Submitted:
01 February 2024
Posted:
02 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Isolation and Purification
2.3. PCR Amplification and Sequencing
2.4. Data and Statistical Analysis
2.4.1. Sequence Editing and Alignment
2.4.2. Polymorphism and Diversity Analysis
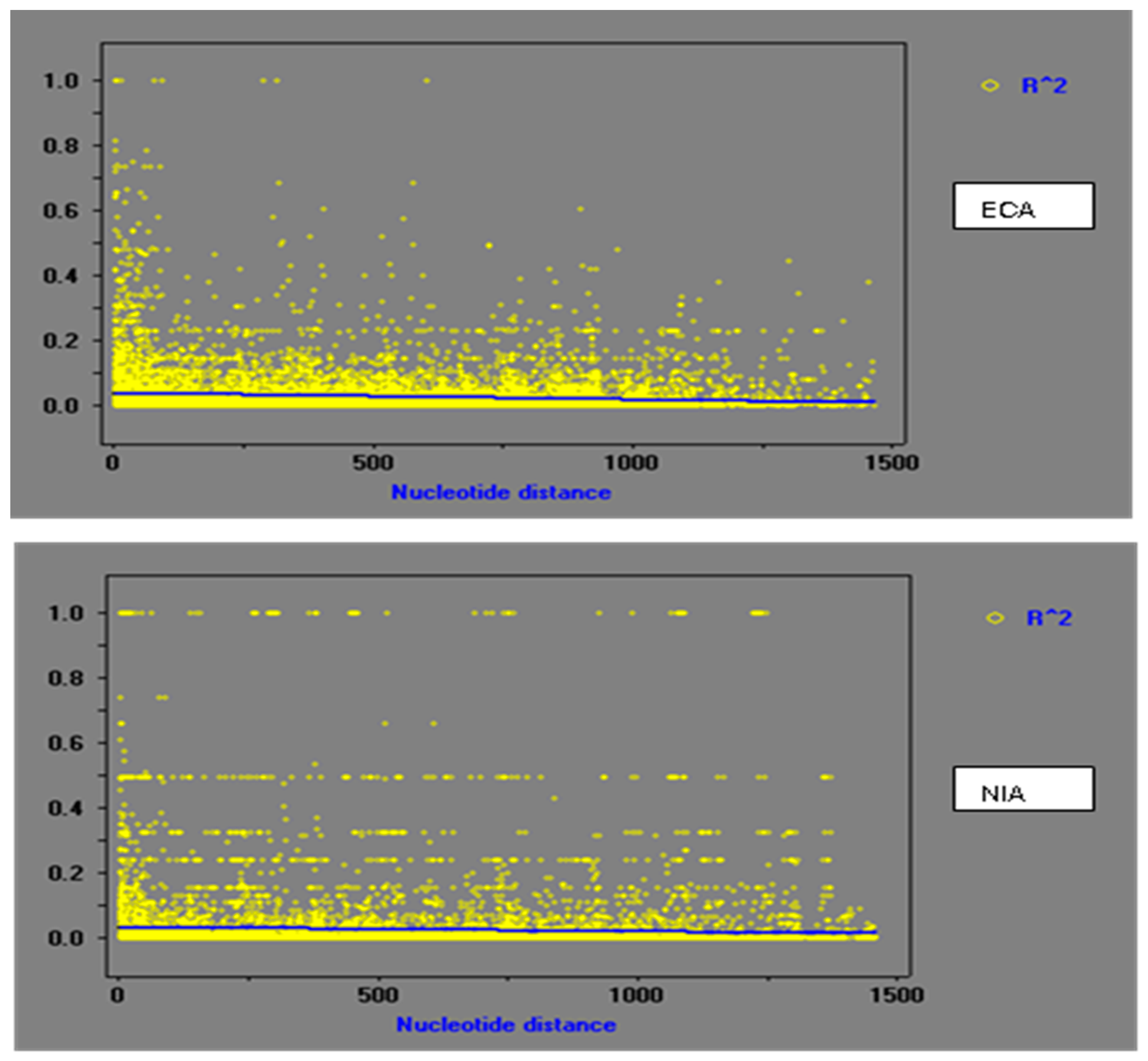
2.4.3. Linkage Disequilibrium
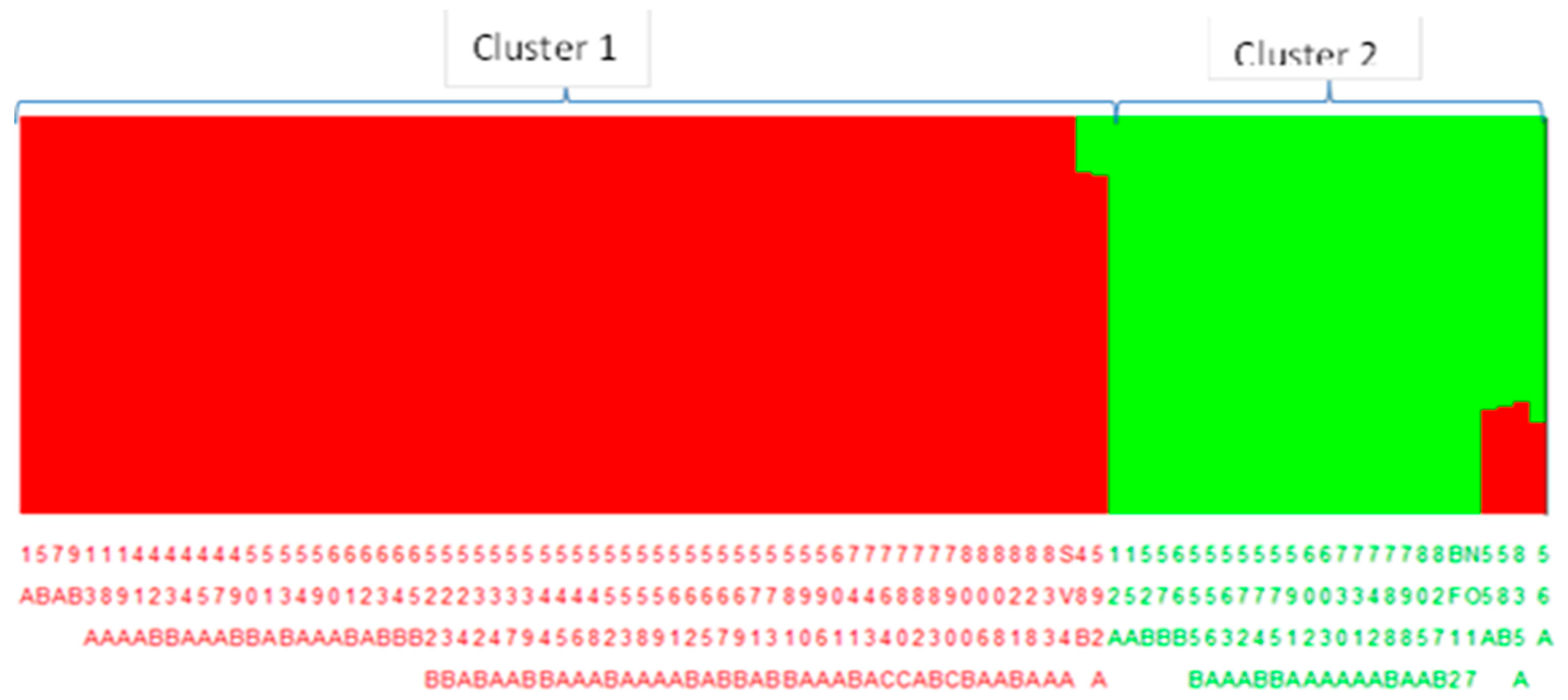
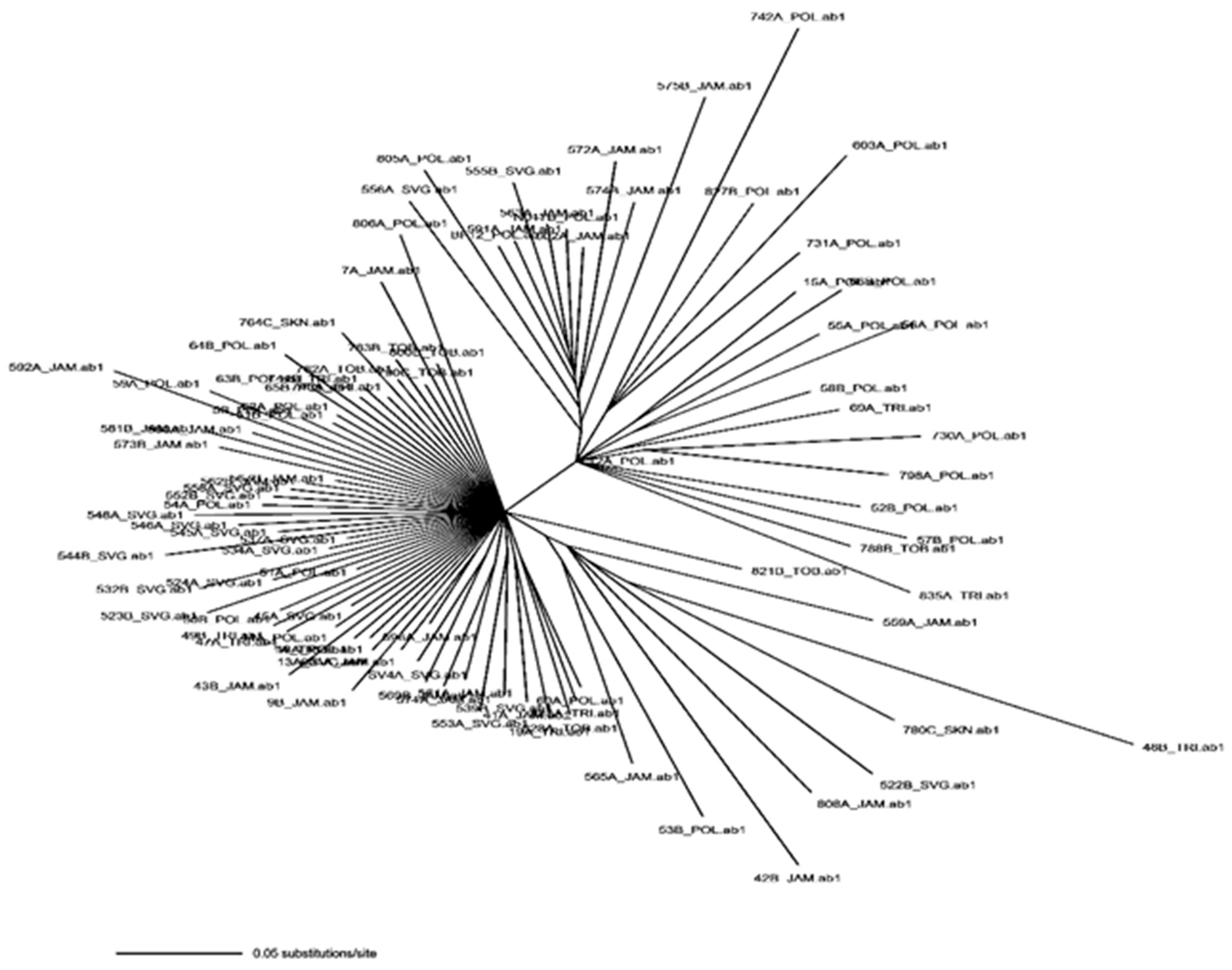
2.4.4. Population Structure and Phylogenetic Analysis
3. Results
3.1. Sequence Analysis
3.2. Identity Analysis
3.3. Population Recombination
3.4. Population Structure and Demographic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zerega, Nyree J. C., Diane C. Ragone, and Timothy J. Motley. "Complex Origins of Breadfruit (Artocarpus Altilis, Moraceae): Implications for Human Migrations in Oceania." American Journal of Botany 91, no. 5 (2004): 760–66. [CrossRef]
- ———. "Breadfruit Origins, Diversity, and Human-Facilitated Distribution." In Darwin’s Harvest: New Approaches to Origins, Evolution, and Conservation of Crop Plants, edited by Timothy J. Motley, Nyree J. C. Zerega, Diane C. Ragone and H. Cross, 213-38. New York: Columbia University Press, 2006.
