Introduction
Archaea are the widest extension of microorganism which supposedly differ from the bacteria and eukarya in their genetic makeup. They are the single celled microbes that are frequently found in the severe conditions like hot springs, hydrothermal vents of deep sea (Bisson-Filho et al., 2018). At the outset archaea are categorized as bacteria but now they are acknowledged as a separate domain of life. They are significant because of their functions in global biogeochemical cycles and their potential uses in biotechnology.
Researchers discovered that archaea, like bacteria and eukarya, belong to the three domains of life after doing phylogenetic analyses on them. They are thought to be a mutated type of microbes descended from the common ancestor of all cellular life form (Martinez-Gutierrez & Aylward, 2021). Archaea are classified into four phyla: Crenarcheota, Korarchaeota, Thaumarchaeota, and Euryarchaeota. Because archaea can sustain in severe environments such as high temperatures, salt, acidity, and pressure, it is thought that archaea played a crucial role in providing an evolution in all over the world.
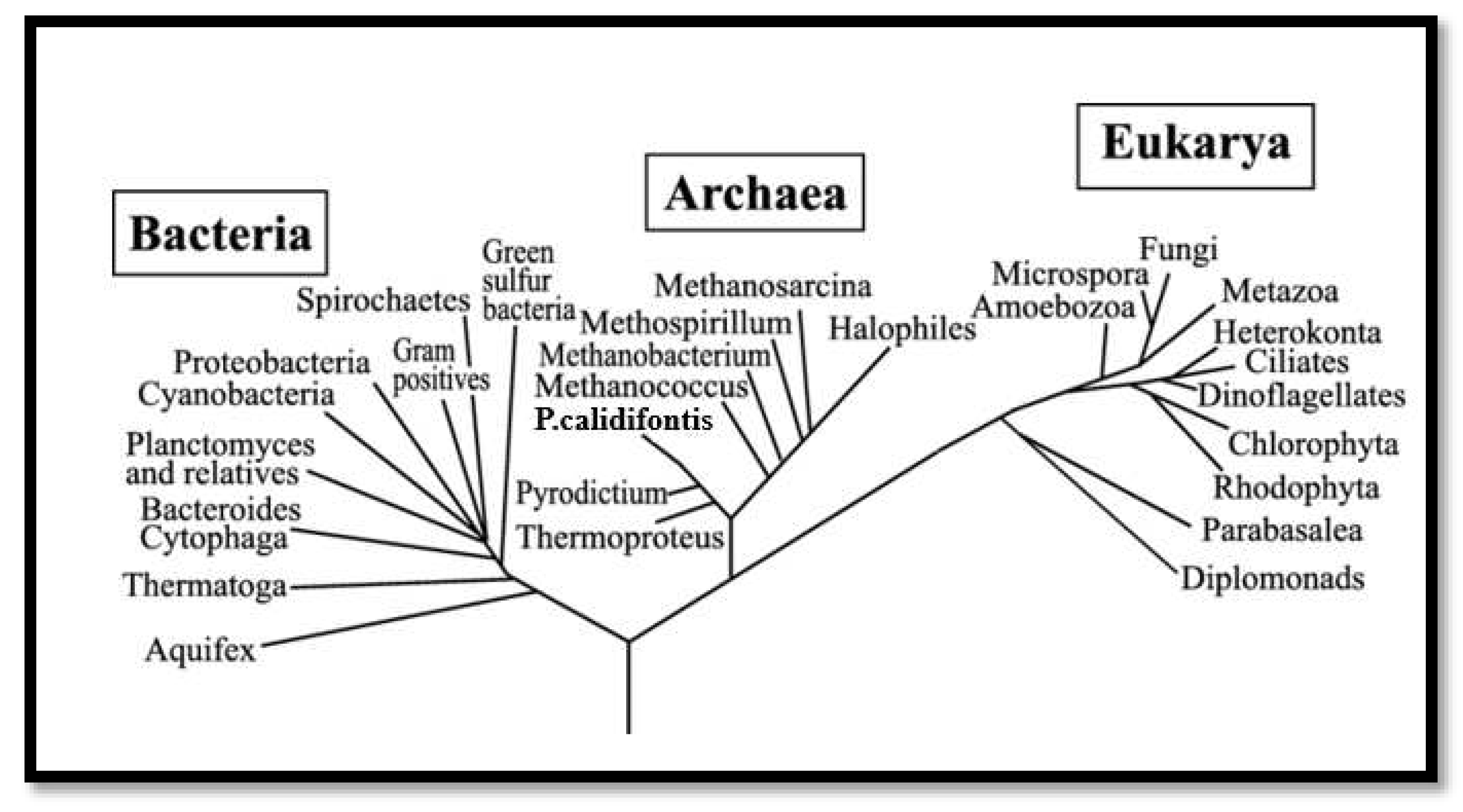
Figure 1.
Phylogenetic analysis centered on genetic factor. The three biotic domains of life are signified as bacteria, archaea, and eukaryote. Three branches of living creatures are linked to the last common ancestor by the ancestral black branch at the base of the phylogenetic tree. In the absence of an outgroup, the root is conjectural (Thomas, 2006).
Figure 1.
Phylogenetic analysis centered on genetic factor. The three biotic domains of life are signified as bacteria, archaea, and eukaryote. Three branches of living creatures are linked to the last common ancestor by the ancestral black branch at the base of the phylogenetic tree. In the absence of an outgroup, the root is conjectural (Thomas, 2006).
Role of HerA in DNA Resection
In all species, DNA end-resection is a vital phase in (HR) repair, and ATPase/Helicases are essential to this procedure. In hyperthermophile archaea, the exo-endonucleases NurA and HerA work in concert with the highly conserved Mre11-Rad50 complex to coordinate the repair of double-stranded DNA breaks. On the other hand, little is known about the HerA-NurA complex's activation and assembly mechanisms (Sung et al., 2014). In accordance with research in biochemistry, human DNA2 can cooperate with the BLM helicase and the ssDNA-binding protein, RPA, to facilitate the 5′ 3′ resection of DNA ends in vitro. RPA is thought to be the endonucleolytic cleaver, whereas DNA2 resects the unwinding DNA and BLM serves as a helicase. (Zhao et al., 2020). Genome integrity is achieved and maintained by a combination of cell processes that guarantee appropriate DNA duplication and repair as well as a genetic transfer from one cell division to the next. A crucial enzyme family for genomic integrity, DNA helicases are molecular motors that unwind double-stranded DNA to create single-stranded substrates (Bochman, 2014). DNA helicases unwind double-stranded DNA and RNA-DNA hybrids into ssDNA templates via ATP hydrolysis.
HerA and Its Mechanism in DNA Double Stranded Break Repair:
Genomic archaeal DNA is constantly susceptible to a wide range of lesions caused by both intrinsic and extrinsic mistakes. The ability to identify damage precisely is crucial for protecting the genome's integrity (Rolfsmeier et al., 2010). DNA damage that is not repaired or is not repaired effectively might result in genomic instability or cell death (Ceccaldi et al., 2016). DNA quality control necessitates MMR, NER, cross-link repair, base excision repair, and double-strand break repair. Important proteins have been increasingly conserved throughout time. BLM is a member of the bacterial, eukaryotic, and archaeal RecQ family of DNA helicases; DNA2 is linked to the bacterial RecB proteins and may function in vitro as both a helicase and a nuclease (Wilkinson et al., 2019). Mre11, Rad50, and the recombinase RadA, a Rad51 paralog, were shown to be similar to eukaryotic HR components in archaea, and several investigations on their functional importance have been published. Short 3′-overhangs at DSB ends are produced by Mre11-Rad50 through 3′- to 5′ exonuclease and endonuclease activity, complicated sensing, and processing. Numerous bacterial genomes, including those of Bacillus halodurans, Clostridium thermocellum, Aquifex aeolicus, and Deinococcus radiodurans, as well as a number of archaeal genomes, contain the HerA and NurA genes (Constantinesco, 2004). The genes typically co-occur in an operon with Mre11 and Rad50, and both of them get activated in response to UV radiation. These genes have been associated with HR in previous studies, and additional research supports the notion that the eukaryotic counterparts of these genes, HerA and NurA, are Exo1/EXO1 and Dna2/DNA2, respectively.
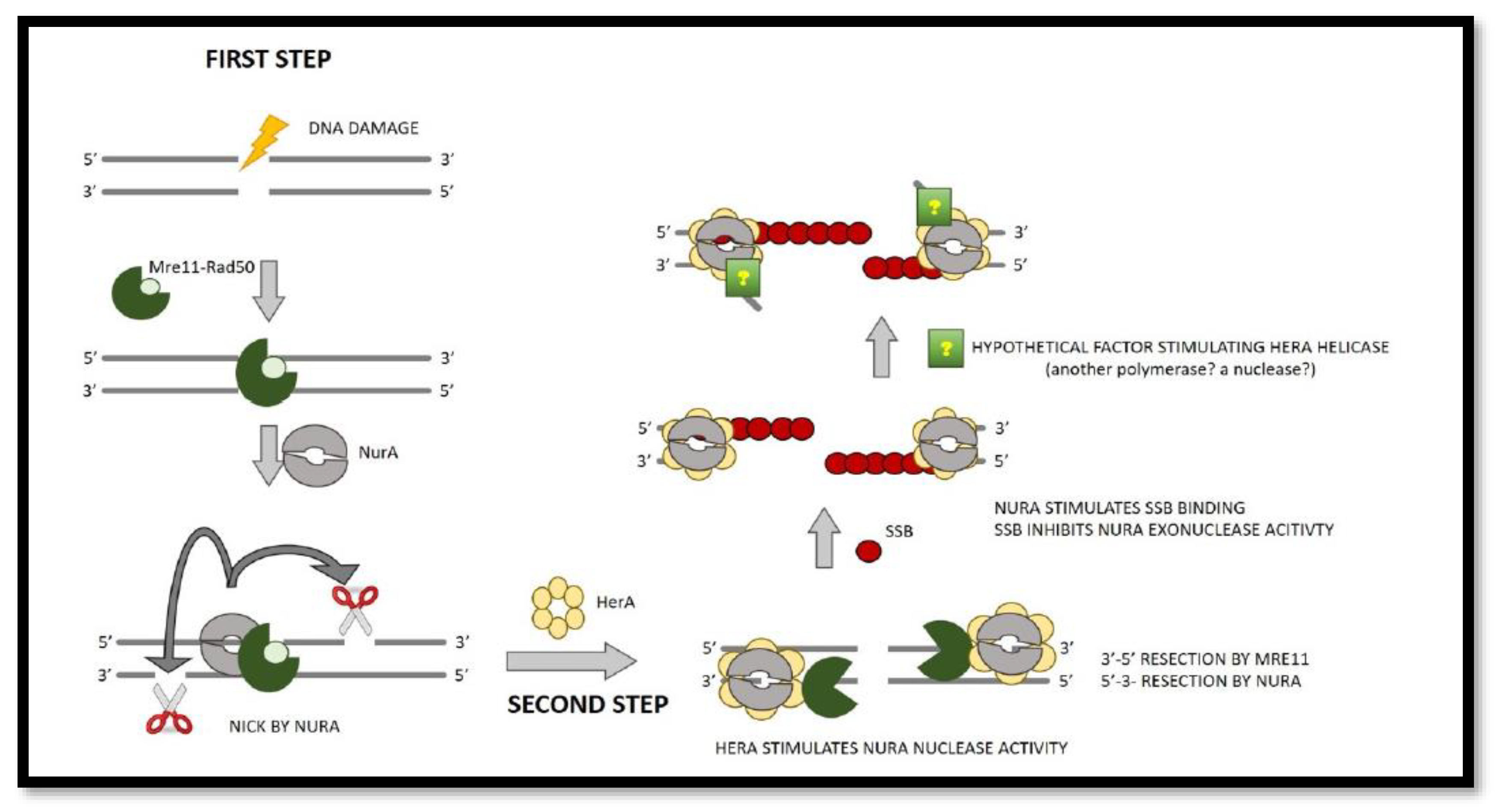
Figure 2.
