Submitted:
30 January 2024
Posted:
31 January 2024
You are already at the latest version
Abstract
Keywords:
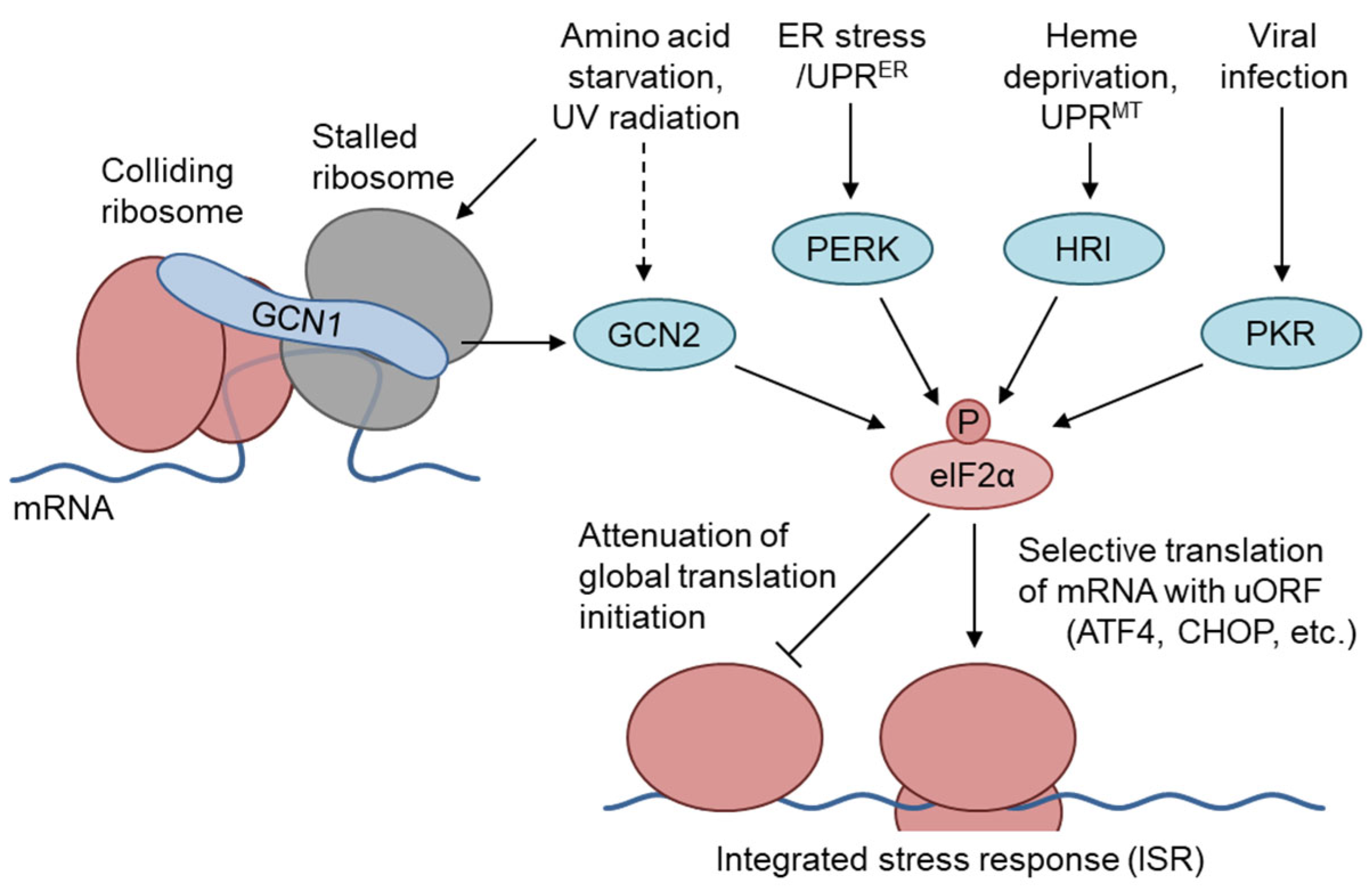
1. The GCN1–GCN2 Pathway Regulates the Amino Acid Starvation Branch of the Integrated Stress Response
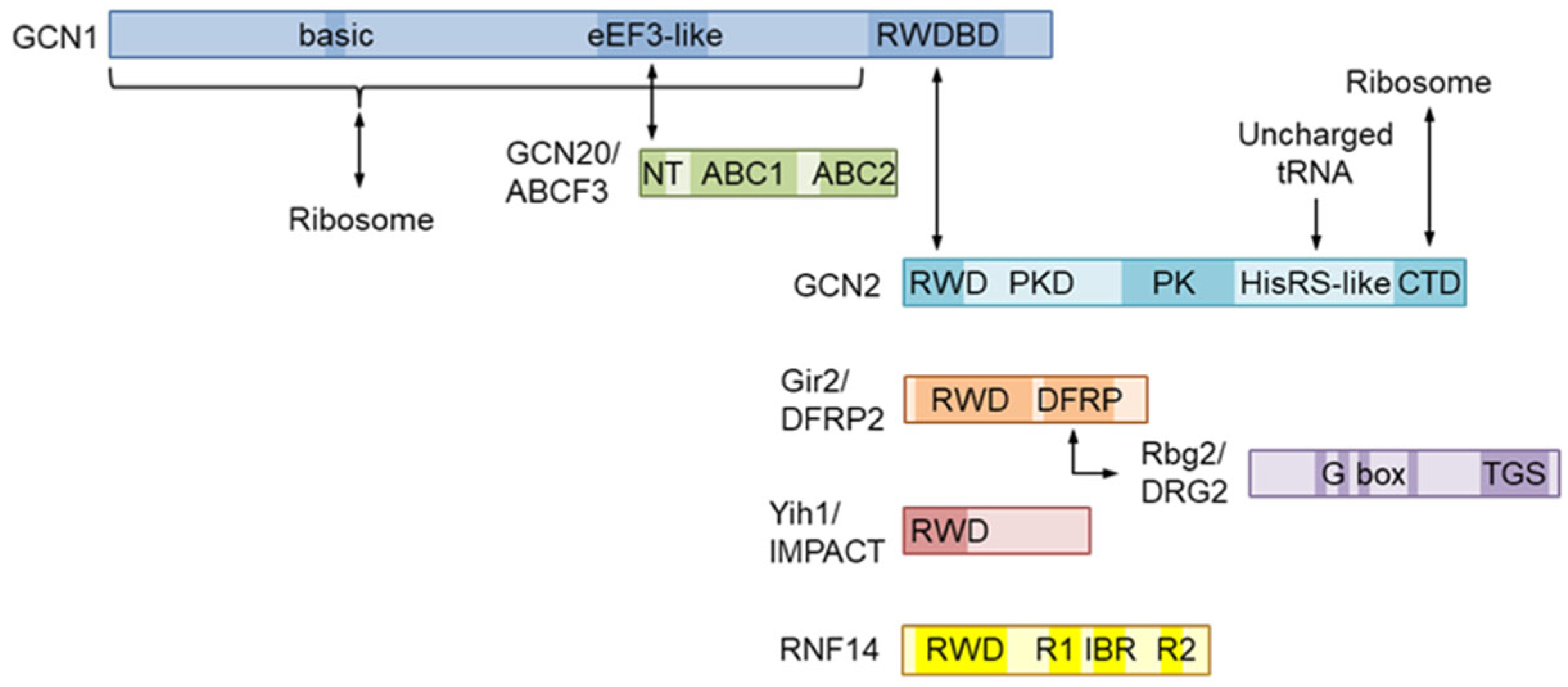
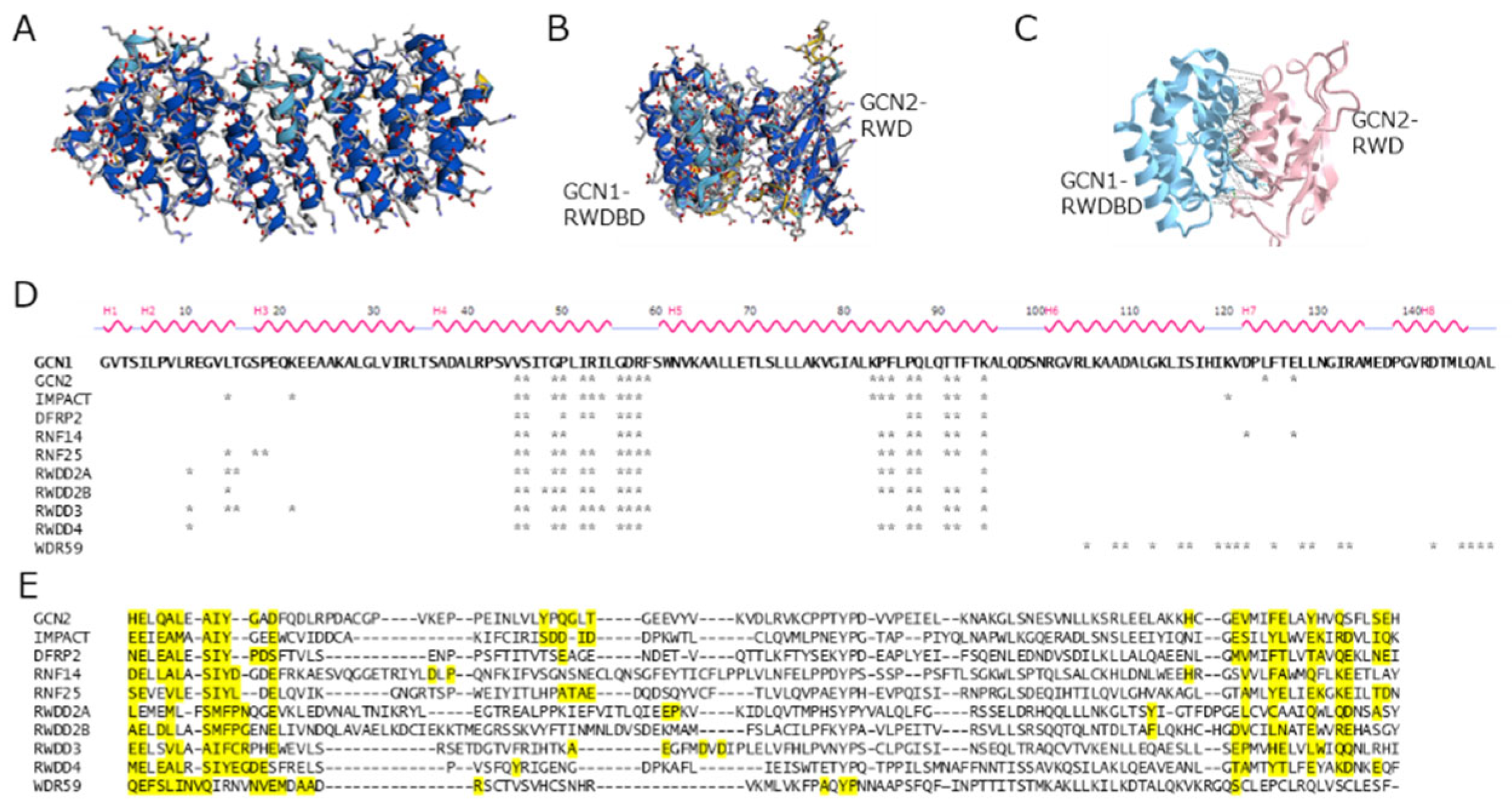
2. Molecular Structure of GCN1 and Its Role in ISR Activation
3. Emerging Role of GCN1 through GCN2-Independent Pathways
3.1. Identification of an Alternative Amino Acid Deprivation Response Distinct from the Canonical GCN2 Pathway in Mammalian Cells
3.2. Role of GCN1-Dependent and GCN2-Independent Mechanisms in Mammalian Development
3.3. GCN2-Independent Pathway in Species Other Than Mammals
4. Function and Structure of the GCN1-Interacting RWD-Domain-Containing Proteins
4.1. GCN2/EIF2AK4
4.2. IMPACT
4.3. DFRP2/RWDD1
4.4. RNF14 and RNF25
4.5. RESUME/RWDD3
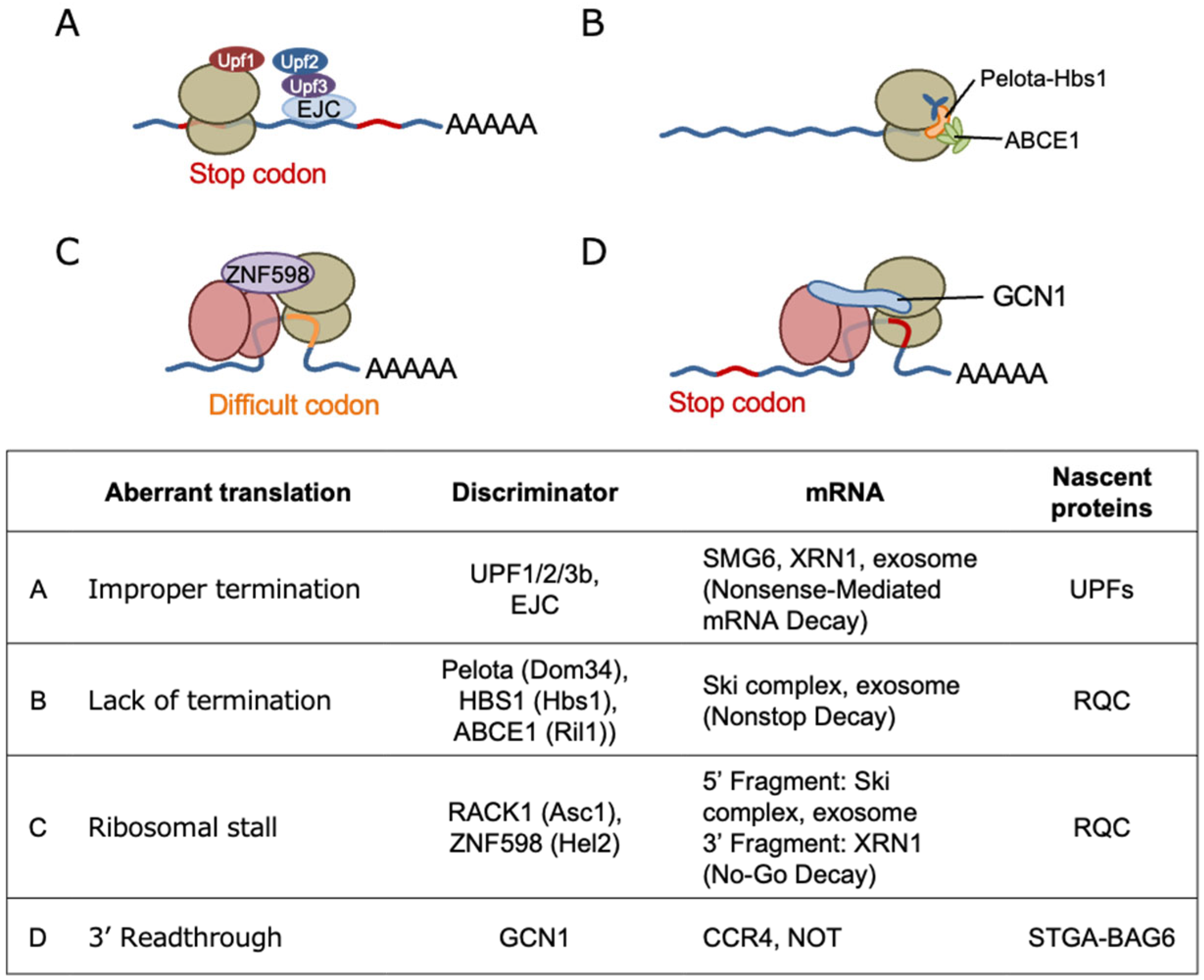
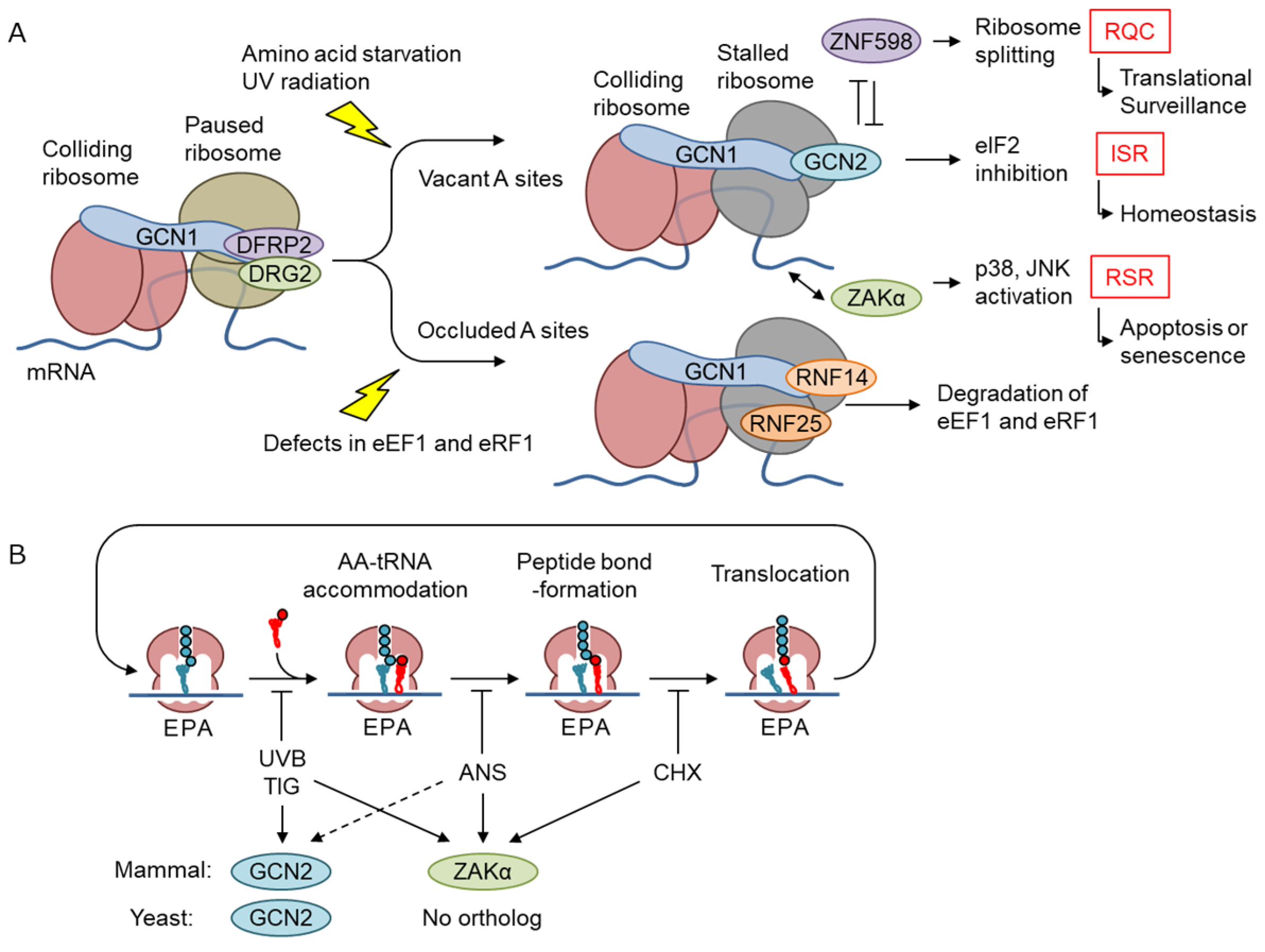
5. Emerging Role of GCN1 in Co-Translational Protein Quality Control
5.1. Quality Control Mechanisms for Stalled Ribosomes
5.2. GCN2 Branch of the ISR
