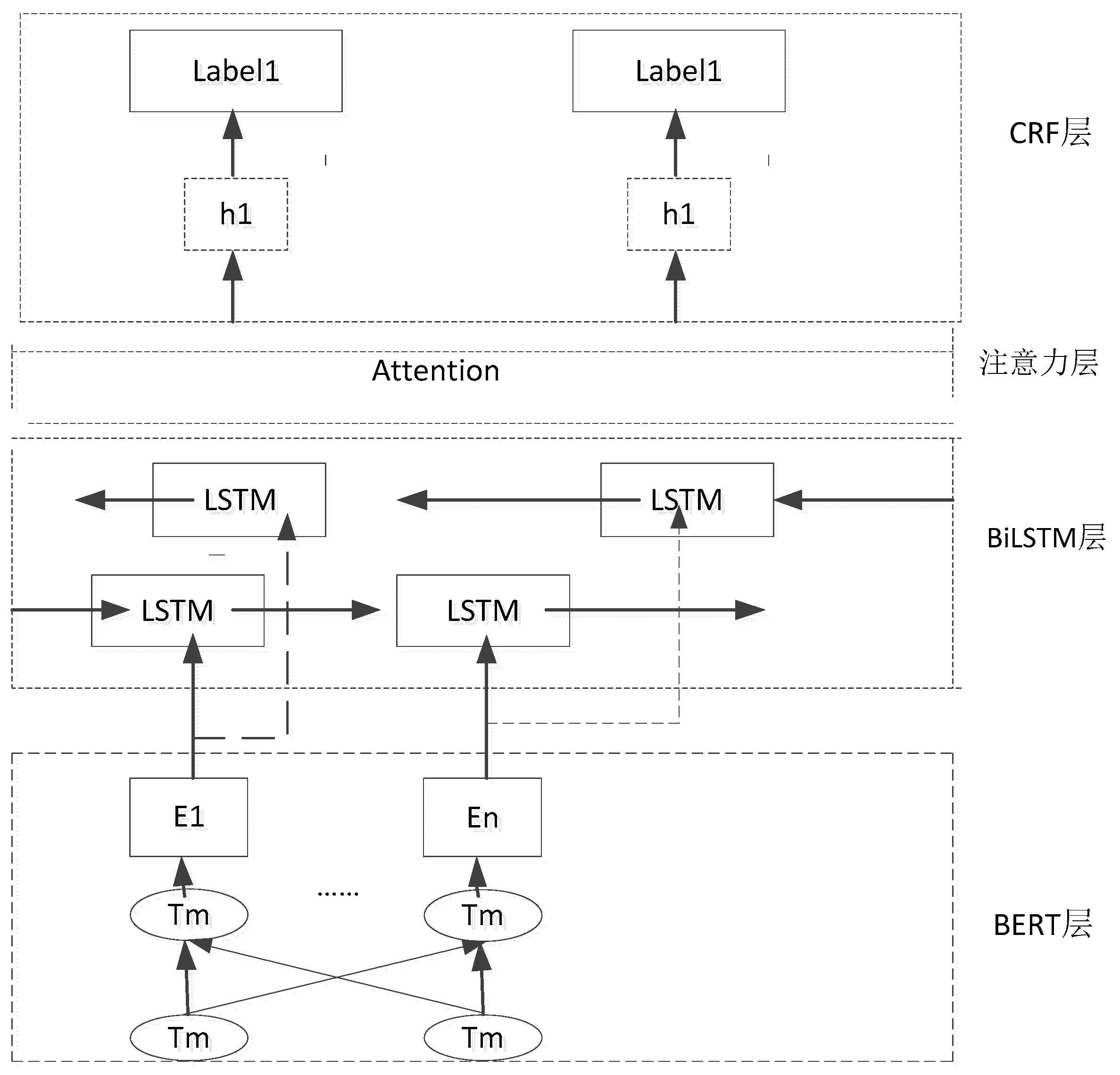
The BERT-bilSTM-CRF model framework is shown in
Figure 2, which is composed of four modules. The BERT layer generates dynamic word vectors by pre-training the external input text data, and the word vector information becomes the input of the BiLSTM layer to complete the bidirectional training, so as to further extract text features
[8]. The attention mechanism mainly extracts the feature information that plays a decisive role in entity recognition from the output results of the BiLSTM layer, assigns the weights, and uses the weight check to directly evaluate which embeddings are the preferred embeddings for specific downstream tasks. Finally, the CRF layer can effectively constrain the dependence between the predicted labels and model the label sequence. In this way, the global optimal sequence
[9] can be obtained.