End resection model in SSO (two phases). Mre11/Rad50 complexes (seen in dark and light green, respectively) are the first to recognize and bind the double stranded DNA. When NurA (grey dimer) nicks a DNA strand with a free 5′ end, the resection process begins. The second stage is to: To create ssDNA in a bidirectional manner, the ssDNA exonuclease activities 3′-5′ and 5′-3′ are used. HerA (a yellow hexamer that promotes NurA exonuclease) activates Mre11 and NurA activity. The SSB (red circle) is now attracted to the DNA and binds to the ssDNA generated by Mre11 and NurA, resulting in the formation of a nucleoprotein filament that suppresses NurA function.
Figure 2.
End resection model in SSO (two phases). Mre11/Rad50 complexes (seen in dark and light green, respectively) are the first to recognize and bind the double stranded DNA. When NurA (grey dimer) nicks a DNA strand with a free 5′ end, the resection process begins. The second stage is to: To create ssDNA in a bidirectional manner, the ssDNA exonuclease activities 3′-5′ and 5′-3′ are used. HerA (a yellow hexamer that promotes NurA exonuclease) activates Mre11 and NurA activity. The SSB (red circle) is now attracted to the DNA and binds to the ssDNA generated by Mre11 and NurA, resulting in the formation of a nucleoprotein filament that suppresses NurA function.
NurA and HerA are hypothesized to be key proteins in archaeal homologous recombination systems for DNA end resection. Thermus thermophilus HB8 proteins have significant sequence similarities with archaeal NurA and HerA. Researchers examined the cellular activity of NurA and HerA in T. thermophilus HB8 using disruptant mutants of these two proteins, either with or without agents that damage DNA. Deleted NurA and HerA had no effect on cell growth or genomic stability since neither gene was required for survival. Surprisingly, these T. thermophilus HB8 disruptors resisted mitomycin C and UV treatment. Furthermore, DNA replication inhibitor and which is vulnerable to oxidative stress which is not affected between disruptants and the wild type (Fujii et al., 2018).
When NurA and/or HerA were disrupted in T. thermophilus HB8, tolerance to UV exposure and MMC treatment was greater than in WT. Furthermore, NurA and HerA enzymatic activity in T. thermophilus HB8 cells seems to interact with or impede UV and MMC-induced DNA repair. NurA and HerA in D. radiodurans are equally resistant to UV and -rays as WT, although NurA is more resistant to MMC treatment than WT. These discrepancies imply that NurA and HerA play distinct roles in T. thermophilus HB8 and D. radiodurans responses to UV and MMC-induced DNA damage (Ahdash et al., 2017).
Modifications in NurA-HerA activities may be attributed to quaternary structural changes between T. thermophilus and other species. In a hexameric form, recombinant T. thermophilus HB8 HerA showed archaeal and Deinococcus HerA characteristics. NurA from T. thermophilus HB8 was only detected in a monomeric form. Deinococcus NurA, on the other hand, exhibits monomer-dimer equilibration, whereas the archaeal NurAs only create homodimers. Although the crystal structure of Thermotoga maritima NurA shows a dimeric form, it is unknown if Thermotoga NurA dimers in solution (Cheng et al., 2015). As a result, in solution, T. thermophilus HB8 NurA may exist in an equilibrium state of monomer and dimer or mostly in a monomeric state.
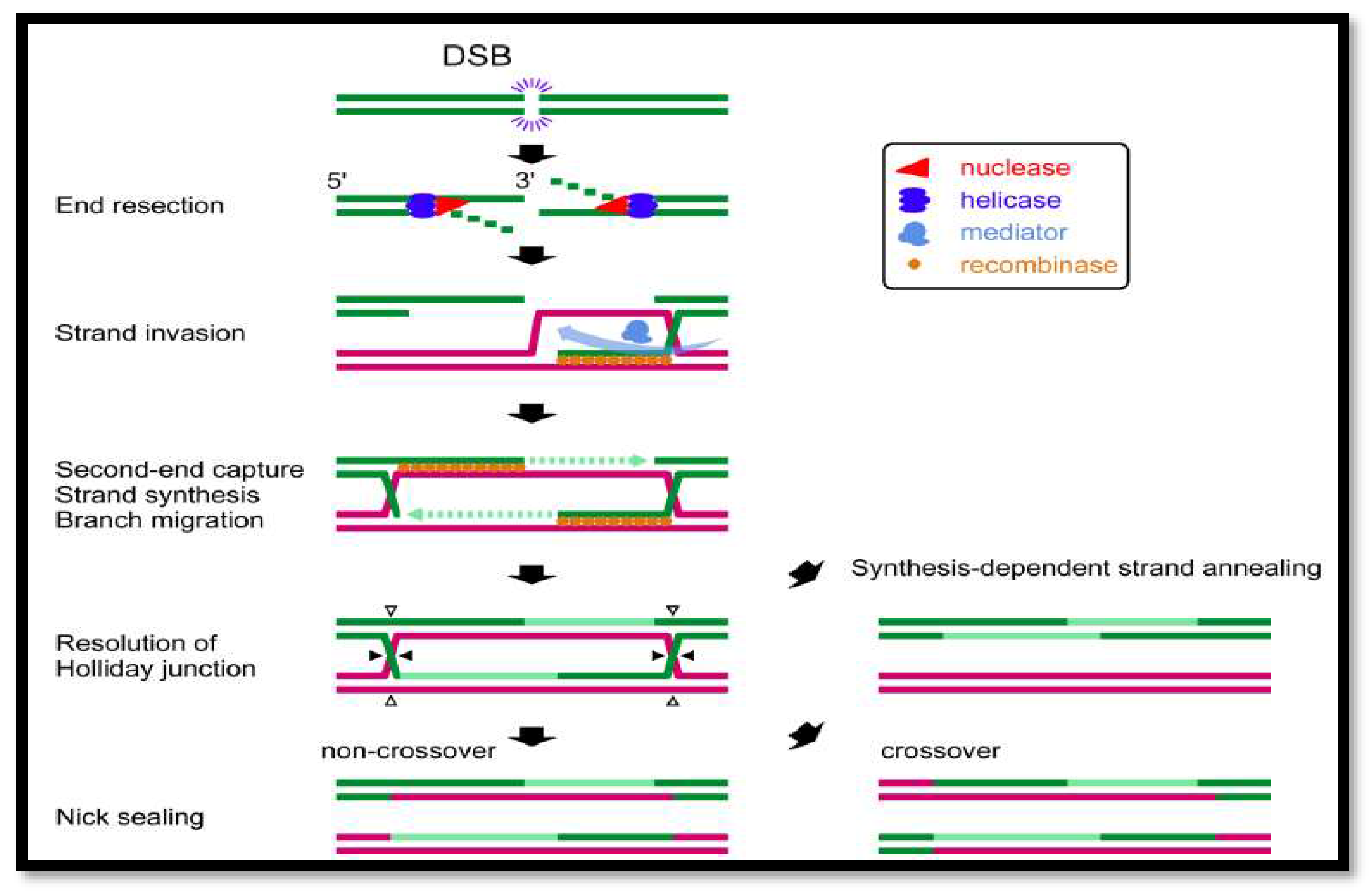
Figure 3.
A summary of DSB repair by homologous recombination. Green lines represent broken strands, whereas magenta lines represent homologous strands. The direction of the incision for resolving Holliday junctions in the fifth step is shown by closed or open triangles. Only when both Holliday junctions are incised in the same direction may non-crossover products be generated.
Figure 3.
A summary of DSB repair by homologous recombination. Green lines represent broken strands, whereas magenta lines represent homologous strands. The direction of the incision for resolving Holliday junctions in the fifth step is shown by closed or open triangles. Only when both Holliday junctions are incised in the same direction may non-crossover products be generated.
Results
The structural insights of the DNA helicase purified from different hyper thermophilic strains, further improves the importance of the archaeal helicases. Previous studies show that HerA forms a hexameric composition C₆ has been analyzed alone in Sulfolobus solfataricus (De Falco et al., 2022). On studying the domain view we have seen that every helicase found in the archaea contains following domains named N-terminal HerA-ATP synthase (HAS) barrel domain (Rzechorzek et al., 2014), a central RecA-like domain, n a-helical insert within the RecA-like domain. Pyrobaculum calidifontis is another archaeal specie that possess all the significant features of the helicase contributing in DNA double stranded break repair.
According to the protparam program analysis it is observed that Pca_HerA (0358), Having molecular weight of 66882.12 has 605 total amino acid in which positive amino acids (Asp + Glu) are 72 and negative amino acids (Arg + Lys) are 87 in number. It isoelectric point is 9.26.
HerA helicase of P. calidifontis has almost five different entries at NCBI named by their old locus number Pcal_0358, Pcal_1070, Pcal_1296, Pcal_0763 and Pcal_1112. Multiple sequence alignments between helicase orthologs and paralogs provided us the information that there are key residues that resembles with the other known helicases of archaea.