5.3. RSR
6. Function of GCN1 in Basal-State Translational Regulation
7. Role of GCN1-Mediated Responses in Energy Homeostasis
7.1. Role of the GCN1–GCN2 Pathway in the Regulation of Insulin Secretion and Sensitivity
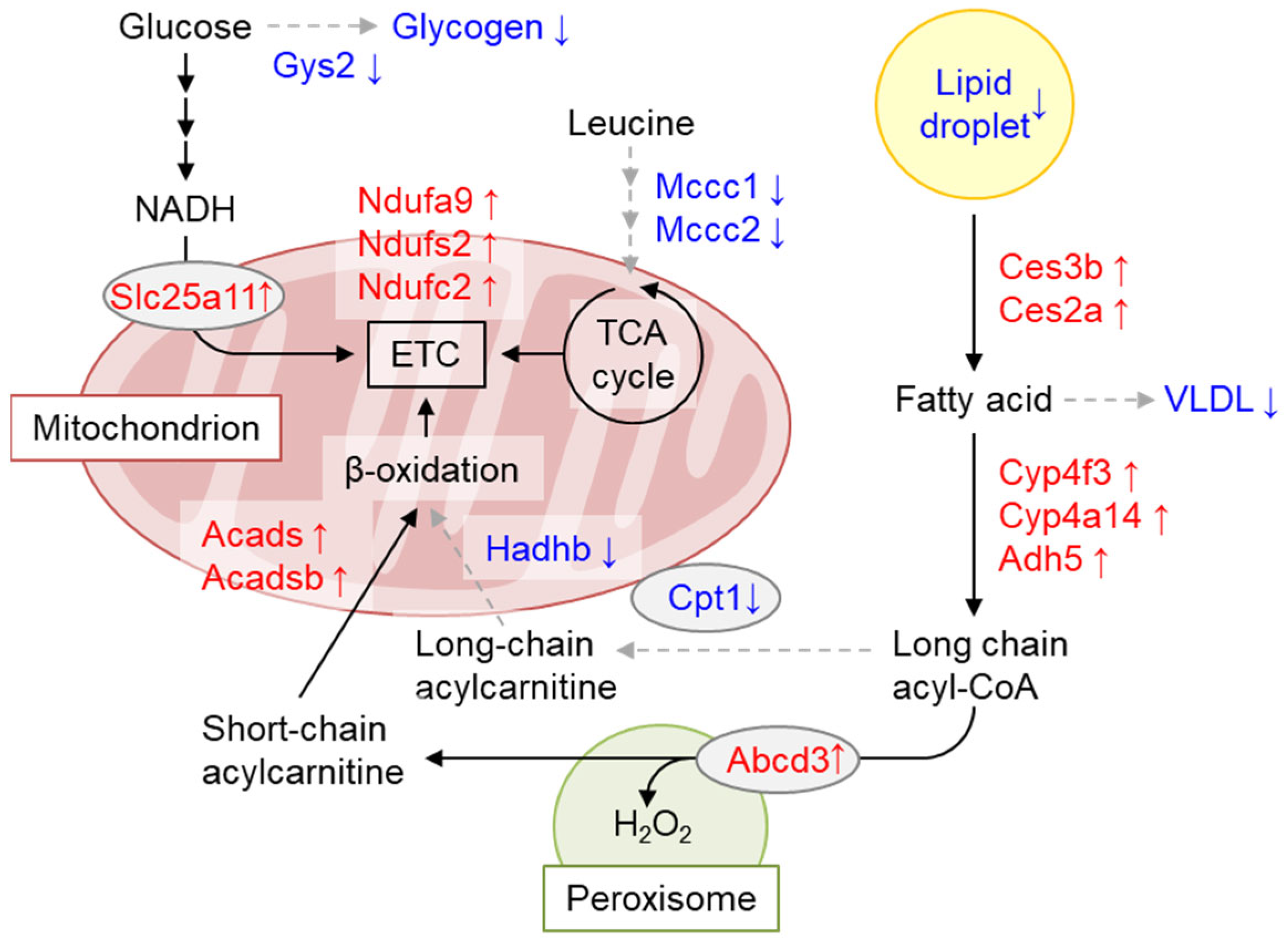
7.2. GCN1 Regulates Energy Storage and Usage
8. Potential Role of GCN1 in Aging and Disease
8.1. Potential Role of GCN1 in Aging
8.2. Potential Role of GCN1 in Neurodegenerative Diseases
9. Future Perspectives
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Castilho BA, Shanmugam R, Silva RC, Ramesh R, Himme BM and Sattlegger E: Keeping the eIF2 alpha kinase Gcn2 in check. Biochim Biophys Acta 1843: 1948-1968, 2014. [CrossRef]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A and Gorman AM: The integrated stress response. EMBO Rep 17: 1374-1395, 2016.
- Taniuchi S, Miyake M, Tsugawa K, Oyadomari M and Oyadomari S: Integrated stress response of vertebrates is regulated by four eIF2alpha kinases. Sci Rep 6: 32886, 2016.
- Baird TD and Wek RC: Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv Nutr 3: 307-321, 2012.
