Submitted:
07 February 2024
Posted:
08 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. TCGA RNA-Sequencing Dataset
2.2. Overall Survival Analysis on TCGA RNA-Sequencing with Multi-Omics Integration in Colorectal Cancer
2.3. ChIP-Sequencing Analysis
2.4. Integrative Analysis
2.5. Deep Learning
2.6. Multivariable Model Built on Methylation Status Outcome
2.7. Statistical Analyses
3. Results
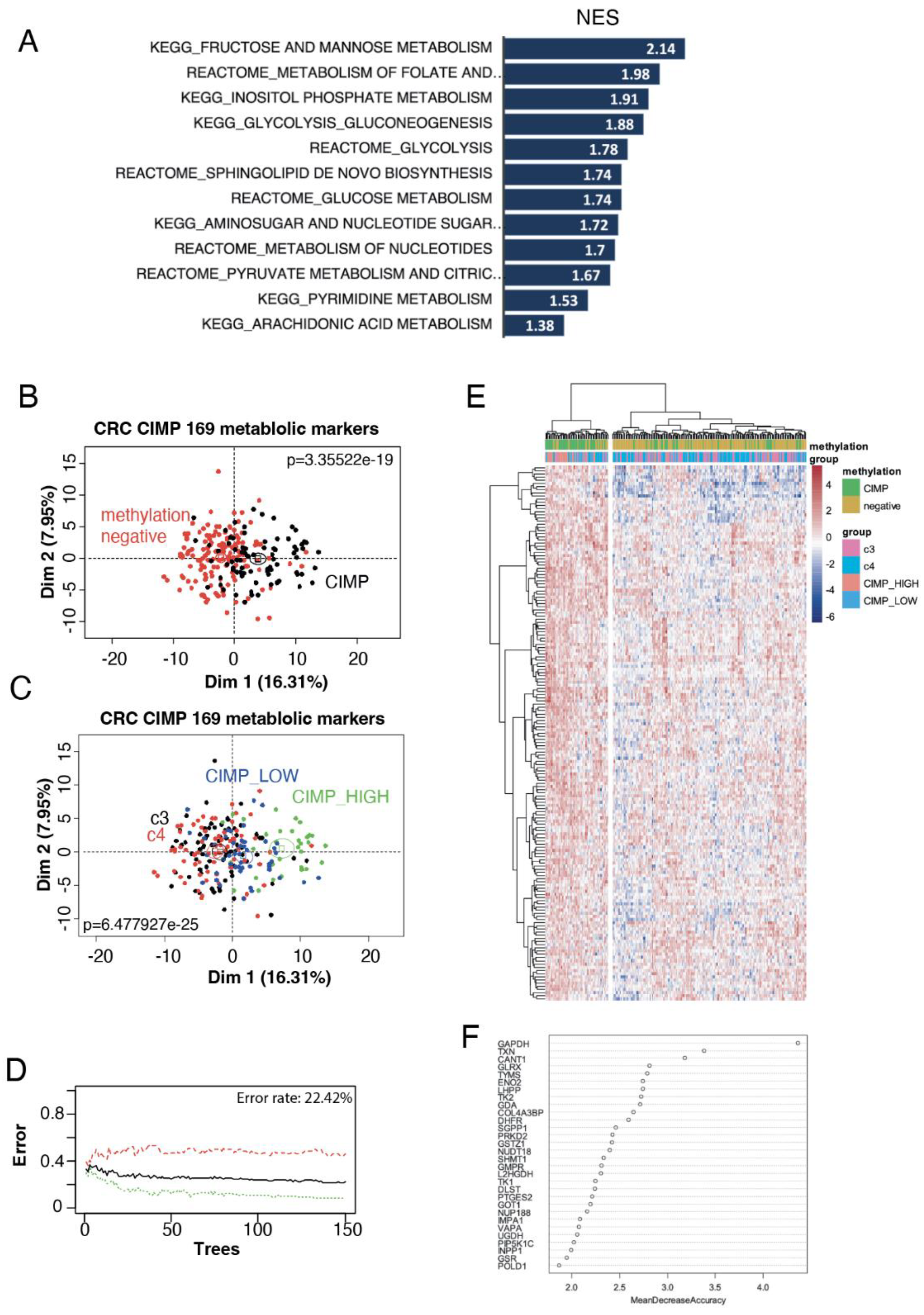
3.1. Hypermetabolism in CIMP CRC Transcriptome
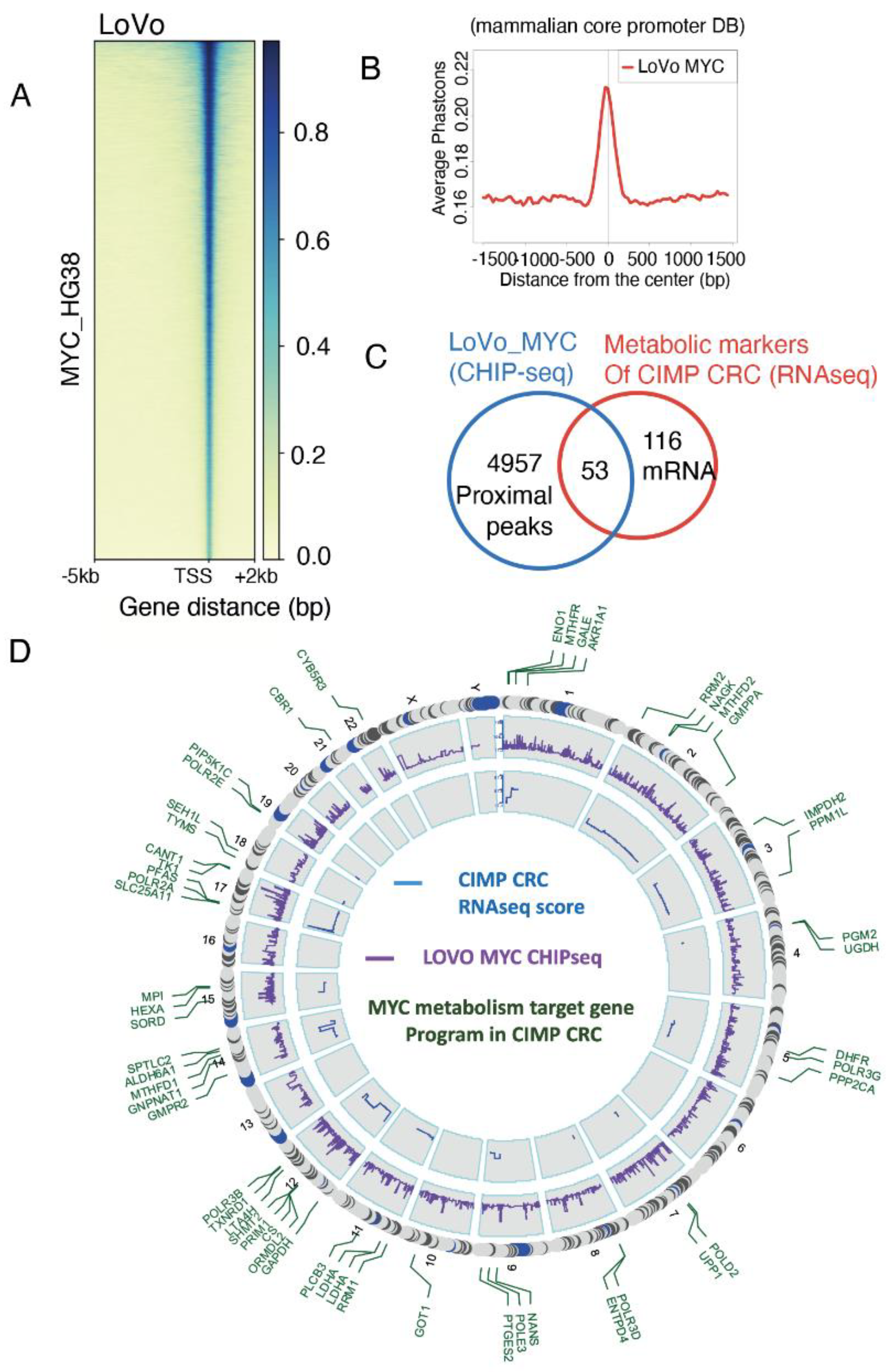
3.2. Myc Regulates one Third of the CIMP-CRC Metabolic Program
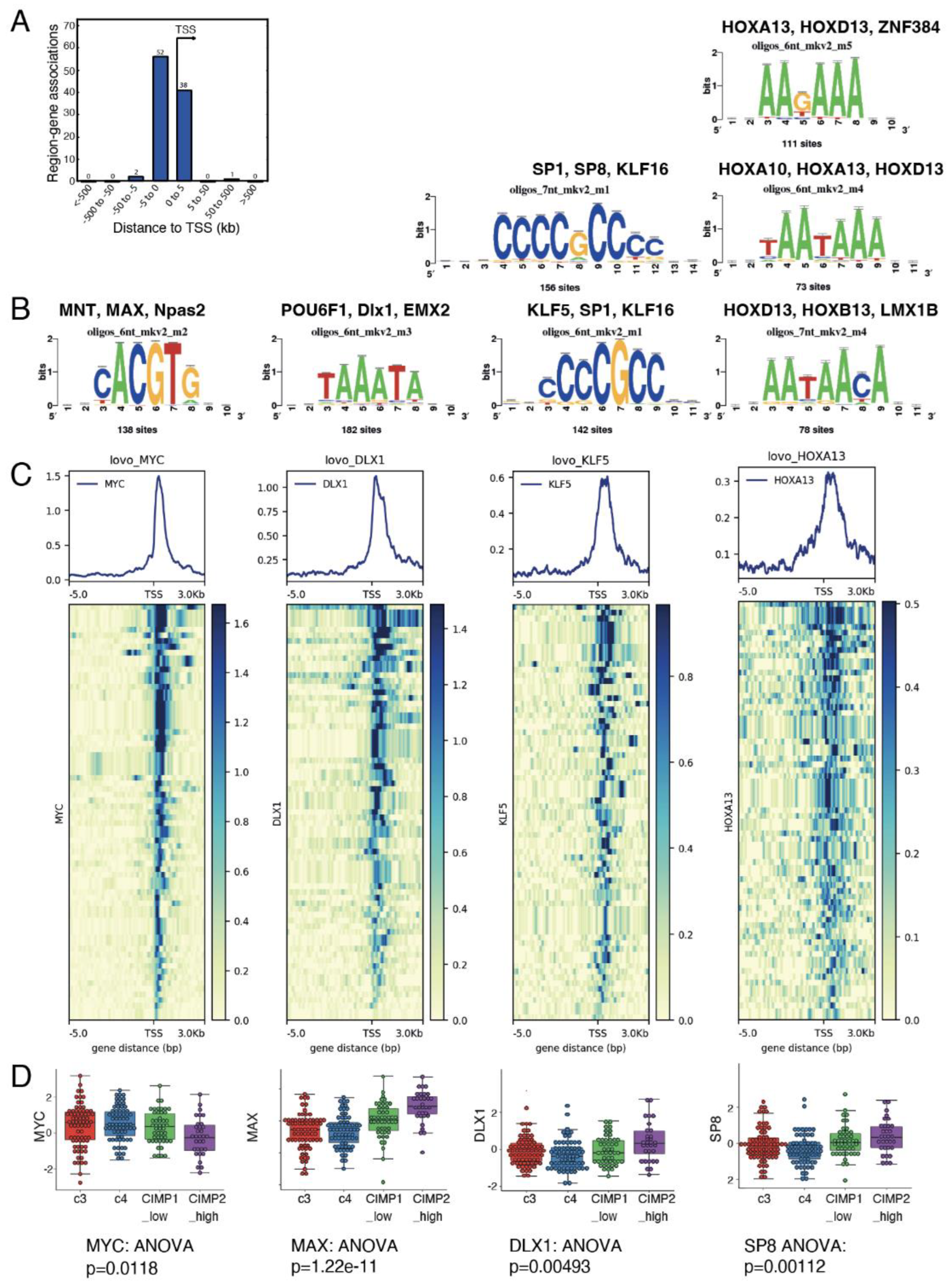
3.3. Genes from the Myc Transcriptional Program Also Have Binding Sites for Other Transcription Factors
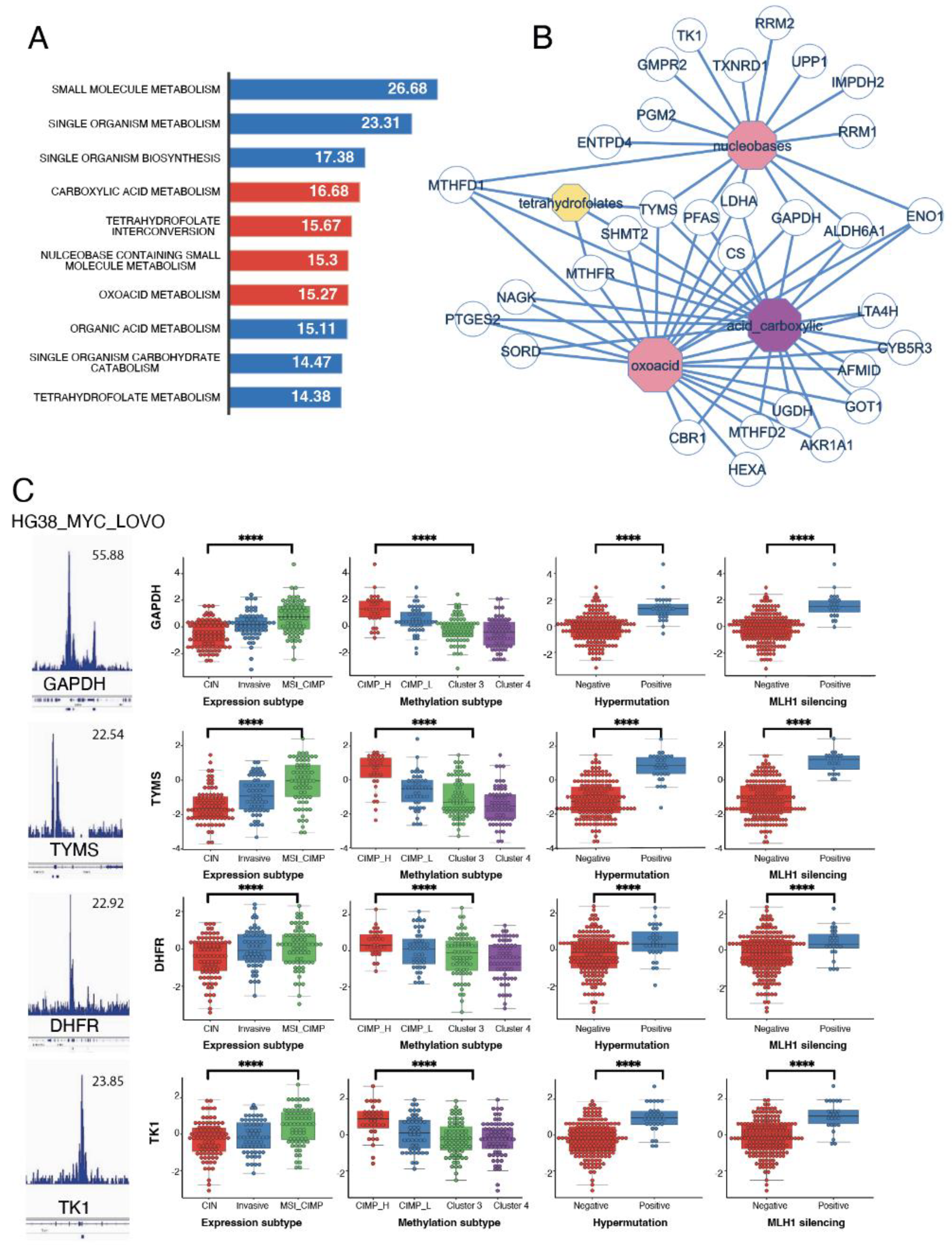
3.4. Metabolism Targets in the Myc Signature Are Associated to Worst Clinical Group in CRC
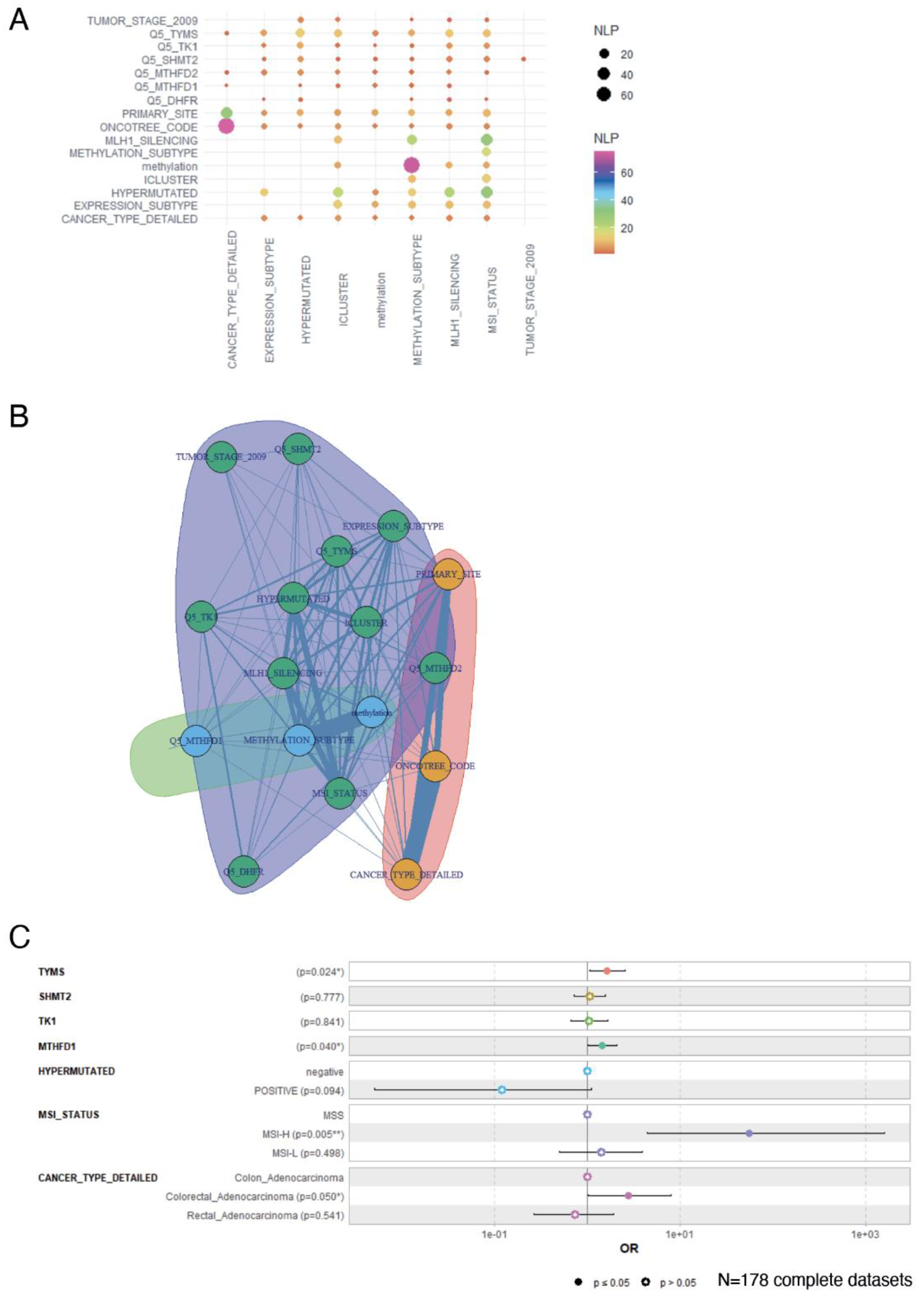
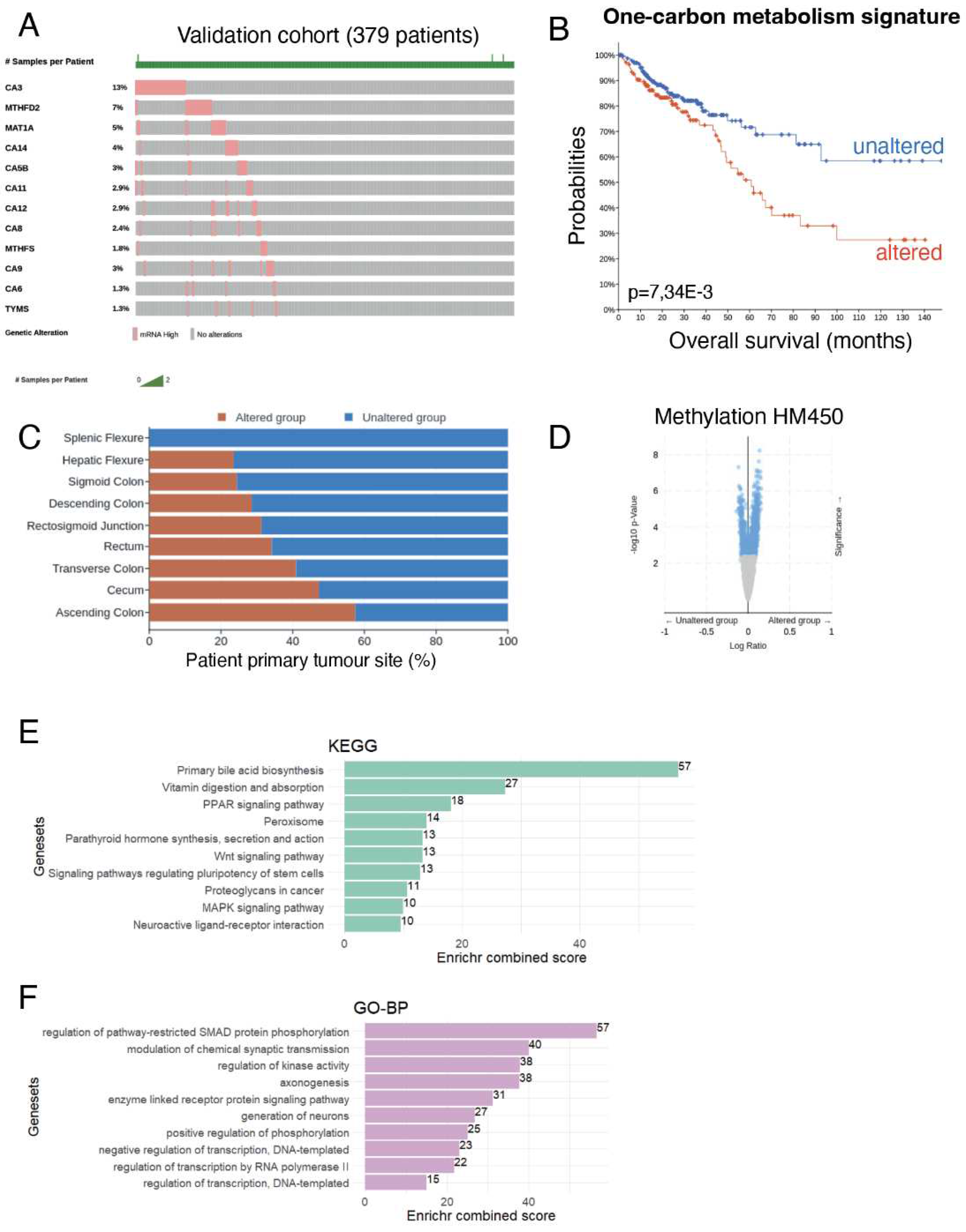
3.5. Overexpression of One Carbon Metabolism Enzymes Are Independent Markers of Methylation Status, MLH1 Silencing, Hypermutation and MSI in Colorectal Cancer
3.6. Activation of 1-C Metabolism Genes Predicts Worst Prognosis Colorectal Cancer Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xi, Y.; Xu, P. Global Colorectal Cancer Burden in 2020 and Projections to 2040. Translational Oncology 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- De Palma, F.D.E.; D’Argenio, V.; Pol, J.; Kroemer, G.; Maiuri, M.C.; Salvatore, F. The Molecular Hallmarks of the Serrated Pathway in Colorectal Cancer. Cancers 2019, 11, 1017. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Deng, Z.; Lang, X.; Jiang, J.; Xie, K.; Lu, S.; Hu, Q.; Huo, Y.; Xiong, X.; Zhu, N.; et al. Meta-Analysis of the Prognostic and Predictive Role of the CpG Island Methylator Phenotype in Colorectal Cancer. Dis Markers 2022, 2022, 4254862. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Gao, X.; Zhang, Y.; Hoffmeister, M.; Brenner, H. Different Definitions of CpG Island Methylator Phenotype and Outcomes of Colorectal Cancer: A Systematic Review. Clinical Epigenetics 2016, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Fedorova, M.S.; Krasnov, G.S.; Lukyanova, E.N.; Zaretsky, A.R.; Dmitriev, A.A.; Melnikova, N.V.; Moskalev, A.A.; Kharitonov, S.L.; Pudova, E.A.; Guvatova, Z.G.; et al. The CIMP-High Phenotype Is Associated with Energy Metabolism Alterations in Colon Adenocarcinoma. BMC Medical Genetics 2019, 20, 52. [Google Scholar] [CrossRef]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell Metabolism 2017, 25, 27–42. [Google Scholar] [CrossRef]
- Newman, A.C.; Maddocks, O.D.K. One-Carbon Metabolism in Cancer. British Journal of Cancer 2017, 116, 1499–1504. [Google Scholar] [CrossRef]
- Asai, A.; Konno, M.; Koseki, J.; Taniguchi, M.; Vecchione, A.; Ishii, H. One-Carbon Metabolism for Cancer Diagnostic and Therapeutic Approaches. Cancer Letters 2020, 470, 141–148. [Google Scholar] [CrossRef]
- Myte, R.; Gylling, B.; Häggström, J.; Schneede, J.; Löfgren-Burström, A.; Huyghe, J.R.; Hallmans, G.; Meyer, K.; Johansson, I.; Ueland, P.M.; et al. One-Carbon Metabolism Biomarkers and Genetic Variants in Relation to Colorectal Cancer Risk by KRAS and BRAF Mutation Status. PLOS ONE 2018, 13, e0196233. [Google Scholar] [CrossRef]
- Mentch, S.J.; Locasale, J.W. One-Carbon Metabolism and Epigenetics: Understanding the Specificity. Annals of the New York Academy of Sciences 2016, 1363, 91–98. [Google Scholar] [CrossRef]
- Hanley, M.P.; Rosenberg, D.W. One-Carbon Metabolism and Colorectal Cancer: Potential Mechanisms of Chemoprevention. Curr Pharmacol Rep 2015, 1, 197–205. [Google Scholar] [CrossRef]