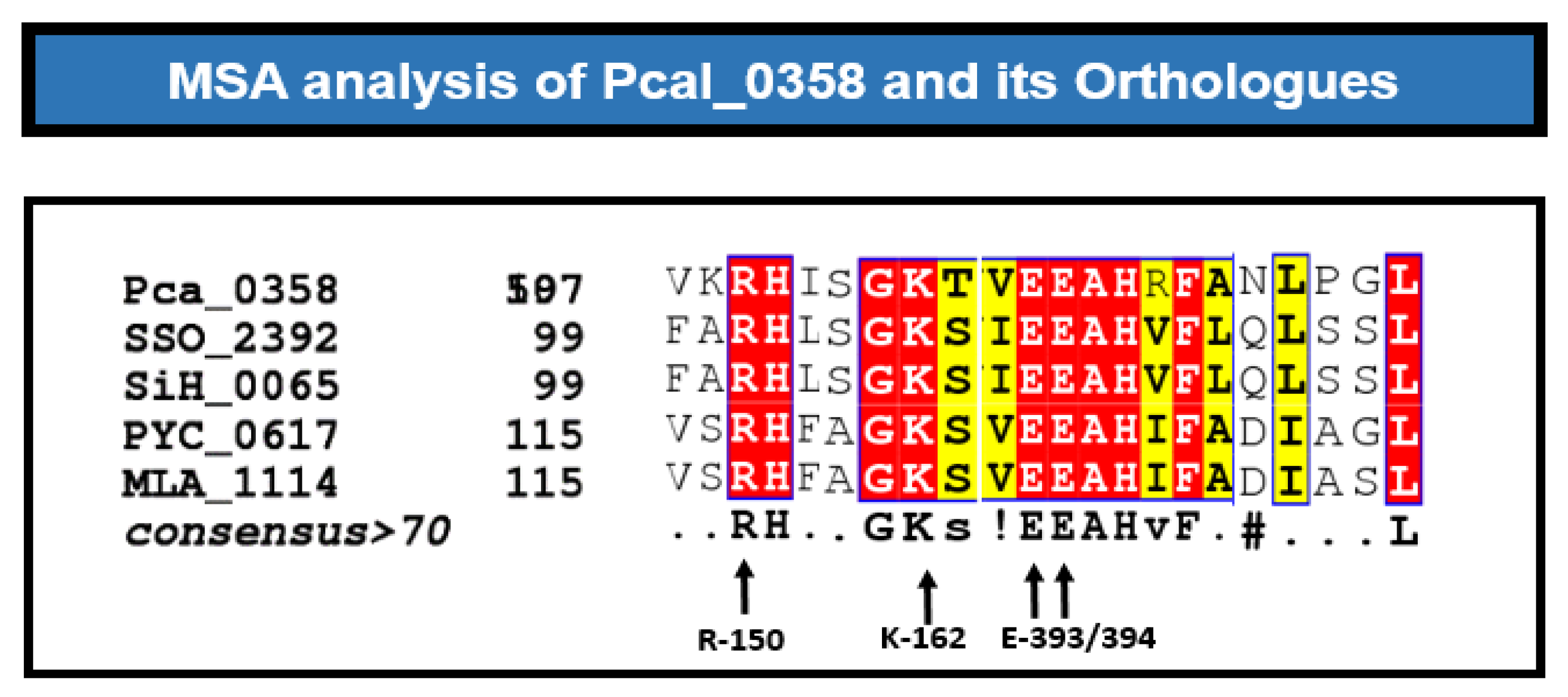
Figure 4.
MSA analysis of Helicase orthologs. Multiple sequence alignment between orthologs named P. calidifontis (Pca_0358), S. solfataricus (SSO_2392), S. shibatae (SiH_0065), P. yayanosii (PYC_0617) and Methanocaldococcus (MLA_1114). At the bottom these were the residues (R-150, K-162, E-393, E-394) that are find conserved in every ortholog of HerA.
Figure 4.
MSA analysis of Helicase orthologs. Multiple sequence alignment between orthologs named P. calidifontis (Pca_0358), S. solfataricus (SSO_2392), S. shibatae (SiH_0065), P. yayanosii (PYC_0617) and Methanocaldococcus (MLA_1114). At the bottom these were the residues (R-150, K-162, E-393, E-394) that are find conserved in every ortholog of HerA.
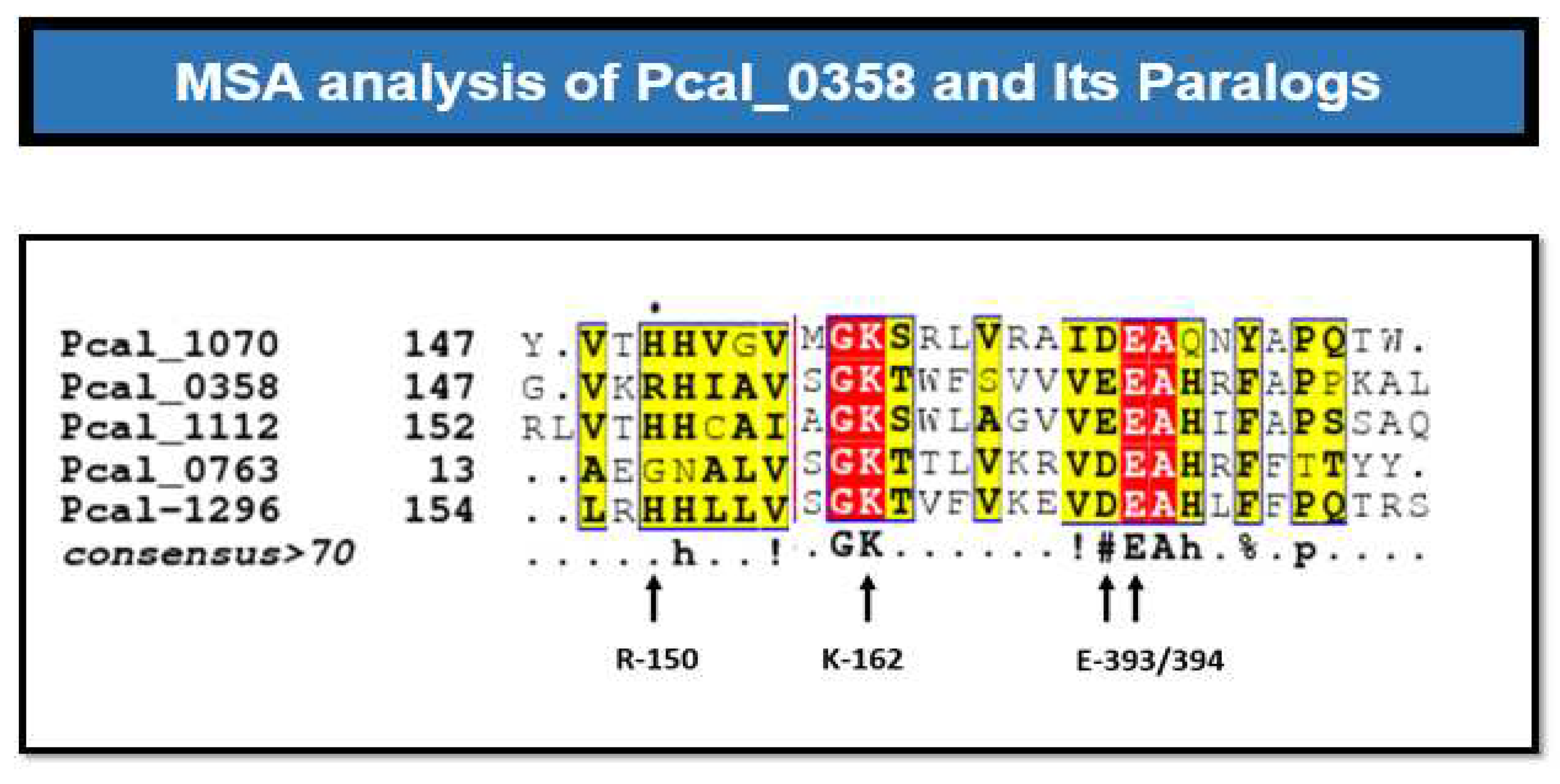
Figure 5.
MSA analysis of P. calidifontis paralogs. This multiple sequence alignment is performed between all the paralogs of the Pca_HerA (0358). The residues mention down below are not conserved in all and also don’t have chemistry conservation.
Figure 5.
MSA analysis of P. calidifontis paralogs. This multiple sequence alignment is performed between all the paralogs of the Pca_HerA (0358). The residues mention down below are not conserved in all and also don’t have chemistry conservation.
Gene neighborhood of helicase protein reveals that HerA (Helicase) works in a complex with other proteins including mre-11, Rad-50 and NurA. Mre-11 and Rad-50 works as a break recognition machinery in the DNA. These two genes will activate the helicase protein to start unwinding the DNA strands and NurA will proceed its nuclease activity.
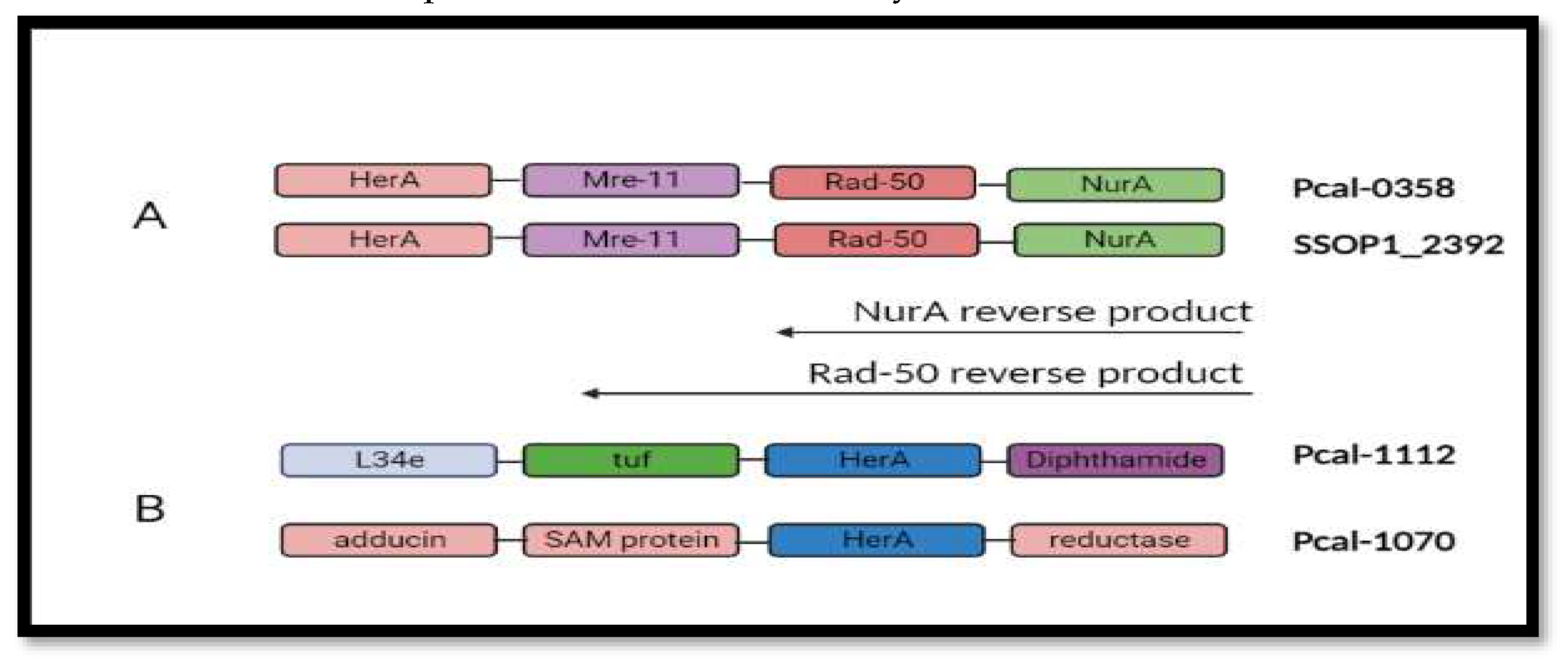
Figure 6.
Gene neighborhood analysis. (A) showing all the DNA repair machinery genes included on single Operonic region between SSOP1_2392 and Pca_0358. (B) Neighborhood analysis in all the paralogs of Pca-HerA.
Figure 6.
Gene neighborhood analysis. (A) showing all the DNA repair machinery genes included on single Operonic region between SSOP1_2392 and Pca_0358. (B) Neighborhood analysis in all the paralogs of Pca-HerA.