- B'Chir W, Maurin AC, Carraro V, et al.: The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res 41: 7683-7699, 2013.
- Iurlaro R and Munoz-Pinedo C: Cell death induced by endoplasmic reticulum stress. FEBS J 283: 2640-2652, 2016.
- Pitale PM, Gorbatyuk O and Gorbatyuk M: Neurodegeneration: Keeping ATF4 on a Tight Leash. Front Cell Neurosci 11: 410, 2017.
- Lee HC, Fu CY, Lin CY, et al.: Poly(U)-specific endoribonuclease ENDOU promotes translation of human CHOP mRNA by releasing uORF element-mediated inhibition. EMBO J 40: e104123, 2021. [CrossRef]
- Anda S, Zach R and Grallert B: Activation of Gcn2 in response to different stresses. PLoS One 12: e0182143, 2017.
- Yamazaki H, Kasai S, Mimura J, et al.: Ribosome binding protein GCN1 regulates the cell cycle and cell proliferation and is essential for the embryonic development of mice. PLoS Genet 16: e1008693, 2020. [CrossRef]
- Ye J, Kumanova M, Hart LS, et al.: The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J 29: 2082-2096, 2010. [CrossRef]
- Deng J, Harding HP, Raught B, et al.: Activation of GCN2 in UV-irradiated cells inhibits translation. Curr Biol 12: 1279-1286, 2002. [CrossRef]