- Roberts-Nkrumah, Laura B. "An Overview of Breadfruit (Artocarpus Altilis) in the Caribbean." Acta Horticulturae 757 (2007): 51-60. [CrossRef]
- Powell, Dulcie. "The Voyage of the Plant Nursery, H.M.S.Providence, 1791–1793." Economic Botany 31, no. 4 (1977): 387-431.
- Jones, A. M. P., D. Ragone, N. G. Tavana, D. W. Bernotas, and S. J. Murch. "Beyond the Bounty : Breadfruit (Artocarpus Altilis) for Food Security and Novel Foods in the 21st Century." Ethnobotany Research and Applications 9 (2011): 129-49. [CrossRef]
- Roberts-Nkrumah, Laura B. "Enhancing Breadfruit Contribution to Food Security in the Caribbean through Improved Supply." Paper presented at the Proceedings of the Caribbean Food Crop Society, Frigate Bay, Federation of St. Kitts and Nevis, 12 to 17, July, 2009 2009.
- Ragone, Diane. "Breadfruit : Diversity , Conservation and Potential." Acta Horticulturae 757 (2007): 19-30. [CrossRef]
- Daley, Oral O., Laura B. Roberts-Nkrumah, and Angela T. Alleyne. "Morphological Diversity of Breadfruit [Artocarpus Altilis (Parkinson) Fosberg] in the Caribbean." Scientia Horticulturae 266 (2020): 109278. [CrossRef]
- Ragone, Diane C. "Description of Pacific Island Breadfruit Cultivars." Acta Horticulturae 413 (1995): 93-98. [CrossRef]
