1. Introduction
Genome packaging is an indispensable step of virus assembly. Double-stranded DNA/RNA viruses with genomes larger than 20 kb generally utilize NTPase motor proteins to translocate their genomes into pre-formed capsids [
1]. Interestingly, three disparate groups of viruses, all requiring translocation of DNA through a lipid membrane for its encapsidation, encode putative genome packaging ATPases belonging to the FtsK/HerA superfamily of P-loop NTPases, suggesting that the fundamental principle of energy utilization for the process is highly conserved. These include the
Nucleocytoviricota phylum (also known as nucleocytoplasmic large dsDNA viruses, NCLDVs), membrane containing dsDNA bacteriophages of the PRD-1 lineage, and filamentous ssDNA phages (
Inoviridae) [
2,
3]. The ATPases typically comprise Walker A and Walker B motifs, and a third characteristic motif with a conserved arginine and a polar residue [
2]. The conserved arginine likely acts as an arginine finger that binds to the γ-phosphate of ATP in the active site of the neighboring subunit [
4]. The polar residue, mostly glutamine, senses the triphosphate of ATP and activates its hydrolysis [
5].
Although different mechanisms for the assembly of filamentous phages [
6] and membrane-containing phages [
7] have been postulated, the mechanisms of action of the genome packaging machinery in all three groups of viruses are not well understood, probably due to the difficulties in obtaining the pure recombinant motor proteins. The genome packaging of NCLDVs is unique in the aspect of the huge genomes that must be packaged. Perhaps, the most extensively studied NCLDV is vaccinia virus, the proto-type virus of the
poxviridae family [
8]. Phylogenetic analysis shows that the putative packaging ATPases of the
poxviridae family, represented by the A32L gene product, form a distinct clade within the NCLDVs [
9]. Infection using a conditional lethal mutant of vaccinia virus encoding an inducible A32L gene produced genome deficient, non-infectious virus particles upon repression of A32 protein expression and thus establishing its necessity in the viral genome packaging [
10]. Previous studies with thioredoxin-tagged A32 protein of the Goatpox virus [
11] and Orf virus [
12] have demonstrated their DNA-dependent and divalent metal ion-dependent ATPase activities. However, it is generally preferable for the protein to be produced in its native untagged form for biochemical and structural characterization, to avoid interference by the tag [
13]. As per the established nomenclature, we refer to the gene encoding the putative genome packaging ATPase of vaccinia virus as ‘A32L’ and the corresponding protein as ‘A32’ [
14].
In this study, we show that the viral ATPases correspond to the minimal functional ATPase domains of their bacterial FtsK homologs with some variations. One such variation is a putative coiled-coil motif towards the C-terminus of poxvirus A32 proteins. We report various methods that were attempted to obtain pure recombinant A32 protein of vaccinia virus, including bacterial cytoplasmic expression with a hexahistidine affinity tag, cleavable thioredoxin and GST solubility tags, periplasmic expression, and the baculovirus expression system. We observed that A32 is highly insoluble with a disordered C-terminus. With the thioredoxin-tagged A32, we further show that the coiled-coil motif regulates the DNA dependence of its ATPase activity.
2. Results and Discussion
2.1. Viral Putative Genome Packaging ATPases form a Conserved β-Sheet Core
Bacterial FtsK is a dsDNA translocase involved in chromosome segregation during cell division. It is comprised of three distinct domains, an N terminal transmembrane domain responsible for localization and recruitment of accessory proteins, a C terminal motor domain forming a RecA-like ATP hydrolysis/nucleotide-binding fold, and an intervening linker domain rich in proline and glutamine residues [
15,
16]. The viral putative genome packaging ATPases of filamentous phages (
Inoviridae family), membrane containing dsDNA phages, and NCLDVs including poxviruses show the characteristic conservation of Walker A and Walker B motifs, arginine finger and sensor motif, specific to the Ftsk/HerA superfamily [
2]. These proteins also exhibit strong structural conservation, despite their highly divergent protein sequences (
Figure 1a-b). Secondary structure analysis demonstrates that the viral proteins are much shorter and largely correspond only to the motor domain of their bacterial homolog i.e., FtsK
CΔγ, with few variations like the presence of coiled-coil motif towards the C-terminus of poxviral ATPase or in the region between the Walker A and Walker B motifs of
Asfarviridae and
Phycodnaviridae and distinct transmembrane, cytoplasmic, and extracellular domains in the filamentous phages (
Figure 1c). They invariably form a RecA-like conserved β-sheet core with alternating α-helices, similar to bacterial FtsK (
Figure S1). Thus, the virus-encoded putative genome packaging ATPases could potentially represent the minimal conserved ATPase/translocase domain of the FtsK/HerA superfamily.
2.2. A32 Exhibits Low Expression and Solubility in E. coli Cytoplasm and No Expression in Periplasm
A32L wildtype gene and its codon-optimized synthetic sequence for expression in
E. coli (A32L
CO), were cloned in pET41a and pET28a vectors, respectively, with a C- terminal hexahistidine tag (expected size: ~32 kDa). A32 protein expressed from pET41a plasmid was highly insoluble. A small amount of the soluble protein, eluted at low concentrations of imidazole (< 100 mM), had many impurities (
Figure S2a) and hence, could not be further purified and concentrated. Surprisingly, A32
CO showed negligible expression in
E. coli, with no observable purified protein either in the lysate upon induction or eluted upon binding with Ni
2+-NTA resin (
Figure S2b) (
Table 1).
Periplasmic recombinant protein expression in
E. coli offers several advantages due to the presence of chaperones, and lesser contaminants and proteases compared to its cytoplasm. The periplasm has an oxidative environment facilitating disulfide bond formation [
13,
17]. Since A32 consists of 8 cysteine residues which might be involved in disulfide bond formation, it was cloned in pET22b vector containing pelB signal peptide sequence for periplasmic expression (expected size: ~34.5 kDa with pelB, ~32.5 kDa after pelB cleavage by periplasmic signal peptidases). A protein at the size of ~35 kDa was seen in the periplasm along with some other bacterial proteins (
Figure S2c). However, when compared to the periplasm fraction of the uninduced cells transformed with pET22b-A32L and uninduced and induced cells transformed with empty pET22b vector, it was confirmed that the 35 kDa band was a host protein (
Figure S2d), and not the expected A32 protein, confirming no expression in the
E.coli periplasm (
Table 1).
2.3. A32 Does Not Express Using Baculovirus Expression System
Since pure wildtype A32 protein could not be produced in the
E. coli cytoplasm or periplasm, the Bac-to-Bac baculovirus expression system was attempted to express A32 in Sf9 insect cells for its capability to produce soluble heterologous proteins at high concentrations, including membrane proteins [
18,
19]. As expected, the uninfected cells were relatively smaller in size and continued to divide, eventually forming multiple layers (
Figure S3a, i and iii). In contrast, baculovirus-infected cells were larger, had ceased cell division at 6 dpi (days post-infection) and were eventually lysed at 8 dpi (
Figure S3a, ii and iv). However, SDS-PAGE and western blot analysis using anti-A32 immune sera of Sf9 cells infected with recombinant baculovirus containing A32L gene (Bac
A32) compared with wildtype baculoviruses (Bac
WT) showed that A32 did not express by the baculovirus expression system (
Figure S3b).
Our results indicate that the soluble A32 protein of the vaccinia virus could not be expressed in its native form in bacterial or baculovirus expression systems (results outlined in
Table 1). Hence, the protein was fused with cleavable GST or thioredoxin tags, well-known for their potential to enhance protein solubility in
E. coli expression systems [
20,
21].
2.4. GST-A32 Exhibits Possibly Incorrect Folding and Auto-Cleavage of GST tag
The A32L gene was cloned into the pGEX-6P-1 vector with an N-terminal GST tag (expected size: ~58 kDa). Expression and purification of GST-A32 using glutathione sepharose beads showed that the protein in the soluble fraction did not bind to the resin. Additionally, bands corresponding to the sizes of A32 protein (~ 31 kDa) and GST (~26 kDa) were observed that were not seen in the uninduced cell lysate, indicating a possibility of auto-cleavage of the tag (
Figure S4a). These findings were confirmed by western blotting with anti-GST antibody (
Figure S4b) (
Table 1).