- Heidelberger, C.; Chaudhuri, N.K.; Danneberg, P.; Mooren, D.; Griesbach, L.; Duschinsky, R.; Schnitzer, R.J.; Pleven, E.; Scheiner, J. Fluorinated Pyrimidines, A New Class of Tumour-Inhibitory Compounds. Nature 1957, 179, 663–666. [Google Scholar] [CrossRef] [PubMed]
- Chalabi-Dchar, M.; Fenouil, T.; Machon, C.; Vincent, A.; Catez, F.; Marcel, V.; Mertani, H.C.; Saurin, J.-C.; Bouvet, P.; Guitton, J.; et al. A Novel View on an Old Drug, 5-Fluorouracil: An Unexpected RNA Modifier with Intriguing Impact on Cancer Cell Fate. NAR Cancer 2021, 3, zcab032. [Google Scholar] [CrossRef] [PubMed]
- Kiweler, N.; Delbrouck, C.; Pozdeev, V.I.; Neises, L.; Soriano-Baguet, L.; Eiden, K.; Xian, F.; Benzarti, M.; Haase, L.; Koncina, E.; et al. Mitochondria Preserve an Autarkic One-Carbon Cycle to Confer Growth-Independent Cancer Cell Migration and Metastasis. Nat Commun 2022, 13, 2699. [Google Scholar] [CrossRef]
- Muzny, D.M.; Bainbridge, M.N.; Chang, K.; Dinh, H.H.; Drummond, J.A.; Fowler, G.; Kovar, C.L.; Lewis, L.R.; Morgan, M.B.; Newsham, I.F.; et al. Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) Hallmark Gene Set Collection. Cell Syst 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. Journal of Statistical Software 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York 2009. [CrossRef]
- Breiman, L. Random Forests. Machine Learning 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef] [PubMed]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.; Marshall, B.; Lewis, S. AmiGO Hub; Web Presence Working Group AmiGO: Online Access to Ontology and Annotation Data. Bioinformatics 2009, 25, 288–289. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Enge, M.; Whitington, T.; Dave, K.; Liu, J.; Sur, I.; Schmierer, B.; Jolma, A.; Kivioja, T.; Taipale, M.; et al. Transcription Factor Binding in Human Cells Occurs in Dense Clusters Formed around Cohesin Anchor Sites. Cell 2013, 154, 801–813. [Google Scholar] [CrossRef] [PubMed]