- Robert F, Williams C, Yan Y, et al.: Blocking UV-induced eIF2alpha phosphorylation with small molecule inhibitors of GCN2. Chem Biol Drug Des 74: 57-67, 2009. [CrossRef]
- Andrade MA, Petosa C, O'Donoghue SI, Muller CW and Bork P: Comparison of ARM and HEAT protein repeats. J Mol Biol 309: 1-18, 2001.
- Jumper J, Evans R, Pritzel A, et al.: Highly accurate protein structure prediction with AlphaFold. Nature 596: 583-589, 2021. [CrossRef]
- Varadi M, Anyango S, Deshpande M, et al.: AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50: D439-D444, 2022. [CrossRef]
- Pochopien AA, Beckert B, Kasvandik S, et al.: Structure of Gcn1 bound to stalled and colliding 80S ribosomes. Proc Natl Acad Sci U S A 118, 2021. [CrossRef]
- Nameki N, Yoneyama M, Koshiba S, et al.: Solution structure of the RWD domain of the mouse GCN2 protein. Protein Sci 13: 2089-2100, 2004. [CrossRef]
- Rakesh R, Krishnan R, Sattlegger E and Srinivasan N: Recognition of a structural domain (RWDBD) in Gcn1 proteins that interacts with the RWD domain containing proteins. Biol Direct 12: 12, 2017.