2.2.4. BERT
Bert is built on top of Transformers with powerful language representation and feature extraction capabilities.
[10]Achieving state of the art on several NLP benchmark tasks, the BERT model used in this study is partially consistent with the BERT model in 2.2.3.
3. Experimental Procedure
3.1. Data Preprocessing
Firstly, data preprocessing was carried out, including eliminating duplicate data, correcting typos, and removing redundant Spaces. The samples of the three types of data after preprocessing are shown in
Table 1. In the experiment, all kinds of data were divided into training set, validation set and test set in the form of 6:2:2.
3.2. Construction of corpus
3.2.1. Data annotation
A corpus is a collection of corpora (texts, etc.) collected for special processing of natural languages. It is characterized by representativity, structure, certain scale and detectability. It is a structured, representative and large-scale corpus text collection
[11] specially collected for one or more application goals. The use of deep learning technology for information extraction requires a large number of manually labeled corpora. TCM clinical syndrome differentiation information usually includes symptoms, symptom duration, tongue diagnosis, pulse diagnosis, etc
[12]. In this study, five entity corpora including symptoms, course of disease, tongue texture, tongue coating and pulse condition were constructed according to the published standards of TCM Clinical Basic Symptom Information Classification and Code, Tongue Diagnosis classification and Code Standard, and pulse diagnosis classification and Code standard.
3.2.2. Training annotation
In the work of named entity recognition, each word needs to be labeled. This study uses BIO labeling method, B-begin represents the beginning of the entity, I-inside represents the middle or end of the entity, and O-outside represents the entity that does not belong to the entity.
3.3. Experimental environment
Python 3.8 was used as the main programming language in this study, and the model was built based on PyTorch 2.2 architecture. The computer operating system was Ubuntu20.04, the memory was 64GB, and the NVIDIA A100 80G GPU computing card was equipped.
3.4. Model parameters
In this experiment, the learning rate, batch-size and epoch were mainly adjusted. The learning rate is set as 0.1, 0.01, 0.001 and 0.0001, the batch size is set as 2, 4, 8, 16, 32 and 64, and the iteration number is set as 10, 20, 33, 40, 40, 50, 60, 70, 80, 90 and 100. By modifying these three parameters, the experiment is carried out continuously. Finally, the most appropriate information recognition model and parameter Settings for each type of TCM diagnosis and treatment data were selected.
3.5. Evaluation Metrics
In this study, precision, recall and F1 value were used to evaluate the model effect. TP represents the number of entities correctly identified by the model, FP represents the number of non-relevant entities identified by the model, and FN represents the number of relevant entities not detected by the model.
4. Results
4.1. Entity recognition results
In the entity recognition experiments of symptoms, tongue, tongue coat, pulse condition, disease course and other entities in three types of TCM diagnosis and treatment data, the entity recognition effect of BERT-BiLSTM-CRF model is better than the other two models. The BERT-BiLSTM-CRF model is used for entity recognition of the test data, and the obtained information recognition results are shown in
Table 2.
4.2. Model identification results
Aiming at the relevant annotation information in the text data of cough, a common disease in traditional Chinese medicine, three models are used for entity recognition. In the experiment, the values of learning rate, batch-size, and epoch were modified to compare the recognition effect of the model. The experimental results are shown in
Table 3.
Through experiments, it is found that the BERT-BiLSTM-CRF model has the best effect on entity recognition in traditional Chinese medicine teaching cases, and the F1 value is 0.967. Secondly, the BERT model has an F1 value of 0.926, which achieves good recognition effect. The BERT-BiLSTM-CRF model has the best effect on entity recognition in traditional Chinese medicine literature, with an F1 value of 0.91. The second is the BERT model, and the difference is not very large. The BERT-BiLSTM-CRF model still has the best effect on entity recognition in TCM clinical medical cases, with an F1 score of 0.82, which shows that this method has good robustness in different corpus environments.
5. Discussion
During the experiment, it is observed that introducing the BERT pre-training layer into the BiLSTM-CRF model will greatly increase the training time, but can significantly improve the entity recognition effect. When using the BERT-BiLSTM-CRF model for entity recognition, it can achieve good recognition results for different types of traditional Chinese Medicine text data. The study also shows that the performance of the model is greatly affected by the learning rate and the number of batch samples. Different learning rate Settings will significantly affect the entity recognition effect, and a smaller batch size will reduce the number of samples used in one iteration, but it will also significantly increase the training time of the model. At the same time, by performing entity recognition experiments on the same traditional Chinese medicine text data, it can be observed that the entity recognition effect of the BERT model is slightly lower than that of the BERT-bilSTM-CRF model, but the model training time is significantly shorter. Considering the time factor, the BiLSTM-CRF model shows great advantages when dealing with TCM clinical diagnosis and treatment text data.