- Daley, Oral O., Laura B. Roberts-Nkrumah, Angela T. Alleyne, and Michael C. Gloster. "Folk Nomenclature and Traditional Knowledge of Breadfruit [Artocarpus Altilis (Parkinson) Fosberg] Diversity in Four Anglophone Caribbean Countries." Journal of Ethnobiology and Ethnomedicine 18, no. 1 (2022): 65. [CrossRef]
- Ragone, Diane, Caugau Tavana, Joan M. Stevens, Patricia A. N. N. Stewart, Rebekka Ston E, Mxfthew Cox, Paul Alan Cox, Ii Brigham, Young Unirersily, and Pruoo Ut. "Nomenclature of Breadfruit Cultivars in Samoa: Saliency, Ambiguity and Monomiality." Journal of Ethnobiology 24, no. 1 (2004): 33-49.
- Korir, N. K., J. Han, L. Shangguan, C. Wang, E. Kayesh, Y. Zhang, and J. Fang. "Plant Variety and Cultivar Identification: Advances and Prospects." Critical Reviews in Biotechnology 33, no. 2 (2013): 111-25. [CrossRef]
- Biswas, M. K., Q. Xu, C. Mayer, and X. Deng. "Genome Wide Characterization of Short Tandem Repeat Markers in Sweet Orange (Citrus Sinensis)." Plos One 9, no. 8 (2014): e104182. [CrossRef]
- Hoshino, Andrea Akemi, Juliana Pereira Bravo, Karina Alessandra Morelli, and Paula Macedo Nobile. "Microsatellites as Tools for Genetic Diversity Analysis." In Genetic Diversity in Microorganisms, edited by M. Caliskan, 149-70. Croatia: INTECH Open Access Publisher, 2012.