- Sattlegger E and Hinnebusch AG: Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2alpha kinase GCN2 during amino acid starvation. J Biol Chem 280: 16514-16521, 2005.
- Vazquez de Aldana CR, Marton MJ and Hinnebusch AG: GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2 alpha kinase GCN2 in amino acid-starved cells. EMBO J 14: 3184-3199, 1995. [CrossRef]
- Marton MJ, Vazquez de Aldana CR, Qiu H, Chakraburtty K and Hinnebusch AG: Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2alpha kinase GCN2. Mol Cell Biol 17: 4474-4489, 1997.
- Hinnebusch AG: Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407-450, 2005.
- Tyzack JK, Wang X, Belsham GJ and Proud CG: ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J Biol Chem 275: 34131-34139, 2000.
- Hirose T and Horvitz HR: The translational regulators GCN-1 and ABCF-3 act together to promote apoptosis in C. elegans. PLoS Genet 10: e1004512, 2014.
- Murina V, Kasari M, Takada H, et al.: ABCF ATPases Involved in Protein Synthesis, Ribosome Assembly and Antibiotic Resistance: Structural and Functional Diversification across the Tree of Life. J Mol Biol 431: 3568-3590, 2019. [CrossRef]
- Brown A, Fernandez IS, Gordiyenko Y and Ramakrishnan V: Ribosome-dependent activation of stringent control. Nature 534: 277-280, 2016.
- Kim Y, Sundrud MS, Zhou C, et al.: Aminoacyl-tRNA synthetase inhibition activates a pathway that branches from the canonical amino acid response in mammalian cells. Proc Natl Acad Sci U S A 117: 8900-8911, 2020.
- Sundrud MS, Koralov SB, Feuerer M, et al.: Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science 324: 1334-1338, 2009.
- De Vito A, Lazzaro M, Palmisano I, et al.: Amino acid deprivation triggers a novel GCN2-independent response leading to the transcriptional reactivation of non-native DNA sequences. PLoS One 13: e0200783, 2018. [CrossRef]
- Palmisano I, Della Chiara G, D'Ambrosio RL, et al.: Amino acid starvation induces reactivation of silenced transgenes and latent HIV-1 provirus via down-regulation of histone deacetylase 4 (HDAC4). Proc Natl Acad Sci U S A 109: E2284-2293, 2012. [CrossRef]
- Izquierdo Y, Kulasekaran S, Benito P, et al.: Arabidopsis nonresponding to oxylipins locus NOXY7 encodes a yeast GCN1 homolog that mediates noncanonical translation regulation and stress adaptation. Plant Cell Environ 41: 1438-1452, 2018. [CrossRef]
- Doerks T, Copley RR, Schultz J, Ponting CP and Bork P: Systematic identification of novel protein domain families associated with nuclear functions. Genome Res 12: 47-56, 2002.
- Pereira CM, Sattlegger E, Jiang HY, et al.: IMPACT, a protein preferentially expressed in the mouse brain, binds GCN1 and inhibits GCN2 activation. J Biol Chem 280: 28316-28323, 2005.
- Waller T, Lee SJ and Sattlegger E: Evidence that Yih1 resides in a complex with ribosomes. FEBS J 279: 1761-1776, 2012.
- Wout PK, Sattlegger E, Sullivan SM and Maddock JR: Saccharomyces cerevisiae Rbg1 protein and its binding partner Gir2 interact on Polyribosomes with Gcn1. Eukaryot Cell 8: 1061-1071, 2009.
- Ishikawa K, Ito K, Inoue J and Semba K: Cell growth control by stable Rbg2/Gir2 complex formation under amino acid starvation. Genes Cells 18: 859-872, 2013.
- Oltion K, Carelli JD, Yang T, et al.: An E3 ligase network engages GCN1 to promote the degradation of translation factors on stalled ribosomes. Cell 186: 346-362 e317, 2023. [CrossRef]
- Wu CC, Peterson A, Zinshteyn B, Regot S and Green R: Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 182: 404-416 e414, 2020.
- Lageix S, Zhang J, Rothenburg S and Hinnebusch AG: Interaction between the tRNA-binding and C-terminal domains of Yeast Gcn2 regulates kinase activity in vivo. PLoS Genet 11: e1004991, 2015.
- Wek SA, Zhu S and Wek RC: The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol 15: 4497-4506, 1995.
- Zaborske JM, Narasimhan J, Jiang L, et al.: Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J Biol Chem 284: 25254-25267, 2009. [CrossRef]
- Costa-Mattioli M, Gobert D, Harding H, et al.: Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature 436: 1166-1173, 2005. [CrossRef]
- Maurin AC, Jousse C, Averous J, et al.: The GCN2 kinase biases feeding behavior to maintain amino acid homeostasis in omnivores. Cell Metab 1: 273-277, 2005. [CrossRef]
- Eyries M, Montani D, Girerd B, et al.: EIF2AK4 mutations cause pulmonary veno-occlusive disease, a recessive form of pulmonary hypertension. Nat Genet 46: 65-69, 2014.
- Roffe M, Hajj GN, Azevedo HF, Alves VS and Castilho BA: IMPACT is a developmentally regulated protein in neurons that opposes the eukaryotic initiation factor 2alpha kinase GCN2 in the modulation of neurite outgrowth. J Biol Chem 288: 10860-10869, 2013.
- Kaplan A, Morquette B, Kroner A, et al.: Small-Molecule Stabilization of 14-3-3 Protein-Protein Interactions Stimulates Axon Regeneration. Neuron 93: 1082-1093 e1085, 2017. [CrossRef]
- Sattlegger E, Swanson MJ, Ashcraft EA, et al.: YIH1 is an actin-binding protein that inhibits protein kinase GCN2 and impairs general amino acid control when overexpressed. J Biol Chem 279: 29952-29962, 2004. [CrossRef]
- Cambiaghi TD, Pereira CM, Shanmugam R, et al.: Evolutionarily conserved IMPACT impairs various stress responses that require GCN1 for activating the eIF2 kinase GCN2. Biochem Biophys Res Commun 443: 592-597, 2014. [CrossRef]
- Ferraz RC, Camara H, De-Souza EA, et al.: IMPACT is a GCN2 inhibitor that limits lifespan in Caenorhabditis elegans. BMC Biol 14: 87, 2016. [CrossRef]
- Daugeron MC, Prouteau M, Lacroute F and Seraphin B: The highly conserved eukaryotic DRG factors are required for efficient translation in a manner redundant with the putative RNA helicase Slh1. Nucleic Acids Res 39: 2221-2233, 2011.