Conserved Residue Analysis in 20 HerA Archaeal Species
Analyzing the 25 HerA archaeal orthologues with Pca_HerA, having all the genes on their operonic regions. we have seen that all the orthologues including our concerned archaeal specie have similar conserved residues.
Table 1. Conserved amino acid residues analysis of HerA in 20 archaeal HerA orthologs. This table shows that all 20 amino acids (left) and all the domain-wise conserved residues (right) were shown to be crucially sealed in all HerA orthologues, which are only found in archaea in this examination of 25 archaeal species' conserved residues (see figures and tables).
Structural Analysis Results
Tools that are used primarily for the structural analysis include Chimera, Discovery studio and Phyre2. Predicted structure of the Pca_HerA is made through software named Phyre2 and it shows the 96% similarity with the HerA structure of SSO (PBD ID: 4D2I).
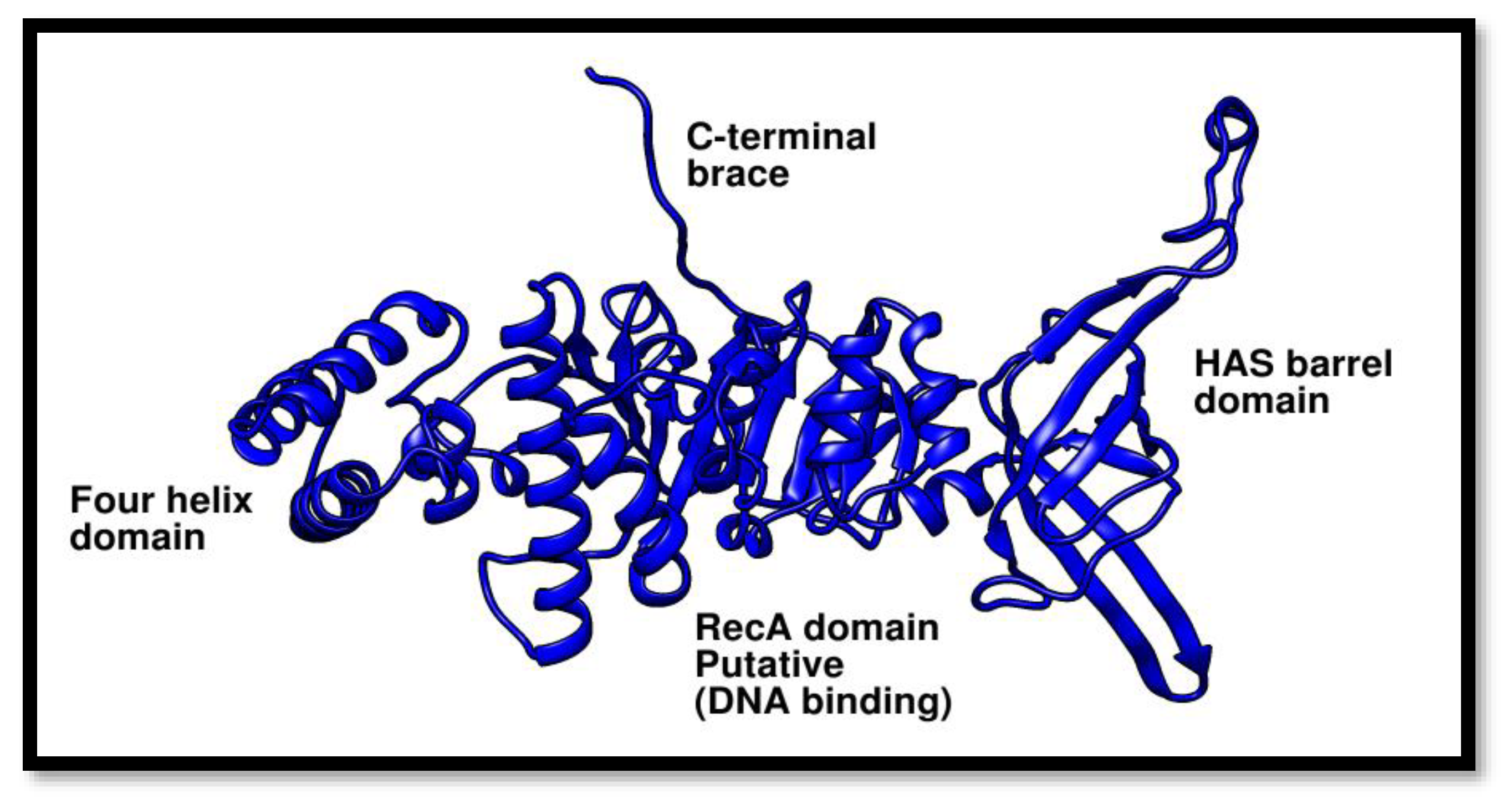
Figure 7.
Phyre2 was used to estimate the structure of the Pca-HerA. This structure demonstrates that the P. calidifontis helicase is made up of the following specific domains: The RecA/DNA binding domain, the four helix bundle domain, and the HAS-barrel domain (N-terminal HerA-ATP Synthase).
Figure 7.
Phyre2 was used to estimate the structure of the Pca-HerA. This structure demonstrates that the P. calidifontis helicase is made up of the following specific domains: The RecA/DNA binding domain, the four helix bundle domain, and the HAS-barrel domain (N-terminal HerA-ATP Synthase).
HerA-ATP Synthase Domain Analysis
The HAS barrel domain, also known as N-terminal HerA-ATP Synthase, is made up of beta barrel sheaths and an alpha-helix. It can be seen that the hydrophobic surface of the barrel domain's alpha helix is accessible for the engagement of the Nur-A nuclease. This barrel domain is composed of 100 amino acids ranging from Lys-2 to Ala-114.
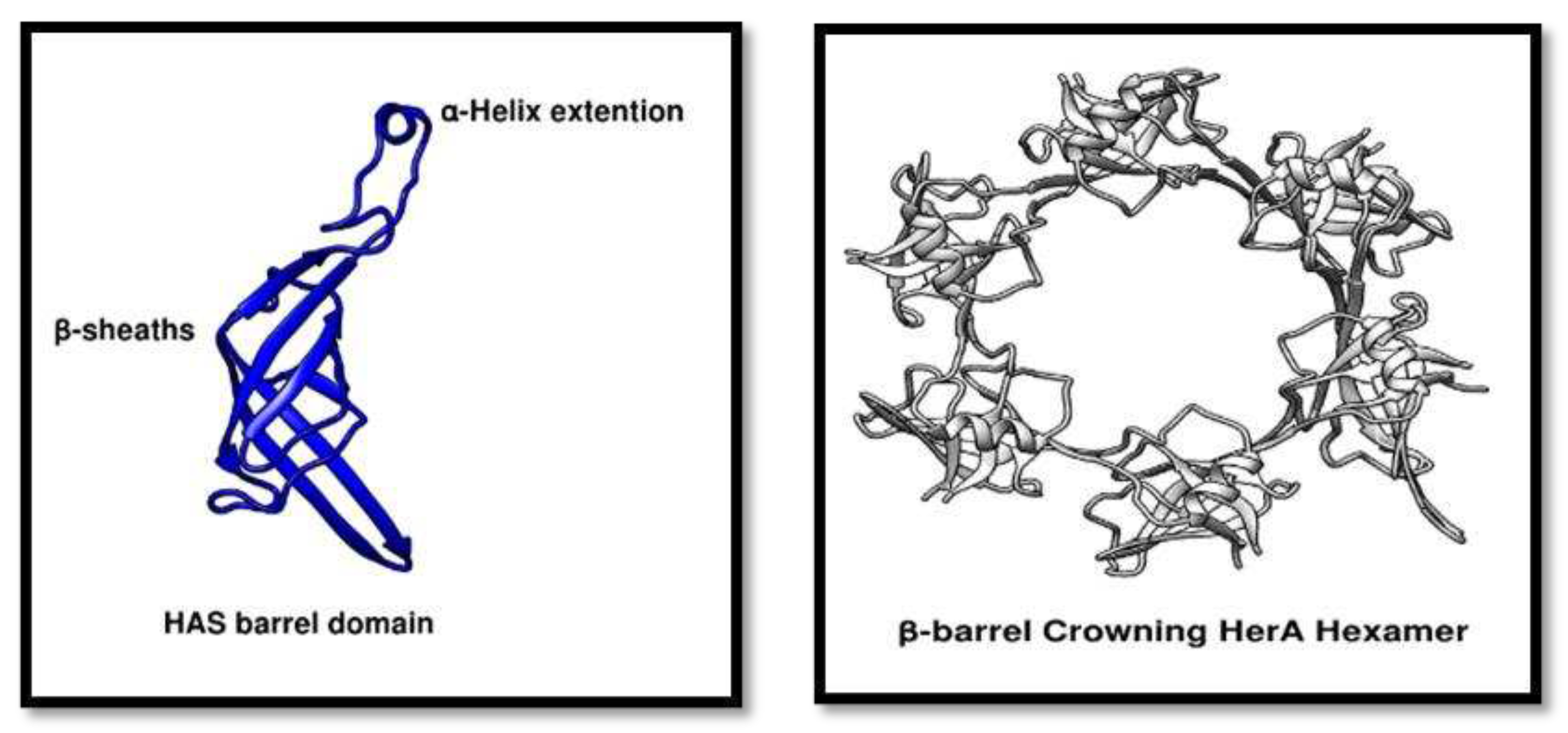
Figure 8.
HAS domain analysis. (A) showing a structural representation of HAS barrel domain containing beta-sheaths and an alpha-helix extension provides the N-terminus point for the attachment of NurA dimer. (B) Cartoon representation of the crowning HerA hexamer. All the pictures were generated through Chimera.
Figure 8.
HAS domain analysis. (A) showing a structural representation of HAS barrel domain containing beta-sheaths and an alpha-helix extension provides the N-terminus point for the attachment of NurA dimer. (B) Cartoon representation of the crowning HerA hexamer. All the pictures were generated through Chimera.