- Kanzaki, Shinya, Keizo Yonemori, Akira Sugiura, and Suranant Subhadrabandhu. "Phylogenetic Relationships between the Jackfruit, the Breadfruit and Nine Other Artocarpus Spp. From Rflp Analysis of an Amplified Region of Cpdna." Scientia Horticulturae 70, no. 1 (1997): 57-66. [CrossRef]
- Zerega, Nyree J. C., Diane C. Ragone, and Timothy J. Motley. "Systematics and Species Limits of Breadfruit (Artocarpus, Moraceae)." Systematic Botany 30, no. 3 (2005): 603-15. [CrossRef]
- Sreekumar, V. B., A. M. Binoy, and Sarah T. George. "Genetic and Morphological Variation in Breadfruit (Artocarpus Altilis (Park.) Fosberg) in the Western Ghats of India Using Aflp Markers." Genetic Resources and Crop Evolution 54 (2007): 1659-65. [CrossRef]
- Witherup, Colby, Diane Ragone, Tyr Wiesner-Hanks, Brian Irish, Brian Scheffler, Sheron Simpson, Francis Zee, M. Iqbal Zuberi, and Nyree J. C. Zerega. "Development of Microsatellite Loci in Artocarpus Altilis (Moraceae) and Cross-Amplification in Congeneric Species." Applications in Plant Sciences 1, no. 7 (2013): 1-6. [CrossRef]
- Zerega, Nyree J. C., Tyr Wiesner-Hanks, Diane Ragone, Brian Irish, Brian Scheffler, Sheron Simpson, and Francis Zee. "Diversity in the Breadfruit Complex (Artocarpus, Moraceae): Genetic Characterization of Critical Germplasm." Tree Genetics and Genomes 11, no. 1 (2015): 1-26. [CrossRef]
- Gardner, E. M., K. M. Laricchia, M. Murphy, D. Ragone, B. E. Scheffler, S. Simpson, E. W. Williams, and N. J. Zerega. "Chloroplast Microsatellite Markers for Artocarpus (Moraceae) Developed from Transcriptome Sequences." Applications in Plant Sciences 3, no. 9 (2015). [CrossRef]
- Zerega, Nyree J. C., and Diane Ragone. "Toward a Global View of Breadfruit Genetic Diversity." Tropical Agriculture, no. Special Issue: International Breadfruit Conference 2015 (2016): 77-91.
- De Bellis, Fabien, Roger Malapa, Valérie Kagy, Stéphane Lebegin, Claire Billot, and Jean-Pierre Labouisse. "New Development and Validation of 50 Ssr Markers in Breadfruit (Artocarpus Altilis, Moraceae) by Next-Generation Sequencing." Applications in Plant Sciences 4, no. 8 (2016): 1600021. [CrossRef]
- Audi, Lauren, Gordon Shallow, Erasto Robertson, Dean Bobo, Diane Ragone, Elliot M. Gardner, Babita Jhurree-Dussoruth, Jacek Wajer, and Nyree J. C. Zerega. "Linking Breadfruit Cultivar Names across the Globe Connects Histories after 230 Years of Separation." Current Biology (2023). [CrossRef]
- Barthe, Stéphanie, Felix Gugerli, Noelle A. Barkley, Laurent Maggia, Céline Cardi, and Ivan Scotti. "Always Look on Both Sides: Phylogenetic Information Conveyed by Simple Sequence Repeat Allele Sequences." Plos One 7, no. 7 (2012): e40699. [CrossRef]
- Estoup, Arnaud, Philippe Jarne, and Jean-Marie Cornuet. "Homoplasy and Mutation Model at Microsatellite Loci and Their Consequences for Population Genetics Analysis." Molecular Ecology 11, no. 9 (2002): 1591-604. [CrossRef]
- Kearse, Matthew, Richard Moir, Amy Wilson, Steven Stones-Havas, Matthew Cheung, Shane Sturrock, Simon Buxton, Alex Cooper, Sidney Markowitz, Chris Duran, Tobias Thierer, Bruce Ashton, Peter Meintjes, and Alexei Drummond. "Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data." Bioinformatics 28, no. 12 (2012): 1647-49. [CrossRef]
- Librado, P., and J. Rozas. "Dnasp V5: A Software for Comprehensive Analysis of DNA Polymorphism Data." Bioinformatics 25, no. 11 (2009): 1451-52. [CrossRef]
- Hudson, Richard R. "A New Statistic for Detecting Genetic Differentiation." Genetics 155, no. 4 (2000): 2011-14. [CrossRef]
- Remington, D. L., J. M. Thornsberry, Y. Matsuoka, L. M. Wilson, S. R. Whitt, J. Doebley, S. Kresovich, M. M. Goodman, and E. S. t. Buckler. "Structure of Linkage Disequilibrium and Phenotypic Associations in the Maize Genome." Proceedings of the National Academy of Sciences of the United States of America 98 (2001): 11479-84. [CrossRef]
- Corander, Jukka, Pekka Marttinen, Jukka Sirén, and Jing Tang. "Enhanced Bayesian Modelling in Baps Software for Learning Genetic Structures of Populations." BMC Bioinformatics 9, no. 1 (2008): 1-14. [CrossRef]
- Corander, Jukka, Pekka Marttinen, and Jing Tang. "Baps : Bayesian Analysis of Population Structure." Structure (2009): 1-27.
- Swofford, D. L. "Paup* Version 4.0 B10. Phylogenetic Analysis Using Parsimony (* and Other Methods)." Sinauer, Sunderland, MA (2002).