- McLean, C.Y.; Bristor, D.; Hiller, M.; Clarke, S.L.; Schaar, B.T.; Lowe, C.B.; Wenger, A.M.; Bejerano, G. GREAT Improves Functional Interpretation of Cis-Regulatory Regions. Nat. Biotechnol. 2010, 28, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Cline, M.S.; Smoot, M.; Cerami, E.; Kuchinsky, A.; Landys, N.; Workman, C.; Christmas, R.; Avila-Campilo, I.; Creech, M.; Gross, B.; et al. Integration of Biological Networks and Gene Expression Data Using Cytoscape. Nat Protoc 2007, 2, 2366–2382. [Google Scholar] [CrossRef] [PubMed]
- Turatsinze, J.-V.; Thomas-Chollier, M.; Defrance, M.; van Helden, J. Using RSAT to Scan Genome Sequences for Transcription Factor Binding Sites and Cis-Regulatory Modules. Nat Protoc 2008, 3, 1578–1588. [Google Scholar] [CrossRef]
- Hu, Y.; Yan, C.; Hsu, C.-H.; Chen, Q.-R.; Niu, K.; Komatsoulis, G.A.; Meerzaman, D. OmicCircos: A Simple-to-Use R Package for the Circular Visualization of Multidimensional Omics Data. Cancer Inform 2014, 13, 13–20. [Google Scholar] [CrossRef]
- Grifoni, D.; Bellosta, P. Drosophila Myc: A Master Regulator of Cellular Performance. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 2015, 1849, 570–581. [Google Scholar] [CrossRef]
- Cliff, T.S.; Wu, T.; Boward, B.R.; Yin, A.; Yin, H.; Glushka, J.N.; Prestegaard, J.H.; Dalton, S. MYC Controls Human Pluripotent Stem Cell Fate Decisions through Regulation of Metabolic Flux. Cell Stem Cell 2017, 21, 502–516.e9. [Google Scholar] [CrossRef]
