Submitted:
29 January 2024
Posted:
31 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Match-up Data
3. Bayesian Neural Network
4. Standard Ocean Colour Models
4.1. OC4
4.2. OCI
5. Evaluation Metrics
6. Results and Discussion
6.1. Maximum Band Ratio Bayesian Ocean Model: SeaWiFS
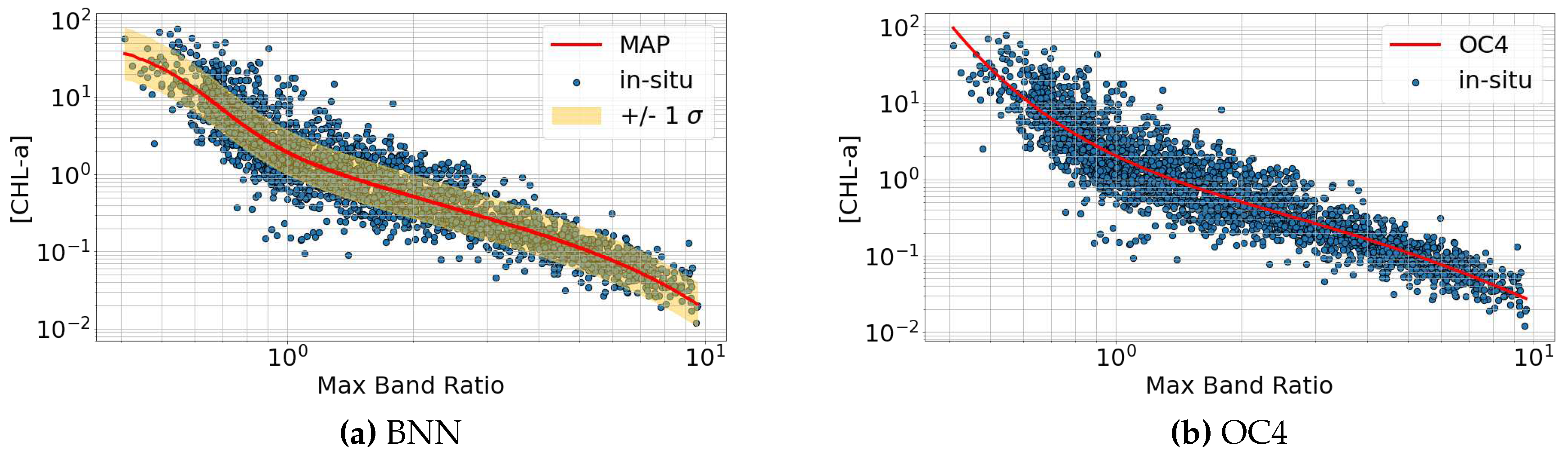