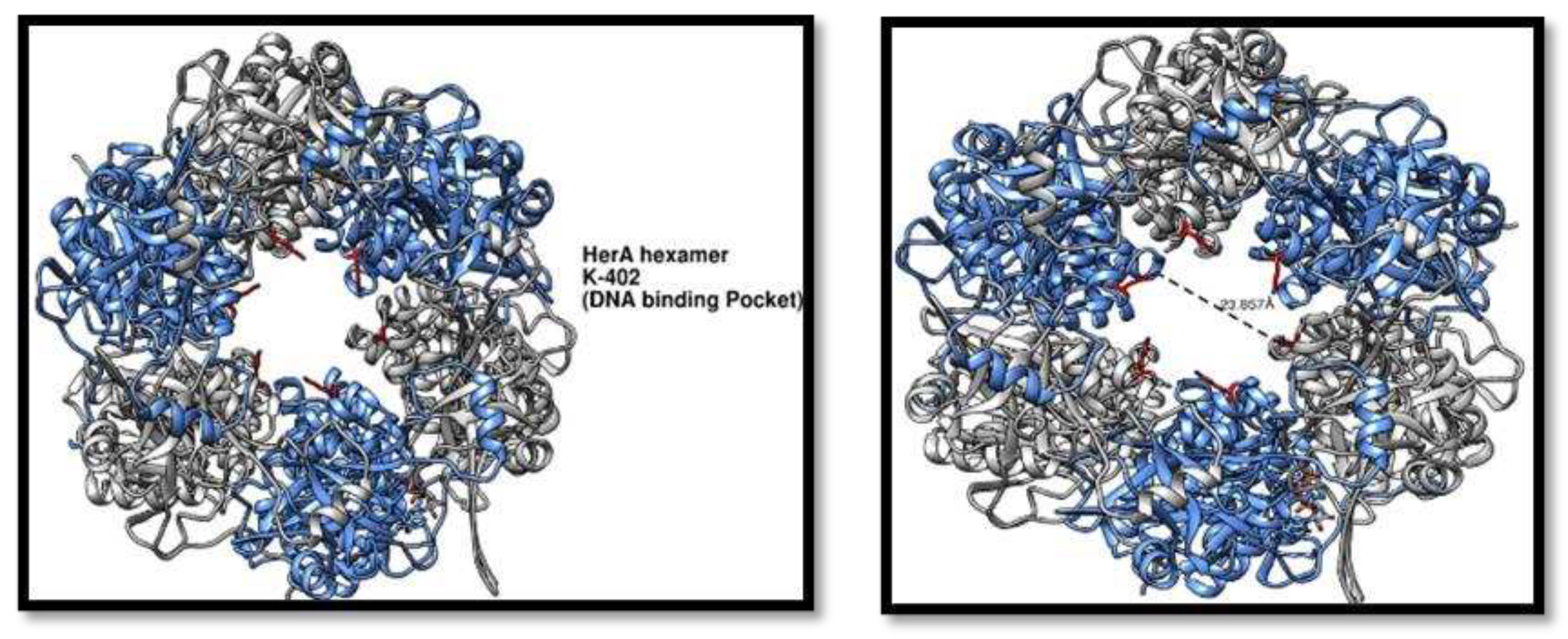
As helicase is involved in the DNA unzipping process its RecA like domain provides a beneficial role in the binding of the DNA. As the helicase make a hexameric structure the residue K-402 of all the hexameric subunits make a ring like pocket that will ultimately provide the entering and binding point for the DNA.
Figure 9.
HerA higher order structure analysis. (A) HerA hexamer showing the DNA binding pocket having the residue K-402 (red) responsible for making the entry point for the DNA. (B) Distance has been calculated for the pocket is 23.857Å.
Figure 9.
HerA higher order structure analysis. (A) HerA hexamer showing the DNA binding pocket having the residue K-402 (red) responsible for making the entry point for the DNA. (B) Distance has been calculated for the pocket is 23.857Å.
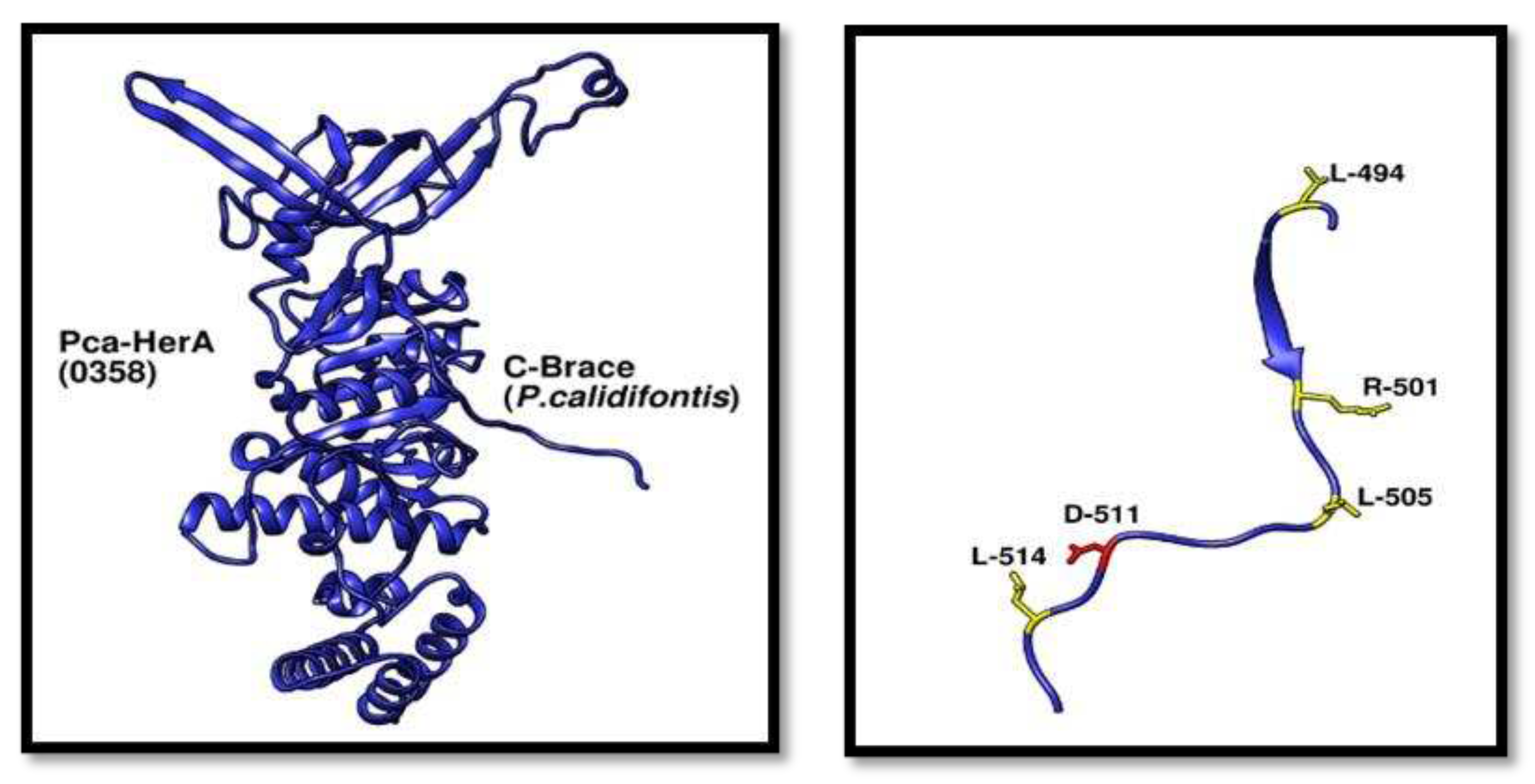
C-Terminal Brace Analysis
This brace plays an investigative role in the study of HerA and NurA complex. As it mainly interacts with the transacting residue mainly from which R-150 is used to study the activity of HerA NurA complex. Other amino acids like D-511 plays a role in the DNA translocation as it assists the ATP hydrolysis that is required for the DNA translocation assay.
Figure 10.
Adjacent C-terminal brace structural view. (A) Showing the monomeric view of Pca_HerA and its C-terminal Brace. (B) C-terminal brace residues analysis and the most important residue D-511 (red) involved in forming salt bridge and if mutated caused severe DNA impairment destruction assay..
Figure 10.
Adjacent C-terminal brace structural view. (A) Showing the monomeric view of Pca_HerA and its C-terminal Brace. (B) C-terminal brace residues analysis and the most important residue D-511 (red) involved in forming salt bridge and if mutated caused severe DNA impairment destruction assay..
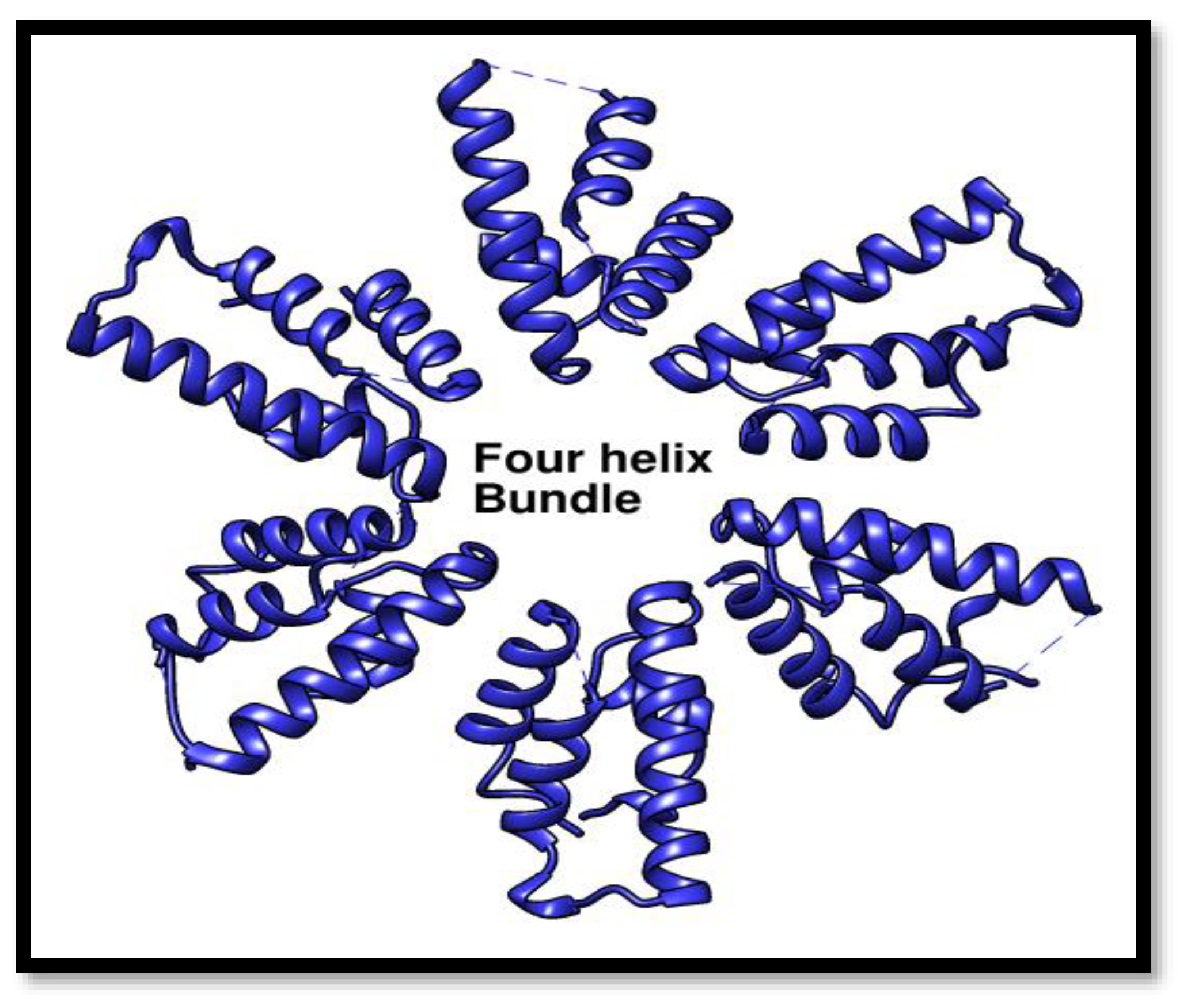
Four Helix Domain
These four helix bundle of helicase is considered to be more crucial as it arranges in a hexameric form and made an entrance point for the B-form DNA to get in and contact with the HerA so that DNA repair process starts.