2.5. Thioredoxin Tag Promotes Solubility and Stability of A32
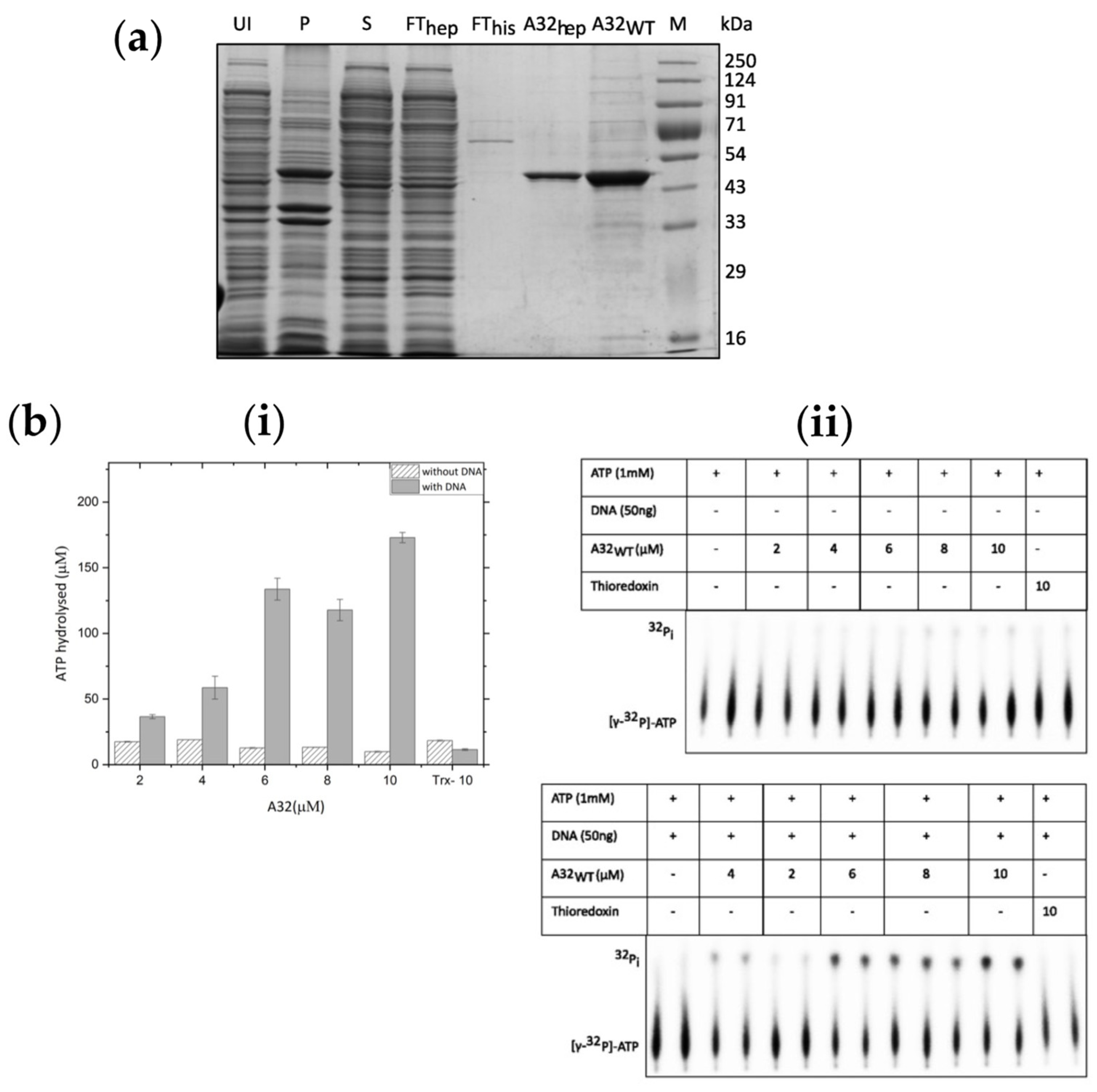
The A32L gene cloned in the pET32b vector resulted in the N-terminal fusion with the thioredoxin tag and a C-terminal hexahistidine tag (A32
WT). A32
WT protein with thioredoxin tag could be obtained in the soluble fraction and was successfully purified by Ni
2+-NTA and heparin affinity chromatographies, albeit with some impurities (concentration of the purified protein was ~1 mg/ml,
Figure 2a). Furthermore, the ATPase activity of the A32
WT protein increased by up to 17-fold in the presence of DNA at the highest protein concentration (
Figure 2b).
We attempted to remove the thioredoxin tag by treating with enterokinase. We observed non-specific cleavage and protein precipitation at both 4°C and 20° C (
Figure S5A). Next, the enterokinase cleavage site was replaced with the NT*-HRV3C cleavage site in the pET32b vector. Although the expression levels of A32 protein from the modified plasmid (A32*
WT) was higher than the native A32
WT protein, the solubility was very low (
Figure S5b). The concentrated, partially purified protein (~ 0.9 mg/ml) was treated with NT*-HRV3C protease at wt/wt ratios 1:1 and 1:2 A32*
WT: NT*HRV3CP. Aliquots were collected at 0, 2 and 10 h post-protease addition. Compared to no protease control, reduction in the concentration of A32*
WT was noted, whereas some protein remained uncut. Because of their similar sizes, NT*-HRV3C protease (35 kDa) and A32
WT (32 kDa) could not be distinguished on the SDS-PAGE gel (
Figure S5c). Western blot with anti-A32 immune sera showed a band around 16 kDa, indicating non-specific cleavage or truncation of the protein upon removal of the thioredoxin tag (
Figure S5d).