- Ishikawa K, Azuma S, Ikawa S, Semba K and Inoue J: Identification of DRG family regulatory proteins (DFRPs): specific regulation of DRG1 and DRG2. Genes Cells 10: 139-150, 2005.
- Westrip CAE, Zhuang Q, Hall C, Eaton CD and Coleman ML: Developmentally regulated GTPases: structure, function and roles in disease. Cell Mol Life Sci 78: 7219-7235, 2021.
- Müller MBD, Kasturi P, Jayaraj GG and Hartl FU: Mechanisms of readthrough mitigation reveal principles of GCN1-mediated translational quality control. Cell 186: 3227-3244.e3220, 2023.
- Xu K, Shimelis H, Linn DE, et al.: Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell 15: 270-282, 2009. [CrossRef]
- Alontaga AY, Ambaye ND, Li YJ, et al.: RWD Domain as an E2 (Ubc9)-Interaction Module. J Biol Chem 290: 16550-16559, 2015. [CrossRef]
- Carbia-Nagashima A, Gerez J, Perez-Castro C, et al.: RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell 131: 309-323, 2007.
- Shan B, Gerez J, Haedo M, et al.: RSUME is implicated in HIF-1-induced VEGF-A production in pituitary tumour cells. Endocr Relat Cancer 19: 13-27, 2012. [CrossRef]
- Gerez J, Fuertes M, Tedesco L, et al.: In silico structural and functional characterization of the RSUME splice variants. PLoS One 8: e57795, 2013. [CrossRef]
- Druker J, Liberman AC, Antunica-Noguerol M, et al.: RSUME enhances glucocorticoid receptor SUMOylation and transcriptional activity. Mol Cell Biol 33: 2116-2127, 2013. [CrossRef]
- Kim KQ and Zaher HS: Canary in a coal mine: collided ribosomes as sensors of cellular conditions. Trends Biochem Sci 47: 82-97, 2022.
- De S and Muhlemann O: A comprehensive coverage insurance for cells: revealing links between ribosome collisions, stress responses and mRNA surveillance. RNA Biol 19: 609-621, 2022.
- Gavin AC, Aloy P, Grandi P, et al.: Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631-636, 2006. [CrossRef]
- Hermjakob H, Montecchi-Palazzi L, Lewington C, et al.: IntAct: an open source molecular interaction database. Nucleic Acids Res 32: D452-455, 2004.
- Gurzeler LA, Link M, Ibig Y, et al.: Drug-induced eRF1 degradation promotes readthrough and reveals a new branch of ribosome quality control. Cell Rep 42: 113056, 2023.
- Meydan S and Guydosh NR: A cellular handbook for collided ribosomes: surveillance pathways and collision types. Curr Genet 67: 19-26, 2021.
- Garzia A, Jafarnejad SM, Meyer C, et al.: The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat Commun 8: 16056, 2017. [CrossRef]
- Matsuo Y, Ikeuchi K, Saeki Y, et al.: Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat Commun 8: 159, 2017. [CrossRef]
- Juszkiewicz S, Speldewinde SH, Wan L, Svejstrup JQ and Hegde RS: The ASC-1 Complex Disassembles Collided Ribosomes. Mol Cell 79: 603-614 e608, 2020.
- Filbeck S, Cerullo F, Pfeffer S and Joazeiro CAP: Ribosome-associated quality-control mechanisms from bacteria to humans. Mol Cell 82: 1451-1466, 2022.
- Hickey KL, Dickson K, Cogan JZ, et al.: GIGYF2 and 4EHP Inhibit Translation Initiation of Defective Messenger RNAs to Assist Ribosome-Associated Quality Control. Mol Cell 79: 950-962 e956, 2020. [CrossRef]
- Sinha NK, Ordureau A, Best K, et al.: EDF1 coordinates cellular responses to ribosome collisions. Elife 9, 2020.
- Juszkiewicz S, Slodkowicz G, Lin Z, Freire-Pritchett P, Peak-Chew SY and Hegde RS: Ribosome collisions trigger cis-acting feedback inhibition of translation initiation. Elife 9, 2020.
- Yan LL and Zaher HS: Ribosome quality control antagonizes the activation of the integrated stress response on colliding ribosomes. Mol Cell 81: 614-628.e614, 2021.
- Fedry J, Silva J, Vanevic M, et al.: Visualization of translation reorganization upon persistent collision stress in mammalian cells. bioRxiv, 2023. [CrossRef]
- Masson GR: Towards a model of GCN2 activation. Biochem Soc Trans 47: 1481-1488, 2019.
- Ishimura R, Nagy G, Dotu I, Chuang JH and Ackerman SL: Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. Elife 5, 2016.
- Darnell AM, Subramaniam AR and O'Shea EK: Translational Control through Differential Ribosome Pausing during Amino Acid Limitation in Mammalian Cells. Mol Cell 71: 229-243 e211, 2018.
- Stoneley M, Harvey RF, Mulroney TE, et al.: Unresolved stalled ribosome complexes restrict cell-cycle progression after genotoxic stress. Mol Cell 82: 1557-1572 e1557, 2022. [CrossRef]
- Wu CC, Zinshteyn B, Wehner KA and Green R: High-Resolution Ribosome Profiling Defines Discrete Ribosome Elongation States and Translational Regulation during Cellular Stress. Mol Cell 73: 959-970.e955, 2019.
- Lee SJ, Swanson MJ and Sattlegger E: Gcn1 contacts the small ribosomal protein Rps10, which is required for full activation of the protein kinase Gcn2. Biochem J 466: 547-559, 2015.
- Snieckute G, Genzor AV, Vind AC, et al.: Ribosome stalling is a signal for metabolic regulation by the ribotoxic stress response. Cell Metab 34: 2036-2046.e2038, 2022. [CrossRef]
- Li J, Gao L, Chen J, et al.: Mitochondrial ROS-mediated ribosome stalling and GCN2 activation are partially involved in 1-nitropyrene-induced steroidogenic inhibition in testes. Environ Int 167: 107393, 2022. [CrossRef]
- Zhao S, Cordes J, Caban KM, et al.: RNF14-dependent atypical ubiquitylation promotes translation-coupled resolution of RNA-protein crosslinks. Mol Cell 83: 4290-4303.e4299, 2023. [CrossRef]
- Ryder L, Arendrup FS, Martínez JF, et al.: Nitric oxide-induced ribosome collision activates ribosomal surveillance mechanisms. Cell Death Dis 14: 467, 2023. [CrossRef]
- Inglis AJ, Masson GR, Shao S, et al.: Activation of GCN2 by the ribosomal P-stalk. Proc Natl Acad Sci U S A 116: 4946-4954, 2019. [CrossRef]
- Farooq Z, Kusuma F, Burke P, et al.: The amino acid sensor GCN2 suppresses terminal oligopyrimidine (TOP) mRNA translation via La-related protein 1 (LARP1). J Biol Chem 298: 102277, 2022. [CrossRef]
- Iordanov MS, Pribnow D, Magun JL, et al.: Ribotoxic stress response: activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the alpha-sarcin/ricin loop in the 28S rRNA. Mol Cell Biol 17: 3373-3381, 1997. [CrossRef]
- Vind AC, Genzor AV and Bekker-Jensen S: Ribosomal stress-surveillance: three pathways is a magic number. Nucleic Acids Res 48: 10648-10661, 2020.