- Motta, R.; Cabezas-Camarero, S.; Torres-Mattos, C.; Riquelme, A.; Calle, A.; Figueroa, A.; Sotelo, M.J. Immunotherapy in Microsatellite Instability Metastatic Colorectal Cancer: Current Status and Future Perspectives. J Clin Transl Res 2021, 7, 511–522. [Google Scholar]
- Kundu, S.; Ali, M.A.; Handin, N.; Conway, L.P.; Rendo, V.; Artursson, P.; He, L.; Globisch, D.; Sjöblom, T. Common and Mutation Specific Phenotypes of KRAS and BRAF Mutations in Colorectal Cancer Cells Revealed by Integrative -Omics Analysis. Journal of Experimental & Clinical Cancer Research 2021, 40, 225. [Google Scholar] [CrossRef]
- Charitou, T.; Srihari, S.; Lynn, M.A.; Jarboui, M.-A.; Fasterius, E.; Moldovan, M.; Shirasawa, S.; Tsunoda, T.; Ueffing, M.; Xie, J.; et al. Transcriptional and Metabolic Rewiring of Colorectal Cancer Cells Expressing the Oncogenic KRASG13D Mutation. Br J Cancer 2019, 121, 37–50. [Google Scholar] [CrossRef]
- Hutton, J.E.; Wang, X.; Zimmerman, L.J.; Slebos, R.J.C.; Trenary, I.A.; Young, J.D.; Li, M.; Liebler, D.C. Oncogenic KRAS and BRAF Drive Metabolic Reprogramming in Colorectal Cancer. Mol Cell Proteomics 2016, 15, 2924–2938. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Kang, B.; Petkovich, D.A.; Bhandari, Y.R.; In, J.; Stein-O’Brien, G.; Kong, X.; Xie, W.; Zachos, N.; Maegawa, S.; et al. Aging-like Spontaneous Epigenetic Silencing Facilitates Wnt Activation, Stemness, and BrafV600E-Induced Tumorigenesis. Cancer Cell 2019, 35, 315–328.e6. [Google Scholar] [CrossRef] [PubMed]
- Ushijima, T.; Suzuki, H. The Origin of CIMP, At Last. Cancer Cell 2019, 35, 165–167. [Google Scholar] [CrossRef] [PubMed]
- Augert, A.; Mathsyaraja, H.; Ibrahim, A.H.; Freie, B.; Geuenich, M.J.; Cheng, P.-F.; Alibeckoff, S.P.; Wu, N.; Hiatt, J.B.; Basom, R.; et al. MAX Functions as a Tumor Suppressor and Rewires Metabolism in Small Cell Lung Cancer. Cancer Cell 2020, 38, 97–114.e7. [Google Scholar] [CrossRef] [PubMed]