2.6. A32 Possesses a Structurally Disordered, but Indispensable C-Terminus
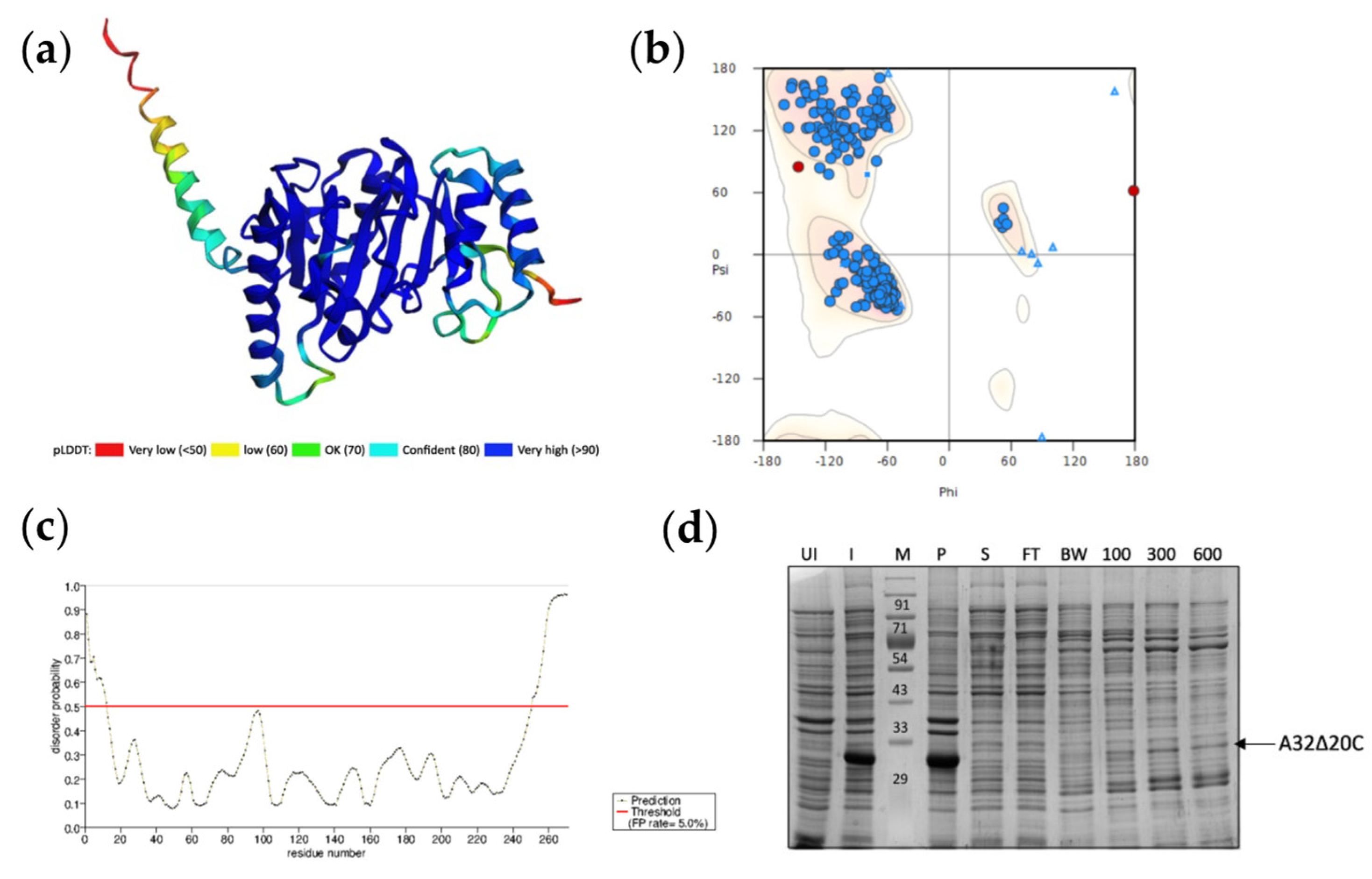
Putative three-dimensional structure of A32 was predicted using the AI-powered ColabFold modification of AlphaFold2 algorithm in no template mode [
22]. A high confidence structure was obtained with an average pLDDT score 89.8 and pTM score 0.865, where the local pLDDT scores of the core ATPase domain was very high (>90). However, the local pLDDT score decreased for the C-terminal helix of A32 (
Figure 3a). Ramachandran plot analysis of the predicted structure shows that the 99.25% of the amino acids are in the favored and allowed regions, with only two outliers i.e., Asn267 and Val268 (
Figure 3b). When analyzed for the possible disordered regions using PrDOS server, the C-terminal 20 aa show high probability for disorder (
Figure 3c). To evaluate if deletion of the disordered 20 aa could result in better folding and stability of the protein, a deletion mutant A32LΔ20C was constructed in pET41a vector and expressed in
E. coli. While the mutant protein was expressed at high concentrations, it was still highly insoluble (
Figure 3d).
Thus, we demonstrate that pure A32 protein could only be obtained when fused with the thioredoxin tag. All our attempts to express the recombinant A32L gene are summarized in
Table 1.
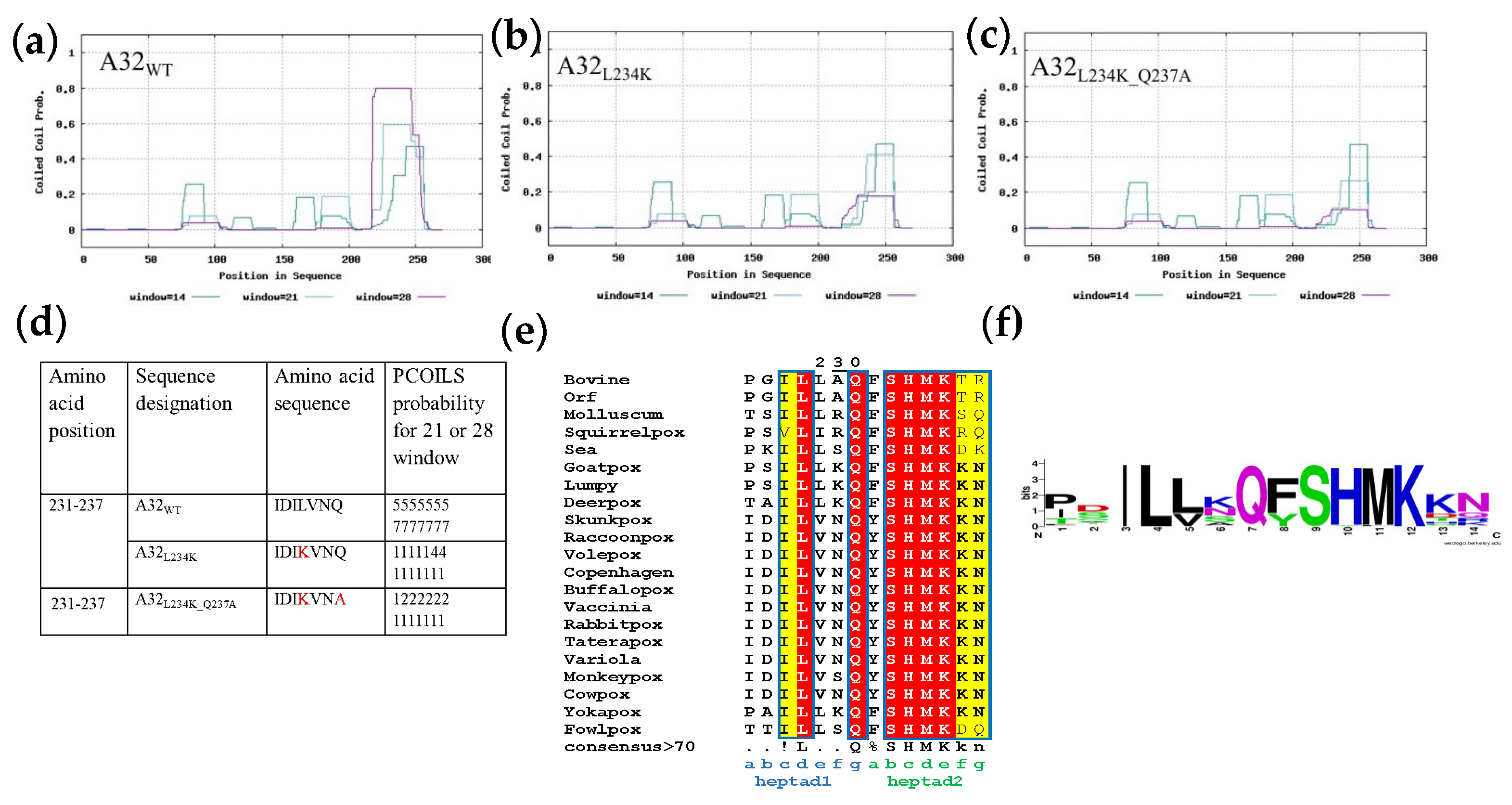
2.7. A32 Forms a Conserved Coiled Coil Motif Towards Its C-Terminus
A coiled-coil motif (CCM) was predicted in the region between aa 231 to 244, next to the disordered C-terminal helix of the A32 protein. To discern if the reduction in stability of A32 upon deletion of C-terminal helix was related to the nearby coiled coil motif, the role of latter in the protein’s activity was evaluated. With the hypothesis that substitution of conserved hydrophobic aa residue(s) with hydrophilic aa might destabilize the coiled-coil motif without interfering with the overall structure, A32 mutants- A32
L234K and A32
L234K_Q237A were designed. Both mutants show much reduced probability to form the CCM as predicted by the PCOILS server (
Figure 4b-4d). The CCM motif selected for mutagenesis is also strictly conserved in other poxvirus family members (
Figure 4e and 4f).