Figure 11.
Four Helix domain the entry point for DNA in repair. An animated depiction of the Pca_HerA four helix bundle domain arranged to create the putative pore entrance, giving a positively charged surface suited for directing DNA into the channel.
Figure 11.
Four Helix domain the entry point for DNA in repair. An animated depiction of the Pca_HerA four helix bundle domain arranged to create the putative pore entrance, giving a positively charged surface suited for directing DNA into the channel.
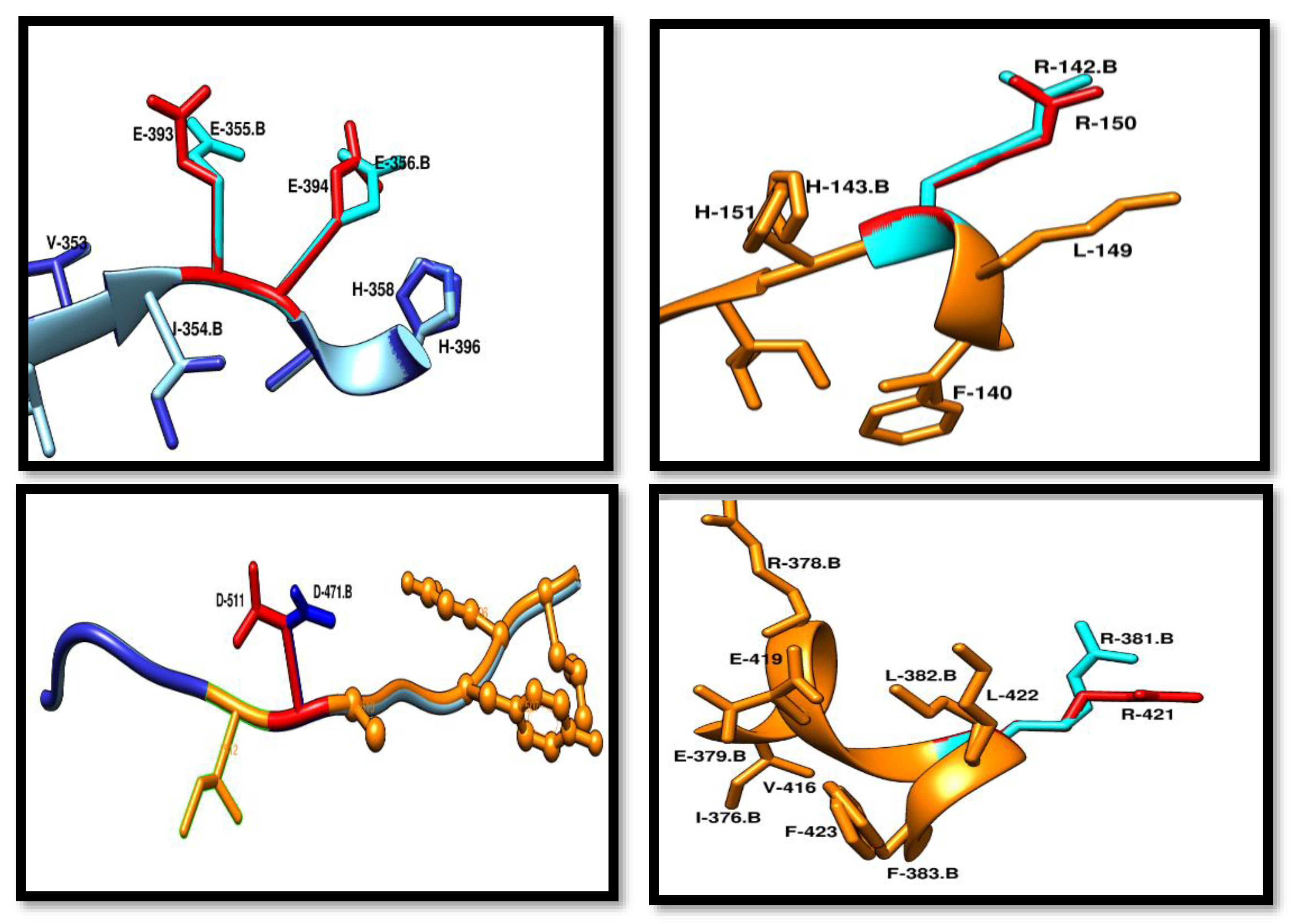
Conserved Residue Analysis of Pca-HerA
By doing the structure based alignment of SSO-2093 and Pca_HerA (0358) results are showing that both the structures have same residue chemistry at specific positions. Residue that are conserved in Pca_HerA (0358) and all the HerA orthologs includes R-150, K-162, K-402, R-421, D-511. Every residue is involved in performing specific type of functions Their mutational analysis shows that a slightest change in the residues will make a major effect on the mechanism of HerA and NurA activity.
Figure 12.
Conserved residues of Pca-HerA in red and the SSO in blue. These residues are walker B motif. R-150 is conserved that is responsible for the interaction with γ-phosphate of the ATP-mimic. D-511 of Pca_HerA in red is responsible for the making of the salt bridge and work as a cleft among the two promoters of helicase. R-421 involved in making a contact through salt bridge with E-394 which is crucial for the water activation before the catalysis interference formation. .
Figure 12.
Conserved residues of Pca-HerA in red and the SSO in blue. These residues are walker B motif. R-150 is conserved that is responsible for the interaction with γ-phosphate of the ATP-mimic. D-511 of Pca_HerA in red is responsible for the making of the salt bridge and work as a cleft among the two promoters of helicase. R-421 involved in making a contact through salt bridge with E-394 which is crucial for the water activation before the catalysis interference formation. .
Structural Orientation of HerA and NurA Complex:
HerA and NurA interacts with each other in the complex for in which the role of Helicase is to Unzip the DNA double strand and NurA will chop of the bases. Their orientation is held when NurA comes in contact with the HAS barrel domain of HerA. It provides the attachment point for both of the proteins.
Figure 13.
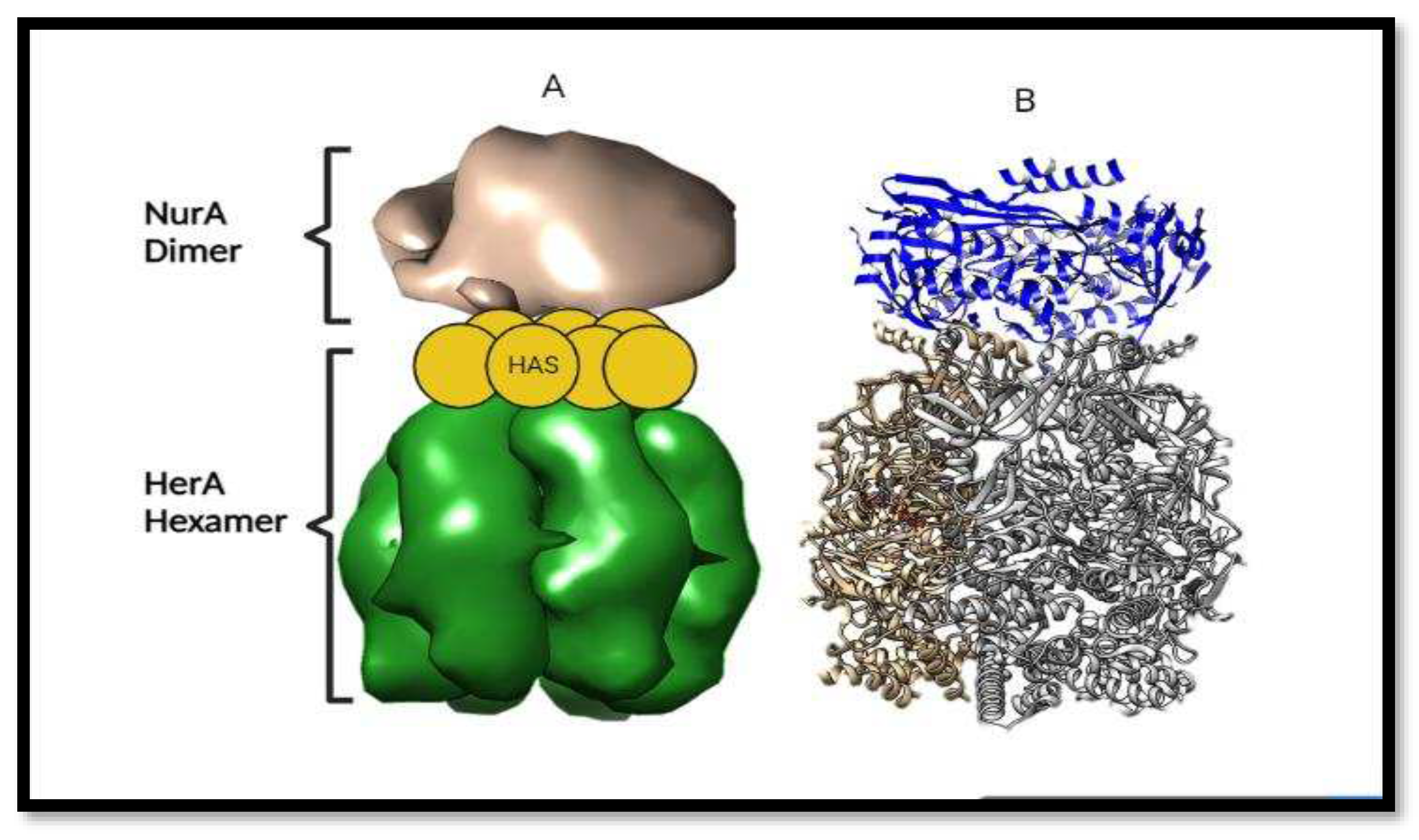
Orientation of HerA and NurA complex. (A) and (B) picture are showing the Adjacent and axial views respectively of the molecular envelope generated by Chimera. In picture (A) HAS barrel domain provides the attachment point for the NurA attachment for the DNA break repair.
Figure 13.
Orientation of HerA and NurA complex. (A) and (B) picture are showing the Adjacent and axial views respectively of the molecular envelope generated by Chimera. In picture (A) HAS barrel domain provides the attachment point for the NurA attachment for the DNA break repair.
Mechanism of HerA and NurA activity in DNA end Re-Sectioning
The process of the DNA repair starts when HerA hexamer allows the entrance of the double stranded DNA, the unzipping of the nucleic acid starts and the NurA dimer present right behind the helicase start chopping off the DNA bases.
Figure 14.