- Juo, Y.Y.; Johnston, F.M.; Zhang, D.Y.; Juo, H.H.; Wang, H.; Pappou, E.P.; Yu, T.; Easwaran, H.; Baylin, S.; van Engeland, M.; et al. Prognostic Value of CpG Island Methylator Phenotype among Colorectal Cancer Patients: A Systematic Review and Meta-Analysis. Annals of Oncology 2014, 25, 2314–2327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, W.; Cao, P. Advances in CpG Island Methylator Phenotype Colorectal Cancer Therapies. Front Oncol 2021, 11, 629390. [Google Scholar] [CrossRef] [PubMed]
- Azwar, S.; Seow, H.F.; Abdullah, M.; Faisal Jabar, M.; Mohtarrudin, N. Recent Updates on Mechanisms of Resistance to 5-Fluorouracil and Reversal Strategies in Colon Cancer Treatment. Biology 2021, 10, 854. [Google Scholar] [CrossRef]
- Bolusani, S.; Young, B.A.; Cole, N.A.; Tibbetts, A.S.; Momb, J.; Bryant, J.D.; Solmonson, A.; Appling, D.R. Mammalian MTHFD2L Encodes a Mitochondrial Methylenetetrahydrofolate Dehydrogenase Isozyme Expressed in Adult Tissues*. Journal of Biological Chemistry 2011, 286, 5166–5174. [Google Scholar] [CrossRef]
- Nilsson, R.; Jain, M.; Madhusudhan, N.; Sheppard, N.G.; Strittmatter, L.; Kampf, C.; Huang, J.; Asplund, A.; Mootha, V.K. Metabolic Enzyme Expression Highlights a Key Role for MTHFD2 and the Mitochondrial Folate Pathway in Cancer. Nat Commun 2014, 5, 3128. [Google Scholar] [CrossRef]
- Koseki, J.; Konno, M.; Asai, A.; Colvin, H.; Kawamoto, K.; Nishida, N.; Sakai, D.; Kudo, T.; Satoh, T.; Doki, Y.; et al. Enzymes of the One-Carbon Folate Metabolism as Anticancer Targets Predicted by Survival Rate Analysis. Sci Rep 2018, 8, 303. [Google Scholar] [CrossRef]