- Wang X, Mader MM, Toth JE, et al.: Complete inhibition of anisomycin and UV radiation but not cytokine induced JNK and p38 activation by an aryl-substituted dihydropyrrolopyrazole quinoline and mixed lineage kinase 7 small interfering RNA. J Biol Chem 280: 19298-19305, 2005. [CrossRef]
- Jandhyala DM, Ahluwalia A, Obrig T and Thorpe CM: ZAK: a MAP3Kinase that transduces Shiga toxin- and ricin-induced proinflammatory cytokine expression. Cell Microbiol 10: 1468-1477, 2008.
- Jandhyala DM, Wong J, Mantis NJ, Magun BE, Leong JM and Thorpe CM: A Novel Zak Knockout Mouse with a Defective Ribotoxic Stress Response. Toxins (Basel) 8, 2016.
- Sauter KA, Magun EA, Iordanov MS and Magun BE: ZAK is required for doxorubicin, a novel ribotoxic stressor, to induce SAPK activation and apoptosis in HaCaT cells. Cancer Biol Ther 10: 258-266, 2010.
- Vind AC, Snieckute G, Blasius M, et al.: ZAKalpha Recognizes Stalled Ribosomes through Partially Redundant Sensor Domains. Mol Cell 78: 700-713 e707, 2020.
- Robinson KS, Toh GA, Rozario P, et al.: ZAKalpha-driven ribotoxic stress response activates the human NLRP1 inflammasome. Science 377: 328-335, 2022.
- Muela-Zarzuela I, Suarez-Rivero JM, Gallardo-Orihuela A, et al.: NLRP1 inflammasome modulates senescence and senescence-associated secretory phenotype. bioRxiv, 2023.
- Snieckute G, Ryder L, Vind AC, et al.: ROS-induced ribosome impairment underlies ZAKalpha-mediated metabolic decline in obesity and aging. Science 382: eadf3208, 2023.
- Doma MK and Parker R: Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561-564, 2006.
- Dimitrova LN, Kuroha K, Tatematsu T and Inada T: Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem 284: 10343-10352, 2009.
- Letzring DP, Wolf AS, Brule CE and Grayhack EJ: Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. RNA 19: 1208-1217, 2013.
- Simms CL, Hudson BH, Mosior JW, Rangwala AS and Zaher HS: An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep 9: 1256-1264, 2014.
- Brandman O and Hegde RS: Ribosome-associated protein quality control. Nat Struct Mol Biol 23: 7-15, 2016.
- Gamble CE, Brule CE, Dean KM, Fields S and Grayhack EJ: Adjacent Codons Act in Concert to Modulate Translation Efficiency in Yeast. Cell 166: 679-690, 2016.
- Joazeiro CAP: Ribosomal Stalling During Translation: Providing Substrates for Ribosome-Associated Protein Quality Control. Annu Rev Cell Dev Biol 33: 343-368, 2017.
- Collart MA and Weiss B: Ribosome pausing, a dangerous necessity for co-translational events. Nucleic Acids Res 48: 1043-1055, 2020.
- Lyu X, Yang Q, Zhao F and Liu Y: Codon usage and protein length-dependent feedback from translation elongation regulates translation initiation and elongation speed. Nucleic Acids Res 49: 9404-9423, 2021.
- Lim HR, Vo MT, Kim DJ, et al.: DRG2 Deficient Mice Exhibit Impaired Motor Behaviors with Reduced Striatal Dopamine Release. Int J Mol Sci 21, 2019. [CrossRef]
- Houston L, Platten EM, Connelly SM, Wang J and Grayhack EJ: Frameshifting at collided ribosomes is modulated by elongation factor eEF3 and by integrated stress response regulators Gcn1 and Gcn20. RNA 28: 320-339, 2022.
- Schuller AP, Wu CC, Dever TE, Buskirk AR and Green R: eIF5A Functions Globally in Translation Elongation and Termination. Mol Cell 66: 194-205.e195, 2017.
- Tesina P, Ebine S, Buschauer R, et al.: Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control. Mol Cell 83: 607-621.e604, 2023. [CrossRef]
- Zeng F, Li X, Pires-Alves M, Chen X, Hawk CW and Jin H: Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells. Cell Rep 37: 109877, 2021.
- Kriachkov V, Ormsby AR, Kusnadi EP, et al.: Arginine-rich C9ORF72 ALS proteins stall ribosomes in a manner distinct from a canonical ribosome-associated quality control substrate. J Biol Chem 299: 102774, 2023. [CrossRef]
- Seo J, Fortuno ES, 3rd, Suh JM, et al.: Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes 58: 2565-2573, 2009.
- Kanno A, Asahara SI, Furubayashi A, et al.: GCN2 regulates pancreatic β cell mass by sensing intracellular amino acid levels. JCI Insight 5, 2020. [CrossRef]
- Liu S, Yuan J, Yue W, et al.: GCN2 deficiency protects against high fat diet induced hepatic steatosis and insulin resistance in mice. Biochim Biophys Acta Mol Basis Dis 1864: 3257-3267, 2018. [CrossRef]
- Bjorkman SH, Marti A, Jena J, et al.: ATF4 Expression in Thermogenic Adipocytes is Required for Cold-Induced Thermogenesis in Mice via FGF21-Independent Mechanisms. bioRxiv, 2023. [CrossRef]
- Miyake M, Zhang J, Yasue A, et al.: Integrated stress response regulates GDF15 secretion from adipocytes, preferentially suppresses appetite for a high-fat diet and improves obesity. iScience 24: 103448, 2021. [CrossRef]
- Yuan J, Li F, Shen X, et al.: Genetic and Pharmacological Inhibition of GCN2 Ameliorates Hyperglycemia and Insulin Resistance in Type 2 Diabetic Mice. Antioxidants (Basel) 11, 2022. [CrossRef]
- Yuan J, Yu Z, Gao J, et al.: Inhibition of GCN2 alleviates hepatic steatosis and oxidative stress in obese mice: Involvement of NRF2 regulation. Redox Biol 49: 102224, 2022. [CrossRef]
- Dehwah MA, Wang M and Huang QY: CDKAL1 and type 2 diabetes: a global meta-analysis. Genet Mol Res 9: 1109-1120, 2010.
- Wei FY, Suzuki T, Watanabe S, et al.: Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J Clin Invest 121: 3598-3608, 2011.