- Hudson, R. R., M. Slatkin, and W. P. Maddison. "Estimation of Levels of Gene Flow from DNA Sequence Data." Genetics 132, no. 2 (1992): 583. [CrossRef]
- Bligh, William. The Log of H. M. S. Providence 1791-1793. England: Genesis Publications Limited, 1976.
- Roberts-Nkrumah, L.B. The Breadfruit Germplasm Collection at the University of the West Indies, St Augustine Campus. Kingston, Jamaica: University of the West Indies Press, 2018.
- Ragone, Diane Carol. "Collection , Establishment , and Evaluation of a Germplasm Collection of Pacific Island Breadfruit." 1991.
- Chen, Haofeng, Peter L. Morrell, Marlene de la Cruz, and Michael T. Clegg. "Nucleotide Diversity and Linkage Disequilibrium in Wild Avocado (Persea Americana Mill.)." Journal of Heredity 99, no. 4 (2008): 382-89. [CrossRef]
- Ingvarsson, Pär K. "Nucleotide Polymorphism and Linkage Disequilibrium within and among Natural Populations of European Aspen Populus Tremula." Genetics 169, no. 2 (2005): 945-53. [CrossRef]
- Ragone, Diane. "Chromosome Numbers and Pollen Stainability of Three Species of Pacific Island Breadfruit (Artocarpus, Moraceae)." American Journal of Botany 88, no. 4 (2001): 693-96. [CrossRef]
- Creste, Silvana, Augusto Tulmann Neto, Roland Vencovsky, Sebastião de Oliveira Silva, and Antonio Figueira. "Genetic Diversity of Musa Diploid and Triploid Accessions from the Brazilian Banana Breeding Program Estimated by Microsatellite Markers." Genetic Resources and Crop Evolution 51, no. 7 (2004): 723-33. [CrossRef]
- Hedrick, Phillip W. Genetics of Populations. 4th ed. Sadbury, Ma, USA: Jones and Bartlett Publishers, 2011.
- Hartl, Daniel L., and Andrew G. Clark. Principles of Population Genetics. 4th ed. Sunderland, Massachusetts, USA: Sinaer Associates, Inc. Publishers, 2007.
- Garris, Amanda J., Susan R. McCouch, and Stephen Kresovich. "Population Structure and Its Effect on Haplotype Diversity and Linkage Disequilibrium Surrounding the Xa5 Locus of Rice (Oryza Sativa L.)." Genetics 165, no. 2 (2003): 759. [CrossRef]
- Flint-Garcia, Sherry A., Jeffry M. Thornsberry, and Edward S. Buckler IV. "Structure of Linkage Disequilibrium in Plants." Annual Review of Plant Biology 54, no. 1 (2003): 357-74. [CrossRef]
- Bamberg, J. B., and A. H. del Rio. "Genetic Heterogeneity Estimated by Rapd Polymorphism of Four Tuber-Bearing Potato Species Differing by Breeding System." American Journal of Potato Research 81, no. 6 (2004): 377-83. [CrossRef]
- Simko, Ivan, Kathleen G. Haynes, and Richard W. Jones. "Assessment of Linkage Disequilibrium in Potato Genome with Single Nucleotide Polymorphism Markers." Genetics 173, no. 4 (2006): 2237-45. [CrossRef]
- Nordborg, Magnus. "Linkage Disequilibrium, Gene Trees and Selfing: An Ancestral Recombination Graph with Partial Self-Fertilization." Genetics 154, no. 2 (2000): 923-29. [CrossRef]
- Hudson, R. R. "Linkage Disequilibrium and Recombination." In In: Handbook of Statistical Genetics, edited by D. J. Balding, M. Bishop and C. Cannings. Chichester: John Wiley and Sons, Ltd, 2001.
- Liu, Aizhong, and John M. Burke. "Patterns of Nucleotide Diversity in Wild and Cultivated Sunflower." Genetics 173, no. 1 (2006): 321-30. [CrossRef]
- Zhu, Y. L., Q. J. Song, D. L. Hyten, C. P. Van Tassell, L. K. Matukumalli, D. R. Grimm, S. M. Hyatt, E. W. Fickus, N. D. Young, and P. B. Cregan. "Single-Nucleotide Polymorphisms in Soybean." Genetics 163, no. 3 (2003): 1123-34. [CrossRef]