6. Conclusions
In this study, three mainstream entity recognition models are used to conduct entity recognition experiments on three types of common texts in the field of TCM syndrome differentiation diagnosis and treatment. A corpus of five types of entities, including symptoms, course of disease, tongue texture, tongue coating and pulse condition, is constructed for entity recognition experiments. In the experiment, the model parameters suitable for a certain type of data are selected for different models. Through the comparative experiments, it can be found that the degree of text structure and the quality of corpus have an important impact on the effect of entity recognition. In the future research, the entity corpus will be expanded and modified to solve the problems of entity nesting and negative prefixes, and constantly improve the corpus. At the same time, more fusion research methods can be explored, and the TCM knowledge graph technology can be combined to lay a foundation for the next step of knowledge reasoning research.
Funding
This work was supported by National Natural Science Foundation of China (No.82004503) and Science and Technology Project of Hebei Education Department(BJK2024108).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kong Jingjing, Yu Qi, Li Jinghua et al. A review of entity Extraction and its application in the field of Traditional Chinese Medicine [J]. World Science and Technology-Traditional Chinese Medicine Modernization 2012, 24, 2957–2963. [Google Scholar]
- Li C, Xie D. Research on automatic extraction method of admission record information of traditional Chinese medicine electronic medical record [J]. World Science and Technology-Modernization of Traditional Chinese Medicine 2023, 25, 1615–1622. [Google Scholar]
- Xu Lina, Li Yan, Zhong Xinyu, Chen Yueyue, Shuai Yaqi. Named Entity Recognition of Traditional Chinese Medicine Prescription Text Based on Bert [J]. Medical Information 2023, 36, 32–37. [Google Scholar]
- Gao Jiayi, Yang Tao, DONG Haiyan et al. Chinese Journal of Traditional Chinese Medicine Information 2021, 28, 20–24.
- Liu Andong, Peng Lin, Ye Qing, et al. Research progress of Named entity recognition in electronic medical records [J]. Computer Engineering and Application 2023, 59, 39–51. (in Chinese). [Google Scholar] [CrossRef]
- Wang Z, Liu L, Yao K, et al. TCM-SAS: A Semantic Annotation System and Knowledgebase of Traditional Chinese Medicine. In Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); IEEE, 2022; pp. 3727–3732.
- Wu Zong-you, Bai Kun-long, Yang Lin-rui et al. Review on Text Mining of Electronic Medical Records [J]. Journal of Computer Research and Development 2021, 58, 513–527. [Google Scholar]
- Shuai Yaqi, Li Yan, Chen Yueyue et al. Entity recognition of chronic bronchitis Traditional Chinese Medicine Cases Based on BERT-BILSTM-CRF [J]. Modern Information Technology 2023, 7, 145–148+152. [Google Scholar]
- Xie Teng, Yang Junan, Liu Hui. BERT-BiLSTM-CRF Model for Chinese Entity Recognition [J]. Applications of Computer Systems 2020, 29, 48–55. [Google Scholar]
- Devlin J, Chang M W, Lee K, et al. Bert: Pre-training of deep bidirectional transformers for language understanding[J]. arXiv 2018, arXiv:1810.04805.
- Deng N, Fu H, Chen X. Named entity recognition of traditional Chinese medicine patents based on BiLSTM-CRF[J]. Wireless Communications and Mobile Computing 2021, 2021, 1–12. [Google Scholar]
- Sun S, H. Research on key technologies of information extraction in acupuncture and moxibustion field of traditional Chinese Medicine [D]. Dalian University of Technology, 2019. [Google Scholar]
- Yi Junhui, ZHA Qinglin. Review of TCM Symptom Information Extraction [J/OL]. Computer Engineering and Application 2023, 1–15.
- Cao S, Wu Q. MC-TCMNER: A Multi-modal Fusion Model Combining Contrast Learning Method for Traditional Chinese Medicine NER[C]. International Conference on Multimedia Modeling; Springer Nature: Cham, Switzerland, 2024; pp. 341–354.
- Smith, L.N. Cyclical learning rates for training neural networks. In Proceedings of the 2017 IEEE Winter Conference on Applications of Computer Vision (WACV); 2017; pp. 464–472. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).