- Takahashi N, Wei FY, Watanabe S, et al.: Reactive sulfur species regulate tRNA methylthiolation and contribute to insulin secretion. Nucleic Acids Res 45: 435-445, 2017. [CrossRef]
- Miyake K, Yang W, Hara K, et al.: Construction of a prediction model for type 2 diabetes mellitus in the Japanese population based on 11 genes with strong evidence of the association. J Hum Genet 54: 236-241, 2009. [CrossRef]
- Kanno A, Asahara SI, Masuda K, et al.: Compensatory hyperinsulinemia in high-fat diet-induced obese mice is associated with enhanced insulin translation in islets. Biochem Biophys Res Commun 458: 681-686, 2015. [CrossRef]
- Kitakaze K, Oyadomari M, Zhang J, et al.: ATF4-mediated transcriptional regulation protects against β-cell loss during endoplasmic reticulum stress in a mouse model. Mol Metab 54: 101338, 2021. [CrossRef]
- Szabat M, Page MM, Panzhinskiy E, et al.: Reduced Insulin Production Relieves Endoplasmic Reticulum Stress and Induces β Cell Proliferation. Cell Metab 23: 179-193, 2016. [CrossRef]
- Liu J, Kasai S, Tatara Y, et al.: Inducible Systemic Gcn1 Deletion in Mice Leads to Transient Body Weight Loss upon Tamoxifen Treatment Associated with Decrease of Fat and Liver Glycogen Storage. Int J Mol Sci 23, 2022. [CrossRef]
- López-Otín C, Blasco MA, Partridge L, Serrano M and Kroemer G: Hallmarks of aging: An expanding universe. Cell 186: 243-278, 2023.
- Stein KC, Morales-Polanco F, van der Lienden J, Rainbolt TK and Frydman J: Ageing exacerbates ribosome pausing to disrupt cotranslational proteostasis. Nature 601: 637-642, 2022.
- Chen Y, Sun T, Bi Z, Ni JQ, Pastor-Pareja JC and Javid B: Premature termination codon readthrough in Drosophila varies in a developmental and tissue-specific manner. Sci Rep 10: 8485, 2020.
- Zhu X, Zhang H and Mendell JT: Ribosome Recycling by ABCE1 Links Lysosomal Function and Iron Homeostasis to 3' UTR-Directed Regulation and Nonsense-Mediated Decay. Cell Rep 32: 107895, 2020.
- Shen PS, Park J, Qin Y, et al.: Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347: 75-78, 2015.
- Wu Z, Tantray I, Lim J, et al.: MISTERMINATE Mechanistically Links Mitochondrial Dysfunction with Proteostasis Failure. Mol Cell 75: 835-848.e838, 2019. [CrossRef]
- Gehrke S, Wu Z, Klinkenberg M, et al.: PINK1 and Parkin control localized translation of respiratory chain component mRNAs on mitochondria outer membrane. Cell Metab 21: 95-108, 2015. [CrossRef]
- Martin PB, Kigoshi-Tansho Y, Sher RB, et al.: NEMF mutations that impair ribosome-associated quality control are associated with neuromuscular disease. Nat Commun 11: 4625, 2020.
- Ishimura R, Nagy G, Dotu I, et al.: RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345: 455-459, 2014. [CrossRef]
- Sudmant PH, Lee H, Dominguez D, Heiman M and Burge CB: Widespread Accumulation of Ribosome-Associated Isolated 3' UTRs in Neuronal Cell Populations of the Aging Brain. Cell Rep 25: 2447-2456.e2444, 2018.
- Li S, Wu Z, Tantray I, et al.: Quality-control mechanisms targeting translationally stalled and C-terminally extended poly(GR) associated with ALS/FTD. Proc Natl Acad Sci U S A 117: 25104-25115, 2020. [CrossRef]
- Rimal S, Li Y, Vartak R, et al.: Inefficient quality control of ribosome stalling during APP synthesis generates CAT-tailed species that precipitate hallmarks of Alzheimer's disease. Acta Neuropathol Commun 9: 169, 2021. [CrossRef]
- Tatara Y, Yamazaki H, Katsuoka F, et al.: Multiomics and artificial intelligence enabled peripheral blood-based prediction of amnestic mild cognitive impairment. Curr Res Transl Med 71: 103367, 2023. [CrossRef]
- Sorrentino V, Romani M, Mouchiroud L, et al.: Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature 552: 187-193, 2017.
- Roffé M, Hajj GN, Azevedo HF, Alves VS and Castilho BA: IMPACT is a developmentally regulated protein in neurons that opposes the eukaryotic initiation factor 2α kinase GCN2 in the modulation of neurite outgrowth. J Biol Chem 288: 10860-10869, 2013.
- Fessler E, Eckl EM, Schmitt S, et al.: A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 579: 433-437, 2020. [CrossRef]
- Guo X, Aviles G, Liu Y, et al.: Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature 579: 427-432, 2020.
- Fessler E, Krumwiede L and Jae LT: DELE1 tracks perturbed protein import and processing in human mitochondria. Nat Commun 13: 1853, 2022.
- Sekine Y, Houston R, Fessler E, Jae LT, Narendra DP and Sekine S: A mitochondrial iron-sensing pathway regulated by DELE1. bioRxiv, 2022.
- Wang SF, Chen MS, Chou YC, et al.: Mitochondrial dysfunction enhances cisplatin resistance in human gastric cancer cells via the ROS-activated GCN2-eIF2alpha-ATF4-xCT pathway. Oncotarget 7: 74132-74151, 2016. [CrossRef]
- Baker BM, Nargund AM, Sun T and Haynes CM: Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS Genet 8: e1002760, 2012.
- Topf U, Suppanz I, Samluk L, et al.: Quantitative proteomics identifies redox switches for global translation modulation by mitochondrially produced reactive oxygen species. Nat Commun 9: 324, 2018.
| RWD-domain-containing proteins | Gene symbol | Amino acids | RWD domain region |
| eIF-2-alpha kinase GCN2 | EIF2AK4 (GCN2) | 1,649 | 25–137 |
| Protein IMPACT | IMPACT | 320 | 14–116 |
| E3 ubiquitin-protein ligase RNF14 | RNF14 | 474 | 11–137 |
| E3 ubiquitin-protein ligase RNF25 | RNF25 | 459 | 18–128 |
| RWD domain-containing protein 1 | RWDD1 (DFRP2) | 243 | 10–114 |
| RWD domain-containing protein 2A | RWDD2A | 292 | 14–134 |
| RWD domain-containing protein 2B | RWDD2B | 319 | 41–165 |
| RWD domain-containing protein 3 | RWDD3 (RESUME) | 267 | 7–114 |
| RWD domain-containing protein 4 | RWDD4 | 188 | 9–111 |
| WD repeat-containing protein 59 | WDR59 | 974 | 393–494 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