- Fuchs, Eric J., and James L. Hamrick. "Genetic Diversity in the Endangered Tropical Tree, Guaiacum Sanctum (Zygophyllaceae)." Journal of Heredity 101, no. 3 (2010): 284-91. [CrossRef]
- Jones, A. Maxwell P., Susan J. Murch, Jim Wiseman, and Diane Ragone. "Morphological Diversity in Breadfruit (Artocarpus, Moraceae): Insights into Domestication, Conservation, and Cultivar Identification." Genetic Resources and Crop Evolution 60 (2013): 175-92. [CrossRef]
| No. | Sample ID | Cultivar name | Taxon | Ploidy | Sample collection site | Accession grouping |
| 1 | 12A | Huehue | Aa | 3n | UFS | NIA |
| 2 | 13A | Hope Marble | Aa | 3n | UFS | ECA |
| 3 | 15A | Ulu’ea | Aa | 2n | UFS | NIA |
| 4 | 18A | Meitehid | Aa | 3n | UFS | NIA |
| 5 | 19A | White | Aa | 3n | UFS | ECA |
| 6 | 1A | Yellow | Aa | 3n | UFS | ECA |
| 7 | 41A | Macca | Aa | 3n | UFS | ECA |
| 8 | 42B | Yellow Heart | Aa | 3n | UFS | ECA |
| 9 | 43B | Yellow Heart | Aa | 3n | UFS | ECA |
| 10 | 44A | Aveloloa | Aa | 3n | UFS | NIA |
| 11 | 45A | Creole | Aa | 3n | UFS | ECA |
| 12 | 47A | White | Aa | 3n | UFS | ECA |
| 13 | 48B | White | Aa | 3n | UFS | ECA |
| 14 | 49B | White | Aa | 3n | UFS | ECA |
| 15 | 50B | Porohiti | Aa | 3n | UFS | NIA |
| 16 | 51A | Porohiti | Aa | 3n | UFS | NIA |
| 17 | 522B | Captain Bligh | Aa | 3n | SVG | ECA |
| 18 | 523B | Unknown 1 | Aa | 3n | SVG | ECA |
| 19 | 524A | Creole | Aa | 3n | SVG | ECA |
| 20 | 52B | Toneno | Aa | 3n | UFS | NIA |
| 21 | 532B | Sally Young | Aa | 3n | SVG | ECA |
| 22 | 534A | White | Aa | 3n | SVG | ECA |
| 23 | 537A | Hog Pen | Aa | 3n | SVG | ECA |
| 24 | 539B | Dessert | Aa | 3n | SVG | ECA |
| 25 | 53B | Roiha’a | Aa | 3n | UFS | NIA |
| 26 | 544B | Kashee Bread | Aa | 3n | SVG | ECA |
| 27 | 545A | Hope Marble | Aa | 3n | SVG | ECA |
| 28 | 546A | Lawyer Caine | Aa | 3n | SVG | ECA |
| 29 | 548A | Dessert | Aa | 3n | SVG | ECA |
| 30 | 54A | Tapeha’a | Aa | 3n | UFS | NIA |
| 31 | 552B | Sally Young | Aa | 3n | SVG | ECA |
| 32 | 553A | White | Aa | 3n | SVG | ECA |
| 33 | 555B | Waterloo/Cotton | Aa | 3n | SVG | ECA |
| 34 | 556A | Soursop | Aa | 3n | SVG | ECA |
| 35 | 558A | Liberal | Aa | 3n | SVG | ECA |
| 36 | 559A | Yellow Heart | Aa | 3n | JAM | ECA |
| 37 | 55A | Piipiia | Aa x Am | 3n | UFS | NIA |
| 38 | 561A | Timor | Aa | 3n | JAM | ECA |
| 38 | 562B | Yellow Heart | Aa | 3n | JAM | ECA |
| 40 | 563A | Couscous | Aa | 3n | JAM | ECA |
| 41 | 565A | Timor | Aa | 3n | JAM | ECA |
| 42 | 567B | Yellow Heart | Aa | 3n | JAM | ECA |
| 43 | 569B | Yellow Heart | Aa | 3n | JAM | ECA |
| 44 | 56A | Meinpadahk | Aa x Am | 3n | UFS | NIA |
| 45 | 571A | Timor | Aa | 3n | JAM | ECA |
| 46 | 572A | Couscous | Aa | 3n | JAM | ECA |
| 47 | 573B | White Heart | Aa | 3n | JAM | ECA |
| 48 | 574B | Macca | Aa | 3n | JAM | ECA |
| 49 | 575B | Brambram | Aa | 3n | JAM | ECA |
| 50 | 57B | Momolega | Aa | 2n | UFS | NIA |
| 51 | 581B | Yellow Heart | Aa | 3n | JAM | ECA |
| 52 | 58B | Unknown 2 | Aa | 3n | UFS | NIA |
| 53 | 590A | White Heart | Aa | 3n | JAM | ECA |
| 54 | 591A | Monkey Breadfruit | Aa | 3n | JAM | ECA |
| 55 | 592A | Monkey Breadfruit | Aa | 3n | JAM | ECA |
| 56 | 596A | Banjam | Aa | 3n | JAM | ECA |
| 57 | 59A | Pua’a | Aa | 3n | UFS | NIA |