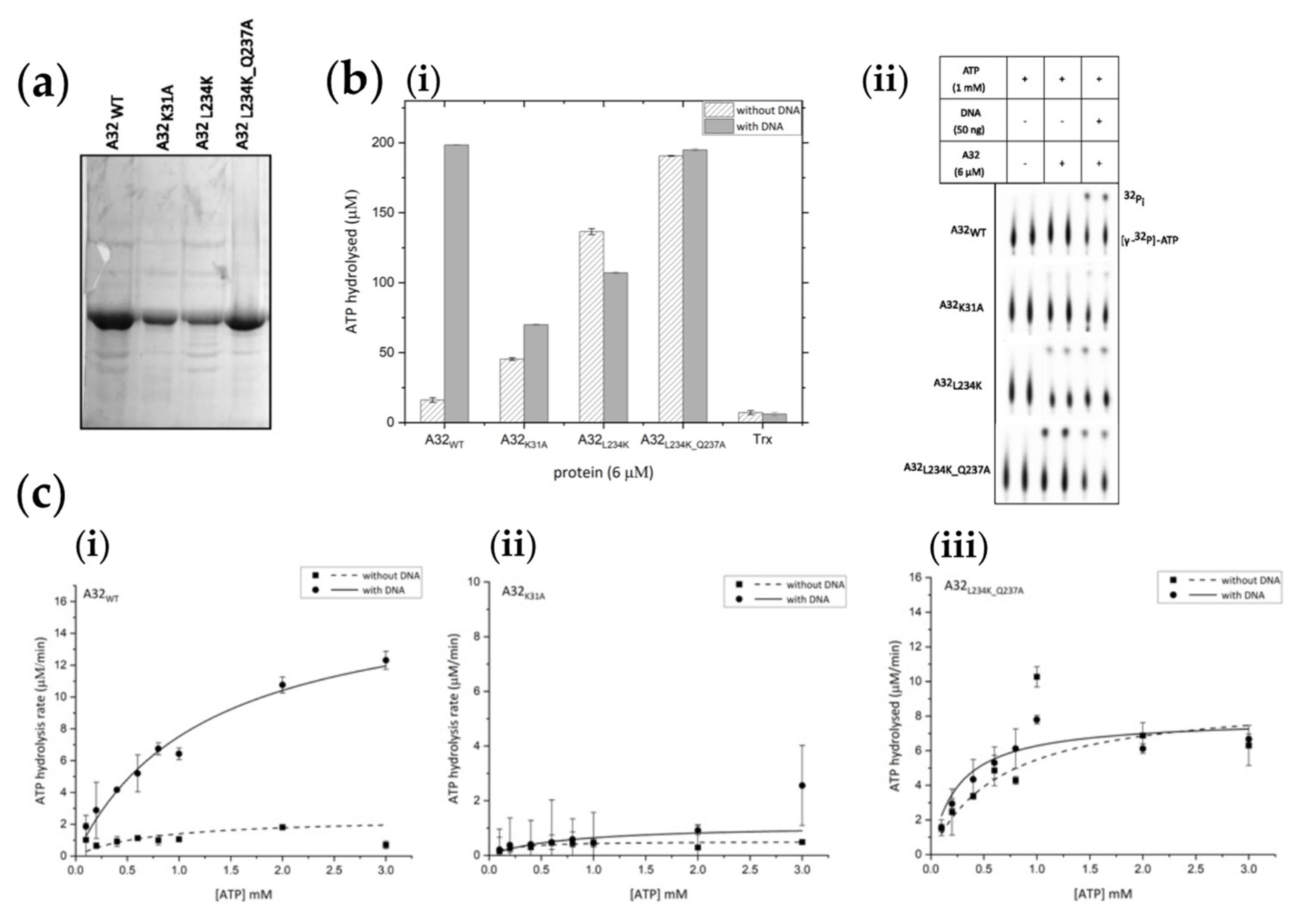
2.8. Coiled-Coil Motif Regulates the DNA-Dependent ATPase Activity of A32
In addition to the CCM mutants A32
L234K and A32
L234K_Q237A, a Walker A lysine mutant (A32
K31A) that should abolish the ATPase activity [
23,
24], was also generated. All three mutants were purified by Ni
2+-NTA and heparin affinity chromatographies and concentrated to about 0.7- 1 mg/ml. Purified proteins contained some impurities along with the protein of interest (
Figure 5a).
A comparative analysis of ATPase activity showed that while A32
K31A possesses negligible ATPase activity, the CCM mutants A32
L234K and A32
L234K_Q237A showed enhanced basal ATPase activities. However, the DNA dependence of the ATPase activity was abolished (
Figure 5b). Thus, the disruption of the CCM of A32 protein resulted in the loss of DNA-dependent regulation of ATPase activity observed in the wildtype protein. These observations were further confirmed by carrying out steady-state kinetics analysis with trace amounts (~ 50 nM) of hot ATP [γ-
32P] and varying concentrations of cold ATP (
Figure 5c and S6). The V
max and K
cat calculated for A32
L234K_Q237A were similar in the absence or presence of DNA, and approximately half the value compared to A32
WT in the presence of DNA. Interestingly, a higher K
m value, indicative of a lower affinity for ATP, was observed for A32
WT with DNA (
Table 2). Thus, the coupling of ATPase regulation to DNA- binding and oligomerization of the A32 protein appears to have mechanistic relevance and calls for further investigations.
3. Materials and Methods
3.1. Sequence and Structure Analysis
Vaccinia virus putative genome packaging ATPase, A32L (Gene ID: 3707685) sequence was retrieved from NCBI. Multiple sequence alignment was performed using Clustal omega [
25]. Three-dimensional structures were predicted with the help of ColabFold v1.5.2-patch modification of AlphaFold2, using 6 recycles, no template and MSA generated using both Uniref and environmental databases [
22,
26]. Transmembrane domains were predicted using DeepTMHMM [
27] and Phobius [
28]. A Combined Transmembrane Topology and Signal Peptide Prediction Method. Coiled-coil motif was predicted using PCOILS [
29], NPS [
30], and paircoil [
31] servers. Experimental structure of Pseudomonas aeruginosa FtsKCΔγ was obtained from PDB (accession ID: 2IUT) [
15]. Predicted ATPase structures of vaccinia virus A32, PRD1 P9 and M13 gpI were aligned using US-Align [
32]. Disordered region in A32 was confirmed from PrDOS server [
33]. Ramachandran plot of A32 structure was generated using WinCoot [
34].
3.2. Recombinant Plasmids Construction
3.2.1. Modification of pET32b Plasmid
pET32b vector (Merck) was modified such that the enterokinase cleavage site was replaced with the human rhinovirus 14 3C protease (NT*-HRV3C protease) cleavage site (5’-CTGGAAGTTCTGTTCCAGGGGCCC-3’ coding for the peptide LEVLFQGP) [
35] through the reverse primer. Recombinant plasmid was constructed by PCR amplification of pET32b vector template using the forward primer, 5’-CCCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGAGC-3’ and the reverse primer, 5’-CCATGGCGGGCCCCTGGAACAGAACTTCCAGGGTACCCAGATCT-3’. The amplified PCR product was inserted into pET32b vector by the standard restriction-ligation cloning procedure. Replacement of protease cleavage site in the modified plasmid pET32b-HRV3C was confirmed by sequencing (Eurofins Genomics India Pvt. Ltd.).
3.2.2. Construction of A32L Expression Plasmids
A32L gene and its deletion mutant A32LΔ20C were PCR-amplified from the genomic DNA of vaccinia virus Western Reserve strain (ATCC® VR-1354TM) and cloned into the bacterial expression vectors (Merck) pET41a, pET22b, pGEX-6P-1, pET32b, pET32b-HRV3C, and the Bac-to-Bac baculovirus expression system vector pFastBac1 (Invitrogen). A synthetic construct of A32L codon optimized for
E. coli, A32LCO (Biomatik) was cloned into pET28a vector. All recombinant plasmids were constructed by standard restriction-ligation cloning methods using primers as described in
Table S1. Walker A motif mutant (A32L
K31A) and coiled-coil motif mutants (A32L
L234K, A32L
L234K_Q237A) were constructed by overlap PCR-based site directed mutagenesis using primers as described in
Table S2 and cloned into pET32b vector. Sequences of all recombinant constructs were verified by sequencing (Eurofins Genomics India Pvt. Ltd.).
3.3. Expression and Purification of A32 Protein in E. coli
All recombinant constructs were transformed into competent E. coli BL21-CodonPlus (DE3)-RIPL cells (Agilent Technologies). Overnight grown primary inoculum was added to LB medium at 1% final concentration, grown at 37º C till O.D.600 was 0.5-0.6, and induced with 0.5 mM IPTG at 20°C for 16 h. Induced cells were harvested, and cell pellet (except pET22b-A32L) resuspended in lysis buffer containing 50 mM Tris-HCl pH7.5, 400 mM NaCl, 10% glycerol, 1% triton X-100, 10 mM imidazole, 1 mM Benzamidine hydrochloride, and 1 mM PMSF. The suspension was treated with 1 mg/ml lysozyme at room temperature (25º C) for 20 minutes and cells lysed by sonication. Cell-free extract was obtained by centrifugation at 13,000 g and processed as follows for obtaining the pure protein.