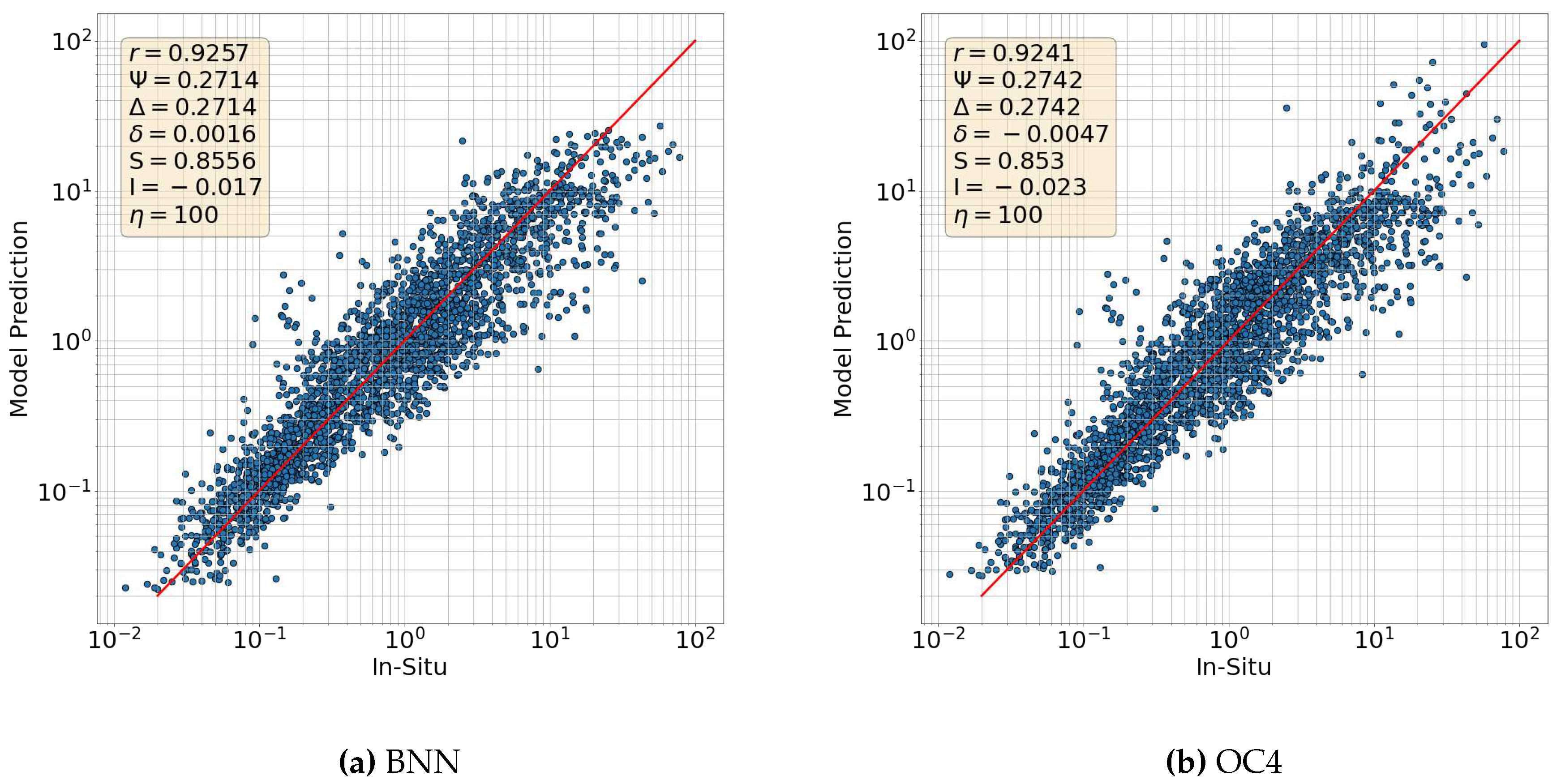
6.2. Comparison with OCI
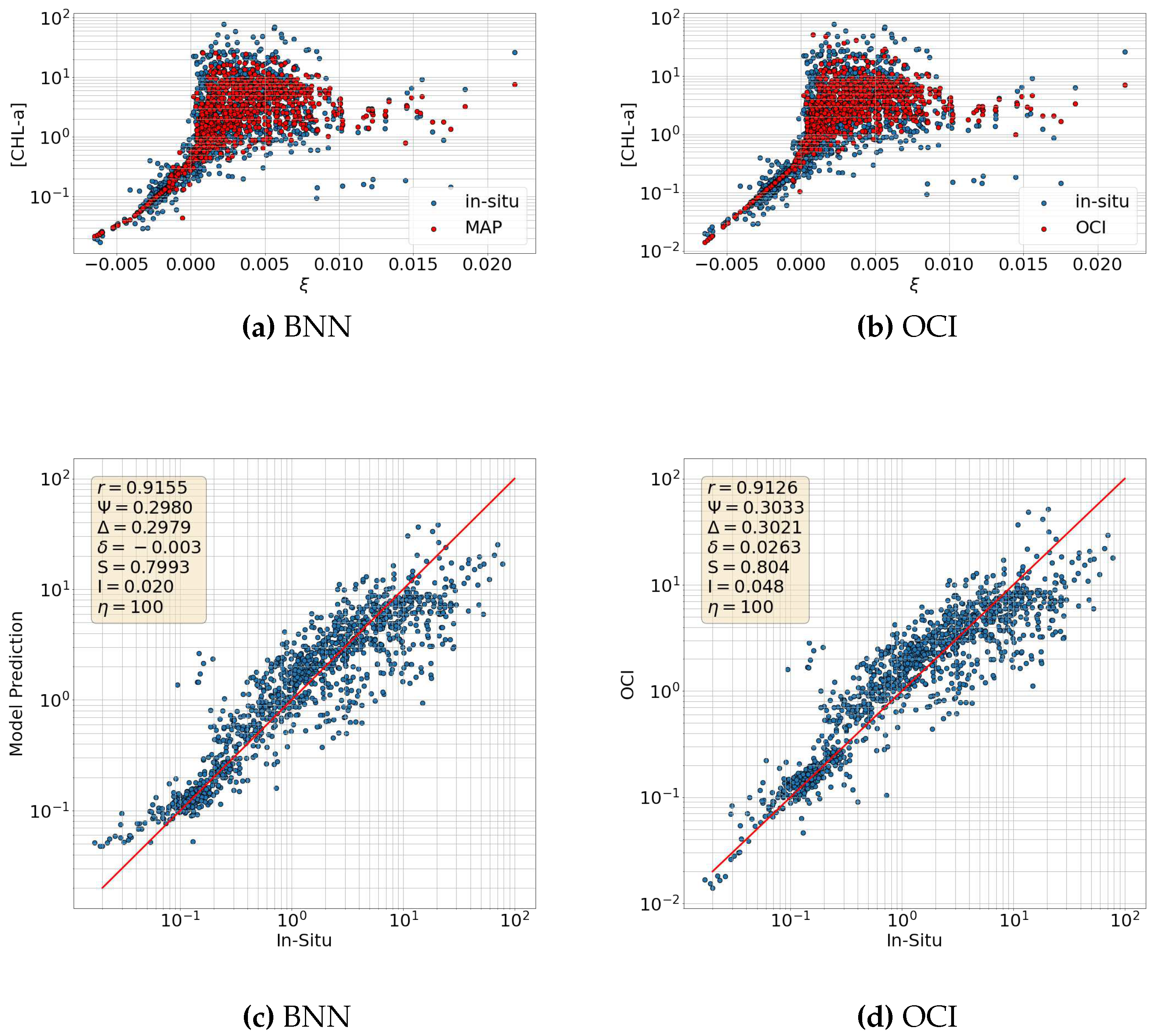
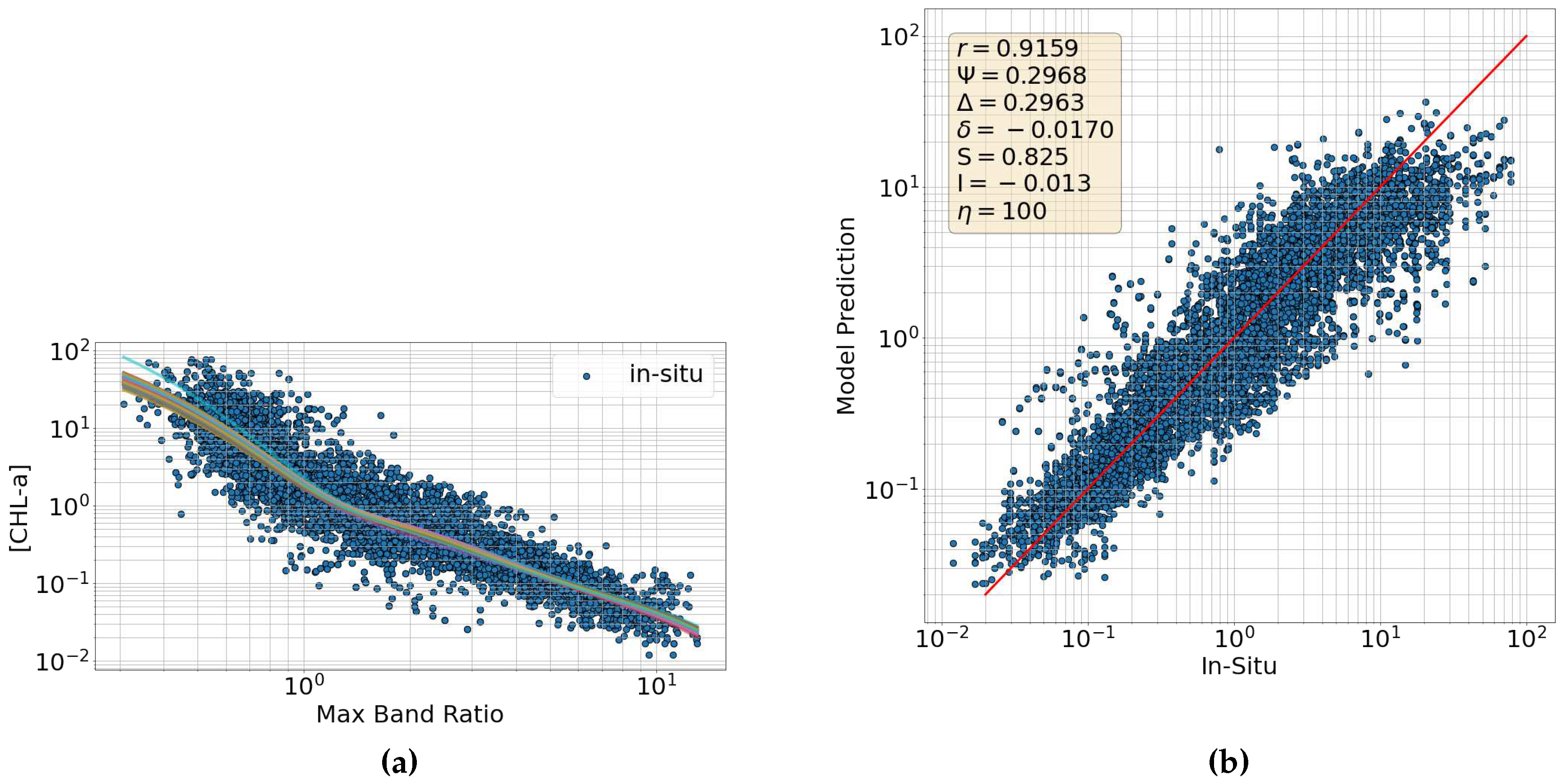