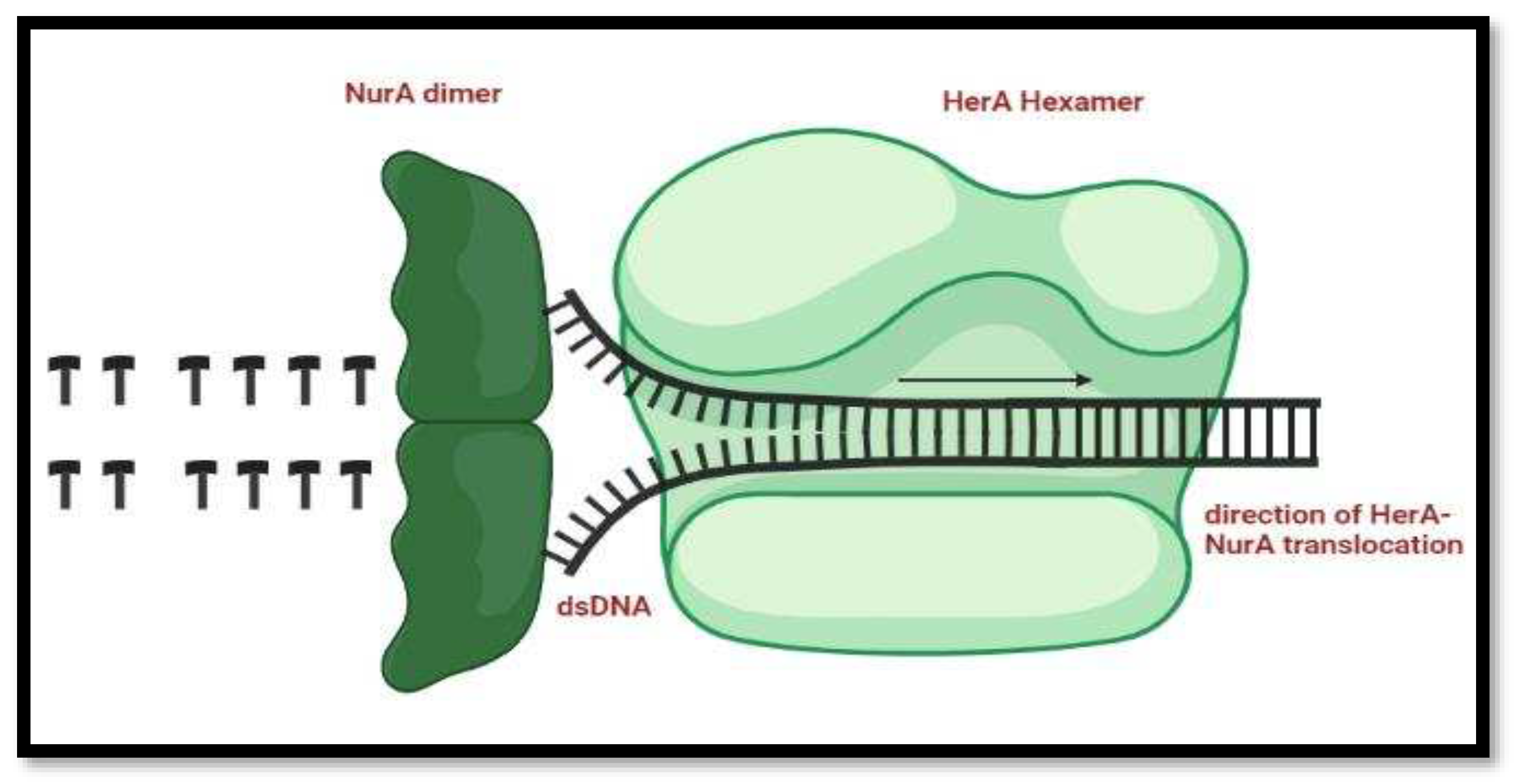
Mechanism of HerA and NurA in the DNA replication process. Double-stranded DNA is routed through HerA and unraveled by NurA's ploughshare activity. In the model, the NurA dimer engages both DNA strands, resulting in total substrate breakdown.
Figure 14.
Mechanism of HerA and NurA in the DNA replication process. Double-stranded DNA is routed through HerA and unraveled by NurA's ploughshare activity. In the model, the NurA dimer engages both DNA strands, resulting in total substrate breakdown.
Summary of Structural Analysis
All of the work that has been done to determine the structure, domains, chemistry of the residues and their representation is shown in the big picture. Here all the results are presented in one frame.
Figure 15.
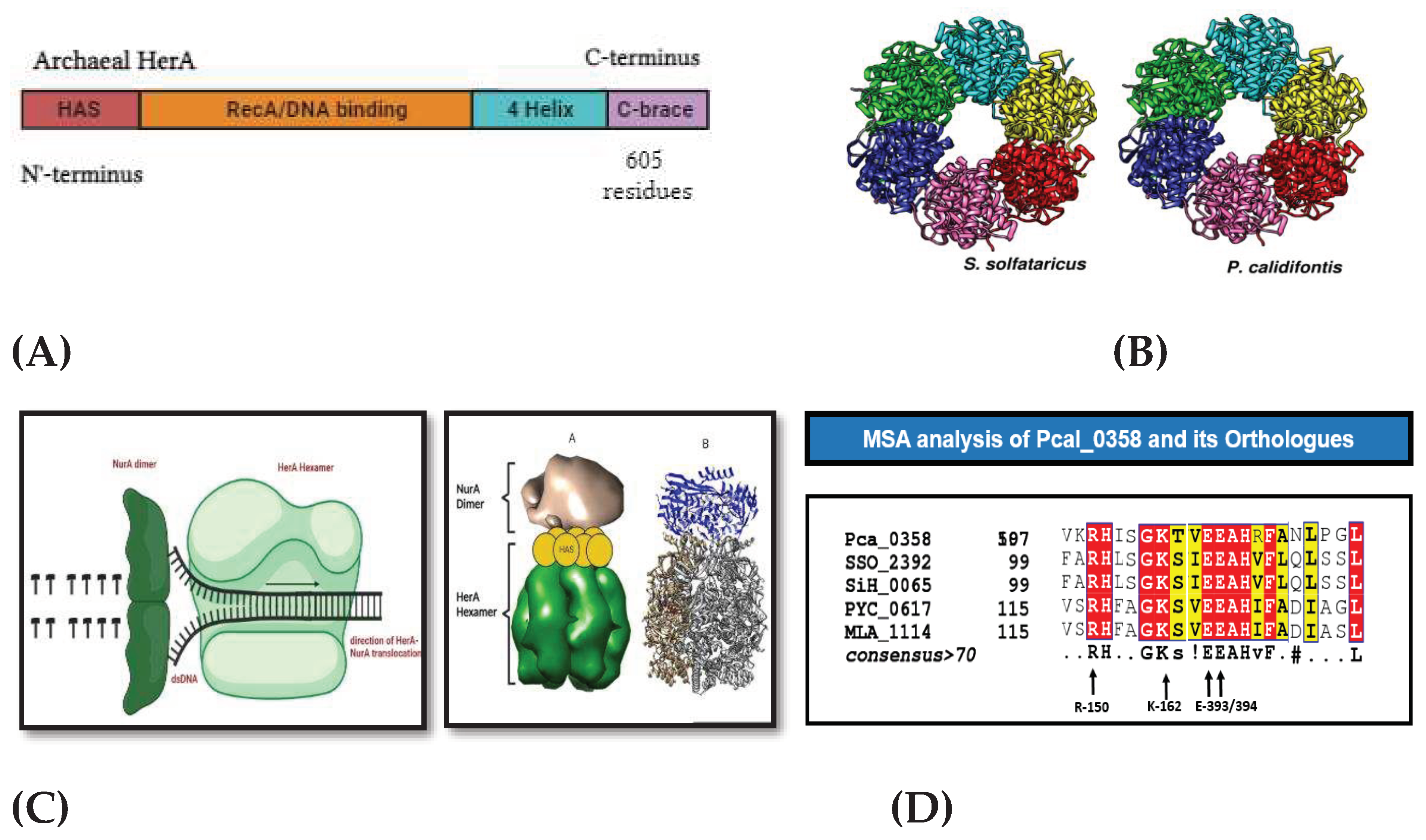
Summary of structural analysis. Picture (A) is showing the domain architecture of the Pca-HerA helicase in Pyrobaculum calidifontis. (B) two structures were shown HerA hexameric structure in S. solfataricus (SSO) and P. calidifontis (Pca) in rainbow colors. These structures were seen resembled in both the helicases of archaea suggesting that these are the orthologs of each other. (C) Mechanism of HerA and NurA in Double stranded DNA repair was shown with their structural orientation in ribbon form. The NurA dimer in the (blue) color and the HerA hexamer in (Grey) color is seen, showing that the HAS domain provides the attachment point for the NurA dimer and starts the process. (D) Sequence based alignment showing the conserved residues, important for proceeding positioning at the nucleotide-binding site to interact with the ATP terminal phosphate during hydrolysis.
Figure 15.
Summary of structural analysis. Picture (A) is showing the domain architecture of the Pca-HerA helicase in Pyrobaculum calidifontis. (B) two structures were shown HerA hexameric structure in S. solfataricus (SSO) and P. calidifontis (Pca) in rainbow colors. These structures were seen resembled in both the helicases of archaea suggesting that these are the orthologs of each other. (C) Mechanism of HerA and NurA in Double stranded DNA repair was shown with their structural orientation in ribbon form. The NurA dimer in the (blue) color and the HerA hexamer in (Grey) color is seen, showing that the HAS domain provides the attachment point for the NurA dimer and starts the process. (D) Sequence based alignment showing the conserved residues, important for proceeding positioning at the nucleotide-binding site to interact with the ATP terminal phosphate during hydrolysis.
Discussion
The implications for understanding DNA processing processes, and a comparison with comparable research in the field.Understanding Pca_HerA functional linkages and probable involvement within cellular pathways is aided by looking at the area around the gene. Our findings are consistent with the neighborhood of genes that code for DNA repair mechanisms that has previously been identified. The article titled as “Structure of the hexameric HerA ATPase reveals a mechanism of translocation – coupled DNA – and processing in archaea” defines a details structural analysis of S. solfataricus (Rzechorzek et al., 2014). The structural data acquired indicated a hexameric configuration of the HerA ATPase subunits, emphasizing its oligomer character. This hexameric shape is essential for its activity as an ATP-dependent DNA helicase, allowing it to move along and unwind DNA strands. The work clarified the ATP-binding sites and essential functional motifs in the HerA ATPase, shedding light on its enzymatic activity. As per the structural analysis in defined 4.4.3 reveals the hexameric structure of P. calidifontis showing the DNA binding pocket. The residue that is involved in the binding of the DNA is K402 while in S. solfataricus is K363. This analysis confirms that the structure we predicted is responsible for making the entry point for the DNA.
The suggested approach of translocation-coupled DNA processing by HerA is one of the study's primary results. HerA ATPase activity propels its travel along DNA, causing double-stranded DNA segments to unravel. This translocation-coupled DNA unwinding mechanism guarantees that DNA strands are efficiently separated during replication and repair processes, allowing for precise copying and mending of genetic material (Constantinesco, 2004). In comparison to the structural analysis predicted structure of the Pca_HerA, was similar and is clearly observed in figure 4.4.8. The structural study of the Bipolar DNA Helicase Gene indicates that it has the motifs and domains required for ATP-dependent DNA unwinding. Its unusual collection of functional components suggests that it might have a dual role in both DNA replication and repair pathways. However, experimental confirmation via biochemical assays and functional investigations is required to corroborate these hypotheses and completely understand HerA functional role in genomic stability and integrity. Understanding the inner workings of this bipolar DNA helicase gene helps us comprehend cellular DNA manipulation processes more broadly.