3.3.1. Purification of A32 and A23Δ20C or A32CO from pET41a or pET28a Vectors, Respectively
The cell-free extract was incubated with Ni2+-NTA agarose beads (Genetix, India) pre-equilibrated with binding buffer (50 mM Tris-Cl pH 7.5, 400 mM NaCl, 10% glycerol, 20 mM imidazole, and 5 mM MgCl2) for 3 h. After washing the beads twice with the binding buffer, the protein of interest was eluted by resuspending them sequentially with elution buffer containing 100, 300, and 600 mM imidazole.
3.3.2. Purification of Periplasmic A32 from pET22b Vector
E. coli periplasm was extracted by cold- shock method described previously [
36]. Briefly, cells were washed twice with 50 mM Tris-Cl pH 7.5, and resuspended in extraction buffer (0.2 M MgCl2, 20 mM Tris-Cl pH 7.5). Cell suspension was incubated in 35º C stirring water bath for 10 minutes followed by incubation on ice for 15 minutes. This was repeated twice. Periplasm was obtained by centrifugation at 20,000 g for 20 minutes at 4º C. Cell pellet was resuspended in 50mM Tris-Cl pH 7.5 and lysed by sonication. Soluble cell-free extract and insoluble cellular debris and proteins were separated by centrifugation at 13,000 g. Periplasmic, cytoplasmic, and insoluble fractions were extracted from cells (-/+IPTG) transformed with pET22b or recombinant pET22b-A32 plasmids and analyzed for A32 expression.
3.3.3. Purification of A32 from pGEX-6P-1 Vector
The cell-free extract was incubated with glutathione sepharose 4B beads (Cytiva) pre-equilibrated with binding buffer (50 mM Tris-Cl pH 7.5, 400 mM NaCl, 10 % glycerol, 1 mM DTT) for 3 h. Flowthrough was collected and the beads washed twice with binding buffer. GST-tagged proteins were eluted with elution buffers containing 10 mM and 20 mM reduced glutathione.
3.3.4. Purification of A32 and Its Mutants From pET32b or pET32b-HRV3C Vectors
After separation of the cell-free extract, its NaCl concentration was adjusted to 200 mM and loaded onto HiTrap Heparin HP column (Cytiva) pre-equilibrated with binding buffer (50 mM Tris pH 7.4, 10% glycerol, and 200 mM NaCl). The protein was eluted by gradient elution (200 mM – 1 M NaCl). Peak fractions were pooled and loaded onto the Histrap HP column (Cytiva) pre-equilibrated with binding buffer containing 20 mM imidazole. The protein was eluted by gradient elution (20 – 600 mM). The pooled protein was concentrated, purity checked, and stored at -80°C. Walker A motif mutant A32K31A was purified same as wild type A32 (A32WT). The coiled coil motif mutants A32L234K and A32L234K_Q237A had reduced affinity for heparin therefore, the mutants were first passed through the Histrap column and the eluates were pooled and passed through the heparin column.
3.4. Cleavage of Thioredoxin Tag by Enterokinase or NT*-HRV3C Protease
35 μg purified thioredoxin tagged A32WT protein (expressed from pET32b vector) was treated with 1 unit of Enterokinase (Novagen, Merck). Protease activity was monitored at 14, 17, and 36 h at 4º C or 20º C.
Recombinant construct expressing the NT*- HRV3C protease was obtained from Addgene and the protease purified by Ni
2+-NTA affinity chromatography and size exclusion chromatography as previously described [
35]. Partially purified A32*
WT protein (expressed from pET32b-HRV3C vector) was incubated with NT*-HRV3C protease at 4º C. Protease activity was monitored at 0, 2 and 10 h with either no protease, or in the presence of protease at 1:1, and 1:2 wt/wt ratios (A32*
WT : NT*- HRV3C).
3.5. Expression of A32 Protein in Sf9 Insect Cells Using the Bac-to-Bac System
A32L gene was amplified and cloned into pFastBac1 vector (Invitrogen) followed by transformation into DH10Bac E. coli cells (Invitrogen). Transformed cells were identified by blue-white screening. Bacmids were purified using PureLink™ HiPure Plasmid Midiprep Kit (Invitrogen) according to the manufacturer’s protocol. ~5×105 Sf9 cells (kind gift from Dr. Virupakshi Soppina, IIT Gandhinagar) maintained in Sf-900™ II SFM serum free media (Gibco) at 27º C were transfected with 1 μg A32L-bacmid or empty-bacmid using 8 μl Cellfectin® II Reagent (Invitrogen) according to the manufacturer’s protocol. Cell size enlargement and lysis were seen 6 days post transfection. P0 stock of the obtained baculovirus was used to infect ~1- 2 ×106 sf9 cells in a T25 flask and the P1 viruses subsequently transferred to 3-4×106 cells in a T75 flask to obtain P2 virus. 1ml P2 virus was used to infect a 30 ml suspension culture at 3×106 cells/ml and incubated at 27° C with shaking at 90 rpm. Cells were harvested before lysis by centrifugation at 8000 g. Total protein was extracted by adding lysis buffer containing 20 mM HEPES pH 7.5, 200 mM NaCl, 5 mM MgCl2, 0.5% NP-40, 7% sucrose, 1x protease inhibitor cocktail (Merck). Cell lysate was analyzed in comparison to empty bacmid-infected sf9 cells for A32 expression.
3.6. ATPase Assay
Purified A32WT or its mutants (2-10 µM) were incubated at 37° C for 30 min in a reaction mixture containing 1 mM cold ATP and trace amounts (~ 50 nM) of ATP [γ-32P] (obtained from BARC, Mumbai), 50 mM Tris–HCl, pH 7.5, 0.1 M NaCl and 5 mM MgCl2 in the presence or absence of 50 ng DNA. Reaction was stopped with 50 mM EDTA followed by thin layer chromatography (PEI-cellulose matrix, Sigma-Aldrich) and autoradiography. Quantification of the data was done by phosphorimaging using Typhoon FLA 9500 (GE Healthcare Life Sciences/Cytiva). Steady-state kinetics was performed by varying the concentration of cold ATP from 0.1 to 3 mM, while keeping the concentration of [γ-32P]ATP at ~ 50 nM and protein concentration at 6 µM. All reactions were performed in duplicates and the data points represent mean with standard deviations. Reproducibility was confirmed by repeating the experiment at least once under identical conditions. From the 32Pi value, the total [Pi] produced was calculated, and the Km and Vmax values were determined by data fitting into the Michaelis–Menten equation using OriginPro 2023b software (Origin labs).
4. Conclusions
Putative genome packaging ATPases of NCLDVs, ssDNA filamentous phages and membrane-containing dsDNA phages, despite their low sequence similarity, share a highly conserved structural fold characteristic of the FtsK/HerA superfamily with few additional motifs in some viruses (
Figure 1) [
2]. In this report, we attempted to purify the putative genome packaging ATPase of vaccinia virus, A32 protein, by various approaches and were able obtain the purified protein with an N-terminal thioredoxin tag. The outcomes of our attempts to obtain the untagged wildtype protein are summarized in
Table 1. We observed that in the absence of thioredoxin tag, the recombinant vaccinia virus A32 protein produced in
E. coli is highly insoluble or prone to precipitation and truncations upon tag cleavage. Our sequence analysis of poxvirus A32 showed the presence of a conserved coiled-coil motif, known to facilitate protein oligomerization [
37]. The CCM has also been reported in the hexameric bacterial FtsK protein [
2]. However, its role in the protein’s activity has not been implicated. The CCM was predicted near the C-terminal disordered region of A32, the deletion of which resulted in further destabilization of the protein. To discern if the CCM has any influence on the ATPase activity of putative genomic packaging ATPase, we purified and compared the activity of the thioredoxin-tagged CCM mutants with thioredoxin-A32 wildtype protein. Our results show that the ATPase activity of thioredoxin-A32 protein is stimulated in the presence of DNA. This finding is consistent with the outcomes reported for A32 homologs of Goatpox virus [
11] and Orf virus [
12]. This DNA-dependence of its activity might be regulated by the CCM, as evidenced by the observation that mutations disrupting the motif led to ATP hydrolysis both in the presence and absence of DNA. Further biochemical and structural studies are needed to understand the mechanistic basis of this regulation, explore the protein’s stoichiometry and how the ATP and DNA bind to A32 and impact its folding; thereby, resulting in the ATP hydrolysis and DNA translocation.