| 58 | 5B | Meitehid | Aa | 3n | UFS | NIA |
| 59 | 601A | Portland Breadfruit | Aa | 3n | JAM | ECA |
| 60 | 602A | Man Bread | Aa | 3n | JAM | ECA |
| 61 | 603A | Ma’afala | Aa | 2n | JAM | NIA |
| 62 | 60A | Unidentified | Aa | - | UFS | - |
| 63 | 61B | Mahani | Aa | 3n | UFS | NIA |
| 64 | 62A | Afara | Aa | 3n | UFS | NIA |
| 65 | 63B | Fafai | Aa | 3n | UFS | NIA |
| 66 | 64B | Yellow | Aa | 3n | UFS | NIA |
| 67 | 65B | Otea | Aa | 3n | UFS | NIA |
| 68 | 66B | Puou | Aa | 2n | UFS | NIA |
| 69 | 69A | Chataigne | Ac | 2n | UFS | ECA |
| 70 | 730A | Masunwa | Aa | 2n | TRI | NIA |
| 71 | 731A | Ma’afala | Aa | 2n | TRI | NIA |
| 72 | 741B | Yellow | Aa | 3n | TRI | ECA |
| 73 | 742A | Ma’afala | Aa | 2n | TRI | NIA |
| 74 | 743A | White | Aa | 3n | TRI | ECA |
| 75 | 764C | Unknown 3 | Aa | 3n | SKN | ECA |
| 76 | 780C | Unknown 4 | Aa | 3n | SKN | ECA |
| 77 | 782A | Unknown 5 | Aa | 3n | TOB | ECA |
| 78 | 783B | Butter Breadfruit | Aa | 3n | TOB | ECA |
| 79 | 788B | Choufchouf | Aa | 3n | TOB | ECA |
| 80 | 790C | Unidentified | Aa | 3n | TOB | ECA |
| 81 | 798A | Ma’afala | Aa | 2n | TRI | NIA |
| 82 | 7A | Timor | Aa | 3n | UFS | ECA |
| 83 | 800 B | White | Aa | 3n | TOB | ECA |
| 84 | 805 A | Pu’upu’u | Aa | 3n | TOB | NIA |
| 85 | 806 A | Meitehid | Aa | 3n | TOB | ECA |
| 86 | 808 A | Timor | Aa | 3n | TOB | ECA |
| 87 | 821 B | White | Aa | 3n | TOB | ECA |
| 88 | 827 B | Ma’afala | Aa | 2n | TOB | ECA |
| 89 | 828 A | Local Yellow | Aa | 2n | TOB | ECA |
| 90 | 833A | Local Yellow | Aa | 3n | TRI | ECA |
| 91 | 835A | Ma’afala | Aa | 2n | TRI | NIA |
| 92 | 9B | Cassava | Aa | 3n | UFS | ECA |
| 93 | BF12 | Unknown 6 | Aa | - | UFS | NIA |
| 94 | NO.17 B | Pu’upu’u | Aa | 3n | UFS | NIA |
| 95 | SV4A | Cocobread | Aa | 3n | UFS | ECA |
| Locus | Primer sequence (5’--3’) | Repeat Motifa |
| MAA40 | F: AGCATTTCAGGTTGGTGAC R: TTGTTCTGTTTGCCTCATC |
(TG)16 |
| MAA54a | F: AACCTCCAAACACTAGGACAAC R: AGCTACTTCCAAAACGTGACA |
(CA)5,(AT)4 |
| MAA71 | F: TTCCTATTTCTTGCAGATTCTC R: AGTGGTGGTAAGATTCAAAGTG |
(CT)11(CA)19 |
| MAA85 | F: TCAGGGTGTAGCGAAGACA R: AGGGCTCCTTTGATGGAA |
(CA)11 |
| MAA96 | F: GGACCTCAAGGATGTGATCTC R: ACACGGTCTTCTTTGGATAGC |
(CA)14(TA)7(TG)3(GT) |
| MAA140 | F: CCATCCCCCATCTTTCCT R: TCCTCGTTTGCCACAGTG |
(CT)25 |
| *MAA178a | F: GATGGAGACACTTTGAACTAGC R: CACCAGGGTTTAAGATGAAAC |
(GT)3,(GT)6,(GT)3,(GA)3,(GA)10 |
| *MAA178b | F: GATGGAGACACTTTGAACTAGC R: CACCAGGGTTTAAGATGAAAC |
(GT)3,(GT)3,(GA)3,(GA)11 |
| MAA182 | F: TACTGGGTCTGAAAAGATGTCT R: CGTTTGCGTTTGGATAAAT |
(CT)19 |
| MAA251 | F: ATCGTCTTTGTCACCACCAC R: ATAGCCGAGTAACTGGATGGA |
(ATC)10 |
| Microsatellite locus | Group | SNP (Parsimony informative) | Nucleotide diversity ( | Nucleotide polymorphism () | Tajima’s D test | Fu’s F test | Harpending index |
| MAA40 | Total | 40 (9) | 0.334 | 0.16 | -1.709 | -2.338 | 0.004 |
| ECA | 50 (7) | 0.323 | 0.204 | -1.269 | -1.266 | 0.002 | |
| NIA | 61 (22) | 0.349 | 0.182 | -1.181 | -2.285 | 0.004 | |
| MAA54A | Total | 50 (7) | 0.328 | 0.154 | -0.536 | -2.468 | 0.046 |
| ECA | 51 (8) | 0.338 | 0.171 | -0.398 | -2.518 | 0.032 | |
| NIA | 56 (30) | 0.193 | 0.1 | -2.069 | -2.609 | 0.065 | |
| MAA71 | Total | 46 (8) | 0.276 | 0.141 | -0.643 | -2.815 | 0.010 |
| ECA | 70 (13) | 0.34 | 0.239 | 0.911 | -1.432 | 0.002 | |
| NIA | 37 (19) | 0.185 | 0.108 | -1.29 | -2.221 | 0.021 | |