- Noguchi, K.; Konno, M.; Koseki, J.; Nishida, N.; Kawamoto, K.; Yamada, D.; Asaoka, T.; Noda, T.; Wada, H.; Gotoh, K.; et al. The Mitochondrial One-Carbon Metabolic Pathway Is Associated with Patient Survival in Pancreatic Cancer. Oncol Lett 2018, 16, 1827–1834. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Yang, H.; Yang, R.; Chen, T.; Fu, Y.; Li, Y.; Fang, X.; Zhang, K.; Zhang, J.; Li, H.; et al. The Folate Cycle Enzyme MTHFD2 Induces Cancer Immune Evasion through PD-L1 up-Regulation. Nat Commun 2021, 12, 1940. [Google Scholar] [CrossRef] [PubMed]
- Rodon, J.; Soria, J.-C.; Berger, R.; Miller, W.H.; Rubin, E.; Kugel, A.; Tsimberidou, A.; Saintigny, P.; Ackerstein, A.; Braña, I.; et al. Genomic and Transcriptomic Profiling Expands Precision Cancer Medicine: The WINTHER Trial. Nat Med 2019, 25, 751–758. [Google Scholar] [CrossRef] [PubMed]
| Variable | Level | negative (n=143) | POSITIVE (n=80) | Total (n=223) | p-value |
|---|---|---|---|---|---|
| MSI_STATUS | MSS | 116 (81.1) | 41 (51.9) | 157 (70.7) | |
| MSI-L | 24 (16.8) | 13 (16.5) | 37 (16.7) | ||
| MSI-H | 3 (2.1) | 25 (31.6) | 28 (12.6) | < 1e-04 | |
| missing | 0 | 1 | 1 | ||
| METHYLATION_SUBTYPE | Cluster3 | 74 (51.7) | 0 (0.0) | 74 (33.2) | |
| Cluster4 | 69 (48.3) | 0 (0.0) | 69 (30.9) | ||
| CIMP_H | 0 (0.0) | 32 (40.0) | 32 (14.3) | ||
| CIMP_L | 0 (0.0) | 48 (60.0) | 48 (21.5) | < 1e-04 | |
| ICLUSTER | c1 | 43 (36.8) | 11 (16.7) | 54 (29.5) | |
| c2b | 16 (13.7) | 22 (33.3) | 38 (20.8) | ||
| c3 | 48 (41.0) | 9 (13.6) | 57 (31.1) | ||
| c2a | 10 (8.5) | 24 (36.4) | 34 (18.6) | < 1e-04 | |
| missing | 26 | 14 | 40 | ||
| MLH1_SILENCING | negative | 142 (99.3) | 56 (70.0) | 198 (88.8) | |
| POSITIVE | 1 (0.7) | 24 (30.0) | 25 (11.2) | < 1e-04 | |
| EXPRESSION_SUBTYPE | CIN | 77 (54.6) | 11 (13.9) | 88 (40.0) | |
| Invasive | 36 (25.5) | 25 (31.6) | 61 (27.7) | ||
| MSI_CIMP | 28 (19.9) | 43 (54.4) | 71 (32.3) | < 1e-04 | |
| missing | 2 | 1 | 3 | ||
| HYPERMUTATED | negative | 125 (93.3) | 51 (69.9) | 176 (85.0) | |