Supplementary Materials
The following supporting information can be downloaded at:
www.mdpi.com/xxx/s1, Table S1: List of primers used for A32L-recombinant plasmid construction; Table S2: List of primers used for overlap PCR for construction of A32L mutants; Figure S1: Alphafold predicted three dimensional structures of viral FtsK-like ATPases; Figure S2: Expression of untagged A32 protein in
E. coli; Figure S3: Expression of A32 in recombinant baculovirus-infected sf9 cells; Figure S4: Expression of GST-tagged A32 in
E. coli cytoplasm; Figure S5: Removal of thioredoxin tag of A32; Figure S6: Autoradiographs of steady-state kinetics in the absence or presence of DNA
Author Contributions
Conceptualization, K.K.,T.A., and U.A.; methodology, U.R., T.A, and K.K; software, U.R., T.A.; validation, U.R., and T.A.; formal analysis, U.R., T.A, and K.K.; investigation, U.R. T.A.; resources, K.K.; data curation, U.R., T.A, and K.K.; writing—original draft preparation, T.R., and K.K.; writing—review and editing, U.R., T.A., and K.K.; supervision, K.K.; project administration, K.K.; funding acquisition, K.K. All authors have read and agreed to the published version of the manuscript.
Funding
“This research in KK lab is funded by The Board of Research in Nuclear Sciences, BRNS [58/14/11/2020-BRNS/37188] and Department of Biotechnology, DBT [BT/PR35928/BRB/10/1841/2019], the Department of Science and Technology, SERB [EMR/2016/005155]. “The APC was funded by Indian Institute of Technology Bombay”.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Perlmutter, J.D.; Hagan, M.F. Mechanisms of Virus Assembly. Annu Rev Phys Chem 2015, 66, 217–239. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Makarova, K.S.; Koonin, E. V.; Aravind, L. Comparative Genomics of the FtsK–HerA Superfamily of Pumping ATPases: Implications for the Origins of Chromosome Segregation, Cell Division and Viral Capsid Packaging. Nucleic Acids Res 2004, 32, 5260–5279. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.; Iyer, L.; Aravind, L. Comparative Genomics and Evolutionary Trajectories of Viral ATP Dependent DNA-Packaging Systems. Genome Dyn 2007, 3, 48–65. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; De-Donatis, G.M.; Schwartz, C.; Fang, H.; Li, J.; Guo, P. An Arginine Finger Regulates the Sequential Action of Asymmetrical Hexameric ATPase in the Double-Stranded DNA Translocation Motor. Mol Cell Biol 2016, 36, 2514–2523. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Chanchal; Jain, D. Sensor i Regulated ATPase Activity of FleQ Is Essential for Motility to Biofilm Transition in Pseudomonas Aeruginosa. ACS Chem Biol 2019, 14, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Loh, B.; Kuhn, A.; Leptihn, S. The Fascinating Biology behind Phage Display: Filamentous Phage Assembly. Mol Microbiol 2018, 0–2. [Google Scholar] [CrossRef]
- Butcher, S.J.; Manole, V.; Karhu, N.J. Lipid-Containing Viruses: Bacteriophage PRD1 Assembly. Adv Exp Med Biol 2012, 726, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Boyle, K.; Traktman, P. Poxviruses. In Viral Genome Replication; Springer, Boston, MA, 2009; pp. 225–247 ISBN 978-0-387-89456-0.
- Rodrigues, R. AL; de Souza, F.G.; de Azevedo, B.L.; da Silva, L.C.; Abrahão, J.S. The Morphogenesis of Different Giant Viruses as Additional Evidence for a Common Origin of Nucleocytoviricota. Curr Opin Virol 2021, 49, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Cassetti, M.C.; Merchlinsky, M.; Wolffe, E.J.; Weisberg, A.S.; Moss, B. DNA Packaging Mutant: Repression of the Vaccinia Virus A32 Gene Results in Noninfectious, DNA-Deficient, Spherical, Enveloped Particles. J Virol 1998, 72, 5769. [Google Scholar] [CrossRef]
- Lee, M.L.; Hsu, W.L.; Wang, C.Y.; Chen, H.Y.; Lin, F.Y.; Chang, M.H.; Chang, H.Y.; Wong, M.L.; Chan, K.W. Goatpoxvirus ATPase Activity Is Increased by DsDNA and Decreased by Zinc Ion. Virus Genes 2016, 52, 625–632. [Google Scholar] [CrossRef]
- Lin, F.Y.; Chan, K.W.; Wang, C.Y.; Wong, M.L.; Hsu, W.L. Purification and Functional Motifs of the Recombinant ATPase of Orf Virus. Protein Expr Purif 2011, 79, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant Protein Expression in Escherichia Coli: Advances and Challenges. Front Microbiol 2014, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Condit, R.C.; Moussatche, N.; Traktman, P. In A Nutshell: Structure and Assembly of the Vaccinia Virion. Adv Virus Res 2006, 66, 31–124. [Google Scholar] [CrossRef] [PubMed]
- Massey, T.H.; Mercogliano, C.P.; Yates, J.; Sherratt, D.J.; Löwe, J. Double-Stranded DNA Translocation: Structure and Mechanism of Hexameric FtsK. Mol Cell 2006, 23, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Jean, N.L.; Rutherford, T.J.; Löwe, J. FtsK in Motion Reveals Its Mechanism for Double-Stranded DNA Translocation. Proc Natl Acad Sci U S A 2020, 117, 14202–14208. [Google Scholar] [CrossRef] [PubMed]
- Taherian, E.; Mohammadi, E.; Jahanian-Najafabadi, A.; Moazen, F.; Akbari, V. Cloning, Optimization of Periplasmic Expression and Purification of Recombinant Granulocyte Macrophage-Stimulating Factor in Escherichia Coli BL21 (DE3). Adv Biomed Res 2019, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Kalathur, R.C.; Panganiban, M.; Bruni, R. Heterologous Expression of Membrane Proteins. 2010, 601, 187–202. [CrossRef]
- Anderson, D.; Harris, R.; Polayes, D.; Ciccarone, V.; Donahue, R.; Gerard, G.; Joel, J. Rapid Generation of Recombinant Baculovirus and Expression of Foreign Genes Using the Bac-to-Bac Baculovirus Expression System. Focus (Madison) 1995, 17, 53–58. [Google Scholar]
- LaVallie, E.R.; DiBlasio, E.A.; Kovacic, S.; Grant, K.L.; Schendel, P.F.; McCoy, J.M. A Thioredoxin Gene Fusion Expression System That Circumvents Inclusion Body Formation in the E. Coli Cytoplasm. Bio/Technology 1993 11:2 1993, 11, 187–193. [Google Scholar] [CrossRef]
- Smith, D.B.; Johnson, K.S. Single-Step Purification of Polypeptides Expressed in Escherichia Coli as Fusions with Glutathione S-Transferase. Gene 1988, 67, 31–40. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Ramakrishnan, C.; Dani, V.S.; Ramasarma, T. A Conformational Analysis of Walker Motif A [GXXXXGKT (S)] in Nucleotide-Binding and Other Proteins; 2002; Vol. 15;
- Dellas, N.; Snyder, J.C.; Dills, M.; Nicolay, S.J.; Kerchner, K.M.; Brumfield, S.K.; Lawrence, C.M.; Young, M.J. Structure-Based Mutagenesis of Sulfolobus Turreted Icosahedral Virus B204 Reveals Essential Residues in the Virion-Associated DNA-Packaging ATPase. J Virol 2016, 90, 2729–2739. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol Syst Biol 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Hallgren, J.; Tsirigos, K.D.; Damgaard Pedersen, M.; Juan, J.; Armenteros, A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM Predicts Alpha and Beta Transmembrane Proteins Using Deep Neural Networks. BioRxiv 2022, 487609. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. A Combined Transmembrane Topology and Signal Peptide Prediction Method. J Mol Biol 2004, 338, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Gabler, F.; Nam, S.Z.; Till, S.; Mirdita, M.; Steinegger, M.; Söding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr Protoc Bioinformatics 2020, 72, e108. [Google Scholar] [CrossRef] [PubMed]
- Lupas, A.; Van Dyke, M.; Stock, J. Predicting Coiled Coils from Protein Sequences. Science (1979) 1991, 252, 1162–1164. [Google Scholar] [CrossRef]
- Berger, B.; Wilson, D.B.; Wolf, E.; Tonchev, T.; Milla, M.; Kim, P.S. Predicting Coiled Coils by Use of Pairwise Residue Correlations. Proceedings of the National Academy of Sciences 1995, 92, 8259–8263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shine, M.; Pyle, A.M.; Zhang, Y. US-Align: Universal Structure Alignments of Proteins, Nucleic Acids, and Macromolecular Complexes. Nature Methods 2022 19:9 2022, 19, 1109–1115. [Google Scholar] [CrossRef]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of Disordered Protein Regions from Amino Acid Sequence. Nucleic Acids Res 2007, 35. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta Crystallogr D Biol Crystallogr 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Abdelkader, E.H.; Otting, G. NT*-HRV3CP: An Optimized Construct of Human Rhinovirus 14 3C Protease for High-Yield Expression and Fast Affinity-Tag Cleavage. J Biotechnol 2021, 325, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Shrivastava, R.; Basu, B.; Apte, S.K.; Phale, P.S. Modulation of Glucose Transport Causes Preferential Utilization of Aromatic Compounds in Pseudomonas Putida CSV86. J Bacteriol 2007, 189, 7556–7562. [Google Scholar] [CrossRef] [PubMed]
- Burkhard, P.; Stetefeld, J.; Strelkov, S. V. Coiled Coils: A Highly Versatile Protein Folding Motif. Trends Cell Biol 2001, 11, 82–88. [Google Scholar] [CrossRef]
Figure 1.