| MAA85 | Total | 55 (1) | 0.303 | 0.253 | 1.48 | -0.451 | 0.001 |
| ECA | 60 (3) | 0.286 | 0.251 | -0.426 | -0.329 | 0.002 | |
| NIA | 72 (9) | 0.325 | 0.297 | 1.479 | 0.303 | 0.004 | |
| MAA96 | Total | 66 (13) | 0.305 | 0.171 | 0.021 | -1.969 | 0.002 |
| ECA | 71 (17) | 0.297 | 0.163 | -1.567 | -1.491 | 0.003 | |
| NIA | 90 (27) | 0.324 | 0.209 | 0.172 | -1.693 | 0.005 | |
| MAA140 | Total | 25 (10) | 0.301 | 0.087 | -2.236 | -3.685 | 0.019 |
| ECA | 33 (23) | 0.302 | 0.096 | -2.287 | -4.752 | 0.030 | |
| NIA | 54 (27) | 0.304 | 0.154 | -1.853 | -2.598 | 0.014 | |
| MAA178A | Total | 57 (12) | 0.33 | 0.181 | -1.498 | -1.545 | 0.003 |
| ECA | 62 (12) | 0.337 | 0.202 | -1.392 | -1.502 | 0.003 | |
| NIA | 63 (17) | 0.28 | 0.192 | -1.182 | -1.396 | 0.007 | |
| MAA178B | Total | 83 (18) | 0.259 | 0.176 | -1.081 | -1.874 | 0.001 |
| ECA | 86 (26) | 0.23 | 0.167 | -0.953 | -1.798 | 0.002 | |
| NIA | 106 (36) | 0.277 | 0.206 | -0.978 | -1.715 | 0.004 | |
| MAA182 | Total | 0 (0) | 0 | 0 | 0 | 0 | ND |
| ECA | 10 (0) | 0.383 | 0.247 | -1.084 | 0.167 | 0.082 | |
| NIA | 39 (1) | 0.333 | 0.348 | 0.164 | 0.075 | 0.006 | |
| MAA251 | Total | 64 (50 | 0.355 | 0.269 | -0.805 | -1.356 | 0.001 |
| ECA | 67 (8) | 0.284 | 0.233 | -0.625 | -0.974 | 0.001 | |
| NAC | 71 (14) | 0.397 | 0.3 | -0.927 | -1.222 | 0.004 | |
| Average | Total | 48.6 (8.3) | 0.159 | 0.279 | -0.701 | -1.850 | 0.010 |
| ECA | 56 (11.7) | 0.197 | 0.312 | -0.909 | -1.590 | 0.016 | |
| NIA | 64.9 (20.2) | 0.210 | 0.297 | -0.767 | -1.536 | 0.013 |
| Loci | Accession Group | No. of haplotype | h | RM | ρ | Walls’B |
| MAA40 | ECA | 58 | 0.996 ± 0.004 | 10 | 0.225 | 0.000 |
| NIA | 31 | 0.998 ± 0.008 | 7 | 0.404 | 0.000 | |
| MAA54A | ECA | 28 | 0.836 ± 0.039 | 7 | 0.623 | 0.000 |
| NIA | 13 | 0.843 ± 0.043 | 5 | 1.196 | 0.000 | |
| MAA71 | ECA | 62 | 1.000 ± 0.003 | 9 | 0.164 | 0.125 |
| NIA | 20 | 0.950 ± 0.024 | 8 | 0.559 | 0.071 | |
| MAA85 | ECA | 61 | 0.999 ± 0.003 | 17 | 0.129 | 0.000 |
| NIA | 32 | 1.000 ± 0.008 | 23 | 0.138 | 0.024 | |
| MAA96 | ECA | 56 | 0.992 ± 0.007 | 10 | 0.458 | 0.129 |
| NIA | 31 | 0.998 ± 0.008 | 11 | 0.286 | 0.075 | |
| MAA140 | ECA | 29 | 0.914 ± 0.024 | 1 | 1.173 | 0.375 |
| NIA | 25 | 0.956 ± 0.029 | 5 | 0.534 | 0.207 | |
| MAA178A | ECA | 59 | 0.998 ± 0.003 | 7 | 0.343 | 0.174 |
| NIA | 32 | 1.000 ± 0.008 | 13 | 0.172 | 0.091 | |
| MAA178B | ECA | 62 | 1.000 ± 0.003 | 17 | 0.241 | 0.125 |
| NIA | 32 | 1.000 ± 0.008 | 16 | 0.154 | 0.127 | |
| MAA182 | ECA | 28 | 0.955 ± 0.011 | 1 | 0.171 | 0.000 |
| NIA | 31 | 0.998 ± 0.008 | 15 | 0.087 | 0.044 | |
| MAA251 | ECA | 60 | 0.999 ± 0.003 | 15 | 0.332 | 0.000 |
| NIA | 32 | 1.000 ± 0.008 | 8 | 0.156 | 0.044 | |
| Average ( | ECA | 50.3 ± 15.268 | 0.969 ± 0.055 | 9.400 ± 5.778 | 0.386 ± 0.315 | 0.093 ± 0.121 |
| NIA | 27.900 ± 6.574 | 0.974 ± 0.050 | 11.100 ± 5.724 | 0.369 ± 0.336 | 0.068 ± 0.063 |
| Locus | All Accessions (n = 94) | |
| FSTa | Snnb | |
| MAA40 | -0.0002 | 0.6084 |
| MAA54A | 0.0105 | 0.5533 |
| MAA71 | 0.0147 | 0.6008 |
| MAA85 | 0.0178 | 0.7018*** |
| MAA96 | 0.0332 | 0.6939*** |
| MAA140 | 0.0513 | 0.6024** |
| MAA178A | 0.0035 | 0.5829 |
| MAA178B | 0.0358 | 0.7128** |
| MAA182 | - | - |
| MAA251 | 0.1095 | 0.6720** |
| Average (n=9) | 0.0306 | 0.6365 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