We detected P. calidifontis HerA helicase and NurA nucleases, which are a component of a thermophilic archaea conserved operon that also includes the mre11 and rad50 genes. We have shown that the assembly of the HerA-NurA complex is essential for the processing of DNA ends and that the enzymatic activity of HerA and NurA is highly reciprocally dependent. The physical stability of the HerA and NurA proteins is consistent with previous studies on proteins from S. solfataricus. The preservation of gene order and contextual structure across several species provides additional evidence for the Pca_HerA gene neighborhood's evolutionary importance. Since neighboring conserved genes from several species are present, Pca_HerA is significant within the conserved pathways (Ahdash et al., 2017). This study demonstrates the lateral and axial views of the complicated HerA-NurA molecular envelope. The structural study also reveals that the HerA-NurA helicase-nuclease complex, Mre11, and Rad50 work together to coordinate the repair of double-stranded DNA breaks. The activation and assembly of HerA-NurA, however, are poorly understood.
An important component of comprehending Pca_HerA functional responsibilities is its structural characterization. Our results are in line with the scientific literature that has already been published, indicating that Pca_HerA does iNdeed possess all of the domains indicated in the literature. The four helix binding domain, the RecA DNA binding domain, the HAS barrel domain, and others are also included (Rzechorzek et al., 2014).
Comparing all the orthologs of Pca_HerA in different archaeal species we gathered the results that every archaeal helicase has same characteristics as ours. This structural analysis provides the support for the functional relevance of these domains, reaffirming their significance in Pca_HerA biological processes.
Chemical conservation in orthologous proteins reveals important information on the maintenance of functional properties throughout evolutionary lineages. Our study of the chemical conservation of Pca_HerA and its orthologs reveals the conservation of key residues and motifs required for its function. This chemical consistency suggests Pca_HerA functional importance in diverse species. It also demonstrates that critical residues involved in ATP binding, DNA binding, and crystal structure maintenance are globally conserved. And are critical for DNA interaction and processing.
Previous research has found that the human minichromosome management (MCM) complex, which is made up of MCM subunits 4, 6, and 7, has an inherent ATP-dependent DNA helicase activity. The archaeon Methanobacterium thermoautotrophicum DH (mth) genome only has a single open reading frame that codes for an MCM protein, as opposed to the many MCM genes (at least six) present in eukaryotes (Kelman et al.,2023).
The verification of the information in the literature on the structural domains, gene neighborhood, and chemical conservation of Pca_HerA has significant ramifications. Our comprehension of Pca_HerA biological processes and its contributions to cellular activities including DNA replication and repair are strengthened by this validation. The understanding learned from this investigation offers strong support for Pca_HerA operations.
There is currently no mechanistic evidence to support how the Mre11-Rad50 complex interacts with the HerA-NurA helicase- nuclease. The essential interactions between auxiliary helicases and nucleases, such as HerA-NurA in archaea or Sgs1-Exo1/Dna2 in eukaryotes, and the core Mre11-Rad50 components during DNA metabolism will be clarified by more structural and functional investigations (Constantinesco, 2004).
The adoption of model organisms that are biochemically manageable, like P. calidifontis, should be helpful in the further understanding of these vital Mre11-Rad50-depeNdent DNA repair mechanisms along with the helicase nuclease activity.
To recapitulate this study which reveals that helicase HerA possess all the concerned attributes that are seen in the other archaeal HerA orthologues. All the aspects including neighboring genes, multiple sequence alignment, structure conservation, conserved domain analysis and chemistry conservation of the residues are aligning to each other suggesting that Pca_HerA (0358) can be considered as the helicase and will proceed further for functional assay. HerA functions in DNA replication by unwinding the double-stranded DNA during the replication process. DNA replication is a key biological process that involves the replication of DNA to generate two identical daughter strands. HerA helicase activity is required for the parental DNA strands to be unwound, allowing the DNA polymerase to manufacture new complementary strands. HerA encodes a helicase enzyme that has a variety of roles in archaeal DNA metabolism. Its actions in DNA replication, repair, homologous recombination, and genome stability work together to guarantee the genome's accuracy. The research of HerA activities not only gives insights into archaeal molecular processes, but also helps to our greater knowledge of DNA-related mechanisms in other domains of life.
References
- Ahdash Z, Lau A M, Byrne R T, Lammens K, Stüetzer A, Urlaub H, Booth P J, Reading E, Hopfner K-P & Politis A. Mechanistic insight into the assembly of the HerA–NurA helicase–nuclease DNA end resection complex. Nucleic Acids Research 2017, 45, 12025–12038. [CrossRef]
- Alhmoud J F, Woolley J F, Al Moustafa A-E & Malki M I. DNA Damage/Repair Management in Cancers. Cancers 2020, 12, 1050. [CrossRef]
- Amo T, Paje M L F, Inagaki A, Ezaki S, Atomi H & Imanaka T. Pyrobaculum calidifontis sp. Nov., a novel hyperthermophilic archaeon that grows in atmospheric air. Archaea 2002, 1, 113–121. [CrossRef]
- Anjum R S, Bray S M, Blackwood J K, Kilkenny M L, Coelho M A, Foster B M, Li S Howard, J A Pellegrini, L Albers S-V, Deery M J & Robinson N P. Involvement of a eukaryotic-like ubiquitin-related modifier in the proteasome pathway of the archaeon Sulfolobus acidocaldarius. Nature Communications 2015, 6, 8163. [CrossRef]
- Beltran L C, Cvirkaite-Krupovic V, Miller J, Wang F, Kreutzberger M A B, Patkowski J B, Costa T R D, Schouten S, Levental I, Conticello V P, Egelman E H & Krupovic M. Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery. Nature Communications 2023, 14, 666. [CrossRef]
- Bisson-Filho A W, Zheng J & Garner E. Archaeal imaging: Leading the hunt for new discoveries. Molecular Biology of the Cell 2018, 29, 1675–1681. [CrossRef]
- Bochman M, L. Roles of DNA helicases in the maintenance of genome integrity. Molecular & Cellular Oncology 2014, 1, e963429. [Google Scholar] [CrossRef]
- Ceccaldi R, Rondinelli B & D’Andrea A D. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends in Cell Biology 2016, 26, 52–64. [CrossRef]
- Cheng K, Chen X, Xu G, Wang L, Xu H, Yang S, Zhao Y & Hua Y. Biochemical and Functional Characterization of the NurA-HerA Complex from Deinococcus radiodurans. Journal of Bacteriology 2015, 197, 2048–2061. [CrossRef]
- Constantinesco, F. A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Research 2004, 32, 1439–1447. [Google Scholar] [CrossRef] [PubMed]
- De Falco M & De Felice M. Take a Break to Repair: A Dip in the World of Double-Strand Break Repair Mechanisms Pointing the Gaze on Archaea. International Journal of Molecular Sciences 2021, 22, 13296. [CrossRef]
- De Falco M, Massa F, Rossi M & De Felice M. The Sulfolobus solfataricus RecQ-like DNA helicase Hel112 inhibits the NurA/HerA complex exonuclease activity. Extremophiles 2018, 22, 581–589. [CrossRef]
- De Falco M, Porritiello A, Rota F, Scognamiglio V, Antonacci A, del Monaco G & De Felice M. The Finely Coordinated Action of SSB and NurA/HerA Complex Strictly Regulates the DNA End Resection Process in Saccharolobus solfataricus. International Journal of Molecular Sciences 2022, 23, 2582. [CrossRef]
- Fujii Y, Inoue M, Fukui K, Kuramitsu S & Masui R. Resistance to UV Irradiation Caused by Inactivation of nurA and herA Genes in Thermus thermophilus. Journal of Bacteriology 2018, 200. [CrossRef]
- Greci M D & Bell S D. Archaeal DNA Replication. Annual Review of Microbiology 2020, 74, 65–80. [CrossRef]
- Haber J, E. DNA Repair: The Search for Homology. BioEssays 2018, 40, 1700229. [Google Scholar] [CrossRef]
- Hopkins B B & Paull T T. The P. furiosus Mre11/Rad50 Complex Promotes 5′ Strand Resection at a DNA Double-Strand Break. Cell 2008, 135, 250–260. [CrossRef]
- Kelman Z, Lee J-K & Hurwitz J. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum ⌬H contains DNA helicase activity. 2023. [CrossRef]
- Lemmens L, Maklad H R, Bervoets I & Peeters E. Transcription Regulators in Archaea: Homologies and Differences with Bacterial Regulators. Journal of Molecular Biology 2019, 431, 4132–4146. [CrossRef]
- Marshall C J & Santangelo T J. Archaeal DNA Repair Mechanisms. Biomolecules 2020, 10, 1472. [CrossRef]
- Martinez-Gutierrez C A & Aylward F O. Phylogenetic Signal, Congruence, and Uncertainty across Bacteria and Archaea. Molecular Biology and Evolution 2021, 38, 5514–5527. [CrossRef]
- Rolfsmeier M L, Laughery M F & Haseltine C A. Repair of DNA Double-Strand Breaks following UV Damage in Three Sulfolobus solfataricus Strains. Journal of Bacteriology 2010, 192, 4954–4962. [CrossRef]
- Rzechorzek N J, Blackwood J K, Bray S M, Maman J D, Pellegrini L & Robinson N P. Structure of the hexameric HerA ATPase reveals a mechanism of translocation-coupled DNA-end processing in archaea. Nature Communications 2014, 5, 5506. [CrossRef]
- Sinha A K, Possoz C, Durand A, Desfontaines J-M, Barre F-X, Leach D R F & Michel B. Broken replication forks trigger heritable DNA breaks in the terminus of a circular chromosome. PLOS Genetics 2018, 14, e1007256. [CrossRef]
- Sung S, Li F Park, Y B Kim, J S, Kim A, Song O, Kim J, Che J, Lee S E & Cho Y. DNA end recognition by the Mre11 nuclease dimer: Insights into resection and repair of damaged DNA. The EMBO Journal 2014, 33, 2422–2435. [CrossRef]
- Ui A, Chiba N & Yasui A. Relationship among DNA double-strand break (DSB), DSB repair, and transcription prevents genome instability and cancer. Cancer Science 2020, 111, 1443–1451. [CrossRef]
- White M F & Allers T. DNA repair in the archaea—An emerging picture. FEMS Microbiology Reviews 2018. [CrossRef]
- Wilkinson O J, Carrasco C, Aicart-Ramos C, Moreno-Herrero F & Dillingham M S. Bulk and single-molecule analysis of a novel DNA2-like helicase-nuclease reveals a single-stranded DNA looping motor. Biochemistry. 2019. [CrossRef]
- Zabolotnaya E, Mela I, HeNderson R M & Robinson N P. Turning the Mre11/Rad50 DNA repair complex on its head: Lessons from SMC protein hinges, dynamic coiled-coil movements and DNA loop-extrusion? Biochemical Society Transactions 2020, 48, 2359–2376. [CrossRef]
- Zhao F, Kim W, Kloeber J A & Lou Z. DNA end resection and its role in DNA replication and DSB repair choice in mammalian cells. Experimental & Molecular Medicine 2020, 52, 1705–1714. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).