Sequence and structure analysis of viral FtsK-like genome packaging ATPases. (a) Multiple sequence alignment of putative genome packaging ATPases of NCLDVs, PRD1 lineage (membrane-containing dsDNA viruses) and Inoviridae family (ssDNA filamentous phages). Green- strictly conserved identical residues; yellow- similar residues (b) Structural alignment of representative ATPases of each virus group using AlphaFold predicted structures. E- β strand (blue), L- loop (green), H- α helix (red). NCLDVs representative- Vaccinia virus A32 (yellow), membrane-containing dsDNA viruses’ representative- PRD1 P9 (pink), ssDNA filamentous phages representative- M13 gp1 (orange) (c) secondary structure elements and domain organization of representative viral ATPases. WA- Walker A motif, WB- Walker B motif, R- arginine finger, Q- sensor residue (red), TM- transmembrane domain (pink), CCM- coiled coil motif (brown). Blue- β strands, yellow- α helices. Conserved ATPase core is highlighted in green. Scale bar: 10 aa. Accession no- Vaccinia virus: YP_233037.1, Organic Lake phycodnavirus 1: ADX05856.1, African swine fever virus: YP_009702812.1, M13 phage- NP_510893.1, PRD1 phage- AAX45927.1, FtsKCΔγ PDB ID- 2IUT.
Figure 1.
Sequence and structure analysis of viral FtsK-like genome packaging ATPases. (a) Multiple sequence alignment of putative genome packaging ATPases of NCLDVs, PRD1 lineage (membrane-containing dsDNA viruses) and Inoviridae family (ssDNA filamentous phages). Green- strictly conserved identical residues; yellow- similar residues (b) Structural alignment of representative ATPases of each virus group using AlphaFold predicted structures. E- β strand (blue), L- loop (green), H- α helix (red). NCLDVs representative- Vaccinia virus A32 (yellow), membrane-containing dsDNA viruses’ representative- PRD1 P9 (pink), ssDNA filamentous phages representative- M13 gp1 (orange) (c) secondary structure elements and domain organization of representative viral ATPases. WA- Walker A motif, WB- Walker B motif, R- arginine finger, Q- sensor residue (red), TM- transmembrane domain (pink), CCM- coiled coil motif (brown). Blue- β strands, yellow- α helices. Conserved ATPase core is highlighted in green. Scale bar: 10 aa. Accession no- Vaccinia virus: YP_233037.1, Organic Lake phycodnavirus 1: ADX05856.1, African swine fever virus: YP_009702812.1, M13 phage- NP_510893.1, PRD1 phage- AAX45927.1, FtsKCΔγ PDB ID- 2IUT.
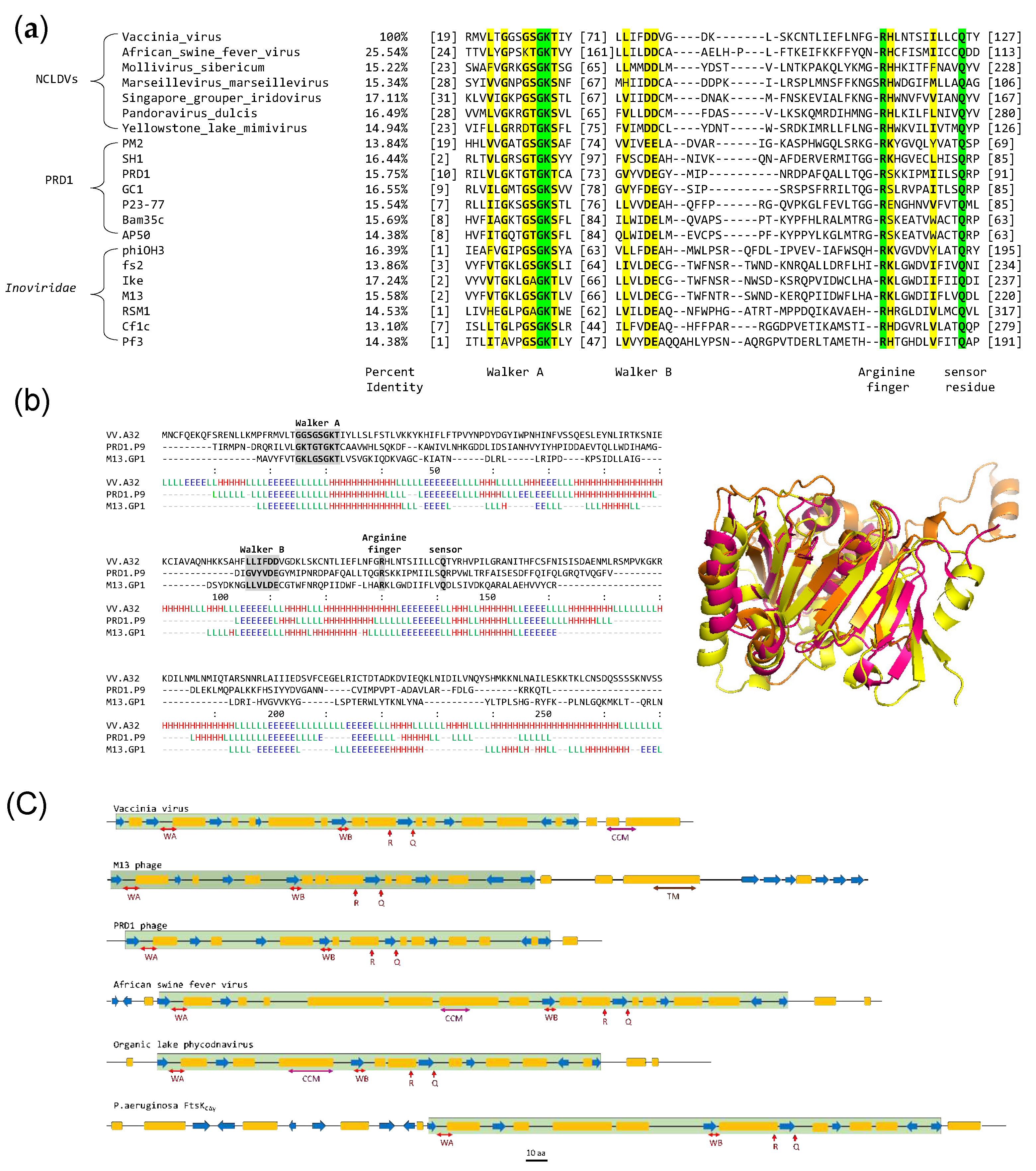
Figure 2.