| POSITIVE | 9 (6.7) | 22 (30.1) | 31 (15.0) | < 1e-04 | |
| missing | 9 | 7 | 16 | ||
| CANCER_TYPE | Colorectal_Adenocarcinoma | 143 (100) | 80 (100) | 223 (100) | < 1e-04 |
| CANCER_TYPE_DETAILED | Colon_Adenocarcinoma | 84 (58.7) | 43 (53.8) | 127 (57.0) | |
| Colorectal_Adenocarcinoma | 15 (10.5) | 23 (28.8) | 38 (17.0) | ||
| Rectal_Adenocarcinoma | 44 (30.8) | 14 (17.5) | 58 (26.0) | 0.001041 | |
| ONCOTREE_CODE | COAD | 84 (58.7) | 43 (53.8) | 127 (57.0) | |
| COADREAD | 15 (10.5) | 23 (28.8) | 38 (17.0) | ||
| READ | 44 (30.8) | 14 (17.5) | 58 (26.0) | 0.001041 | |
| PRIMARY_SITE | 3_-_left_colon | 59 (41.5) | 13 (16.2) | 72 (32.4) | |
| 1_-_right_colon | 24 (16.9) | 42 (52.5) | 66 (29.7) | ||
| 2_-_transverse_colon | 5 (3.5) | 9 (11.2) | 14 (6.3) | ||
| 4_-_rectum | 54 (38.0) | 16 (20.0) | 70 (31.5) | < 1e-04 | |
| missing | 1 | 0 | 1 | ||
| TUMOR_STAGE_2009 | Stage_IIA | 46 (32.6) | 33 (41.8) | 79 (35.9) | |
| Stage_IIIC | 17 (12.1) | 3 (3.8) | 20 (9.1) | ||
| Stage_IIIB | 20 (14.2) | 11 (13.9) | 31 (14.1) | ||
| Stage_I | 31 (22.0) | 15 (19.0) | 46 (20.9) | ||
| Stage_IIIA | 3 (2.1) | 1 (1.3) | 4 (1.8) | ||
| Stage_IV | 22 (15.6) | 12 (15.2) | 34 (15.5) | ||
| Stage_IIB | 2 (1.4) | 3 (3.8) | 5 (2.3) | ||
| Stage_IVA | 0 (0.0) | 1 (1.3) | 1 (0.5) | 0.29398 | |
| missing | 2 | 1 | 3 |
| CRC status | Number of patients | Accuracy | Precision | Recall | F1 score | Cohen Kappa score | AUC: area under curve |
|---|---|---|---|---|---|---|---|
| methylation CIMP | 223 | 0.82 | 0.78 | 0.72 | 0.75 | 0.62 | 0.90 |
| Hypermutation | 207 | 0.95 | 0.87 | 0.83 | 0.85 | 0.82 | 0.98 |
| MLH1 silencing | 223 | 0.96 | 0.86 | 0.80 | 0.83 | 0.81 | 0.99 |
| MSI | 222 | 0.85 | 0.83 | 0.63 | 0.72 | 0.62 | 0.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