Purification and ATPase activity of A32WT (a) Expression and purification of A32WT. UI- uninduced cell lysate; P-insoluble pellet, S-soluble protein supernatant; FThep - unbound flow-through of heparin column; FThis- unbound flow-through of histrap column; A32hep- Pooled A32 fractions from heparin column; A32WT- Purified and concentrated A32 protein after Heparin and Histrap purification; M-marker (b) (i) bar graph and (ii) autoradiograph showing ATPase activity in the presence/absence of DNA as a function of A32WT concentration. Trx denotes 10µM thioredoxin control. Values represent mean of duplicates with standard deviation, normalized with no protein control.
Figure 2.
Purification and ATPase activity of A32WT (a) Expression and purification of A32WT. UI- uninduced cell lysate; P-insoluble pellet, S-soluble protein supernatant; FThep - unbound flow-through of heparin column; FThis- unbound flow-through of histrap column; A32hep- Pooled A32 fractions from heparin column; A32WT- Purified and concentrated A32 protein after Heparin and Histrap purification; M-marker (b) (i) bar graph and (ii) autoradiograph showing ATPase activity in the presence/absence of DNA as a function of A32WT concentration. Trx denotes 10µM thioredoxin control. Values represent mean of duplicates with standard deviation, normalized with no protein control.
Figure 3.
Structural analysis of vaccinia virus A32. (a) Three-dimensional structure predicted by ColabFold modification of AlphaFold2 coloured by pLDDT score. (b) Ramachandran plot of predicted structure. Residues in favored and allowed regions (blue)- 99.25%, Outliers (red)- 0.75% Asn267 and Val268. (c) Disorder prediction using PrDOS- N-terminal 12 aa and C-terminal 20 aa show high probability for disorder. (d) Expression and Ni2+-NTA resin binding of A32Δ20C. UI-uninduced cell lysate; I-induced cell lysate; M-marker; P- insoluble pellet; S-soluble fraction; FT- Flow through; BW-binding buffer wash; 100, 300, and 600-elution with 100, 300, and 600 mM imidazole, respectively.
Figure 3.
Structural analysis of vaccinia virus A32. (a) Three-dimensional structure predicted by ColabFold modification of AlphaFold2 coloured by pLDDT score. (b) Ramachandran plot of predicted structure. Residues in favored and allowed regions (blue)- 99.25%, Outliers (red)- 0.75% Asn267 and Val268. (c) Disorder prediction using PrDOS- N-terminal 12 aa and C-terminal 20 aa show high probability for disorder. (d) Expression and Ni2+-NTA resin binding of A32Δ20C. UI-uninduced cell lysate; I-induced cell lysate; M-marker; P- insoluble pellet; S-soluble fraction; FT- Flow through; BW-binding buffer wash; 100, 300, and 600-elution with 100, 300, and 600 mM imidazole, respectively.
Figure 4.
Assessment of coiled coil motif in A32. Probability plot for coiled-coil motif prediction by PCOILS in (a) A32WT (b) A32L234K and (c) A32L234K_Q237A. The default output of probabilities in the scanning windows of 14 (green), 21 (blue) and 28 (purple) aa residues are shown. (d) The predicted probability of heptad repeats between aa residue 231 to 237 for the scanning windows of 21 or 28. (e) Multiple sequence alignment of coiled coil region in A32 homologs of Chordopoxvirinae subfamily using MUSCLE. Red- strictly conserved residues; yellow-conserved amino acids in majority of the sequences (f) Bit map image for the conservation of coiled-coil motif in Chordopoxvirinae.
Figure 4.
Assessment of coiled coil motif in A32. Probability plot for coiled-coil motif prediction by PCOILS in (a) A32WT (b) A32L234K and (c) A32L234K_Q237A. The default output of probabilities in the scanning windows of 14 (green), 21 (blue) and 28 (purple) aa residues are shown. (d) The predicted probability of heptad repeats between aa residue 231 to 237 for the scanning windows of 21 or 28. (e) Multiple sequence alignment of coiled coil region in A32 homologs of Chordopoxvirinae subfamily using MUSCLE. Red- strictly conserved residues; yellow-conserved amino acids in majority of the sequences (f) Bit map image for the conservation of coiled-coil motif in Chordopoxvirinae.
Figure 5.
Regulation of ATPase activity by coiled coil motif (a) Purified thioredoxin-tagged proteins. A32WT - wild type; A32K31A -Walker A motif mutant; A32L234K -coiled coil motif mutant 1; A32L234K_Q237A - coiled coil motif mutant 2. (b) (i) bar graph and (ii) autoradiograph of comparative ATPase activities of wildtype A32WT and its mutants. Trx denotes 10µM thioredoxin control. Values represent mean of duplicates with standard deviation, normalized with no protein control. (c) Steady- state kinetics analysis of (i) A32WT (ii) A32K31A and (iii) A32L234K_Q237A. Values represent mean of duplicates with standard deviation, normalized with no ATP control.
Figure 5.
Regulation of ATPase activity by coiled coil motif (a) Purified thioredoxin-tagged proteins. A32WT - wild type; A32K31A -Walker A motif mutant; A32L234K -coiled coil motif mutant 1; A32L234K_Q237A - coiled coil motif mutant 2. (b) (i) bar graph and (ii) autoradiograph of comparative ATPase activities of wildtype A32WT and its mutants. Trx denotes 10µM thioredoxin control. Values represent mean of duplicates with standard deviation, normalized with no protein control. (c) Steady- state kinetics analysis of (i) A32WT (ii) A32K31A and (iii) A32L234K_Q237A. Values represent mean of duplicates with standard deviation, normalized with no ATP control.
Table 1.
Summary of various strategies tested to obtain purified A32 protein of vaccinia virus.
Table 1.
Summary of various strategies tested to obtain purified A32 protein of vaccinia virus.
| Feature |
Vector |
Observation/result |
C-terminal 6x-Histidine tag,
codon optimized for E. coli
expression |
pET28a |
Low/no expression |
| C-terminal 8x-Histidine tag |
pET41a |
Low solubility |
Deletion of C-terminal
disordered region |
pET41a |
Higher expression, negligible solubility |
N-terminal Thioredoxin tag,
C-terminal 6x-Histidine tag |
pET32b |
Low solubility, purified and used for ATPase activity analysis |
| Thioredoxin tag cleavage |
pET32b |
Non-specific cleavage, protein precipitation |
| Thioredoxin tag cleavage |
pET32b-HRV3C |
Low solubility, truncation of protein |
| N-terminal GST tag |
pGEX-6P-1 |
Low solubility, inefficient column binding, tag autocleavage |
| Periplasmic expression |
pET22b |
No expression |
| Baculovirus expression system |
pFastBac1 |
No expression |
Table 2.
Steady-state kinetic parameters obtained for ATPase activity of A32 and its mutants.
Table 2.
Steady-state kinetic parameters obtained for ATPase activity of A32 and its mutants.
| Kinetic Parameters |
A32WT
|
A32K31A
|
A32LL234K_Q237A
|
-
DNA |
+
DNA |
-
DNA |
+
DNA |
-
DNA |
+
DNA |
| Vmax (µM min-1) |
2.43 |
16.96 |
0.52 |
1.12 |
9.12 |
7.87 |
| Kcat (min-1) |
0.41 |
2.83 |
0.09 |
0.19 |
1.52 |
1.31 |
| Km (µM) |
0.74 |
1.26 |
0.20 |
0.74 |
0.66 |
0.25 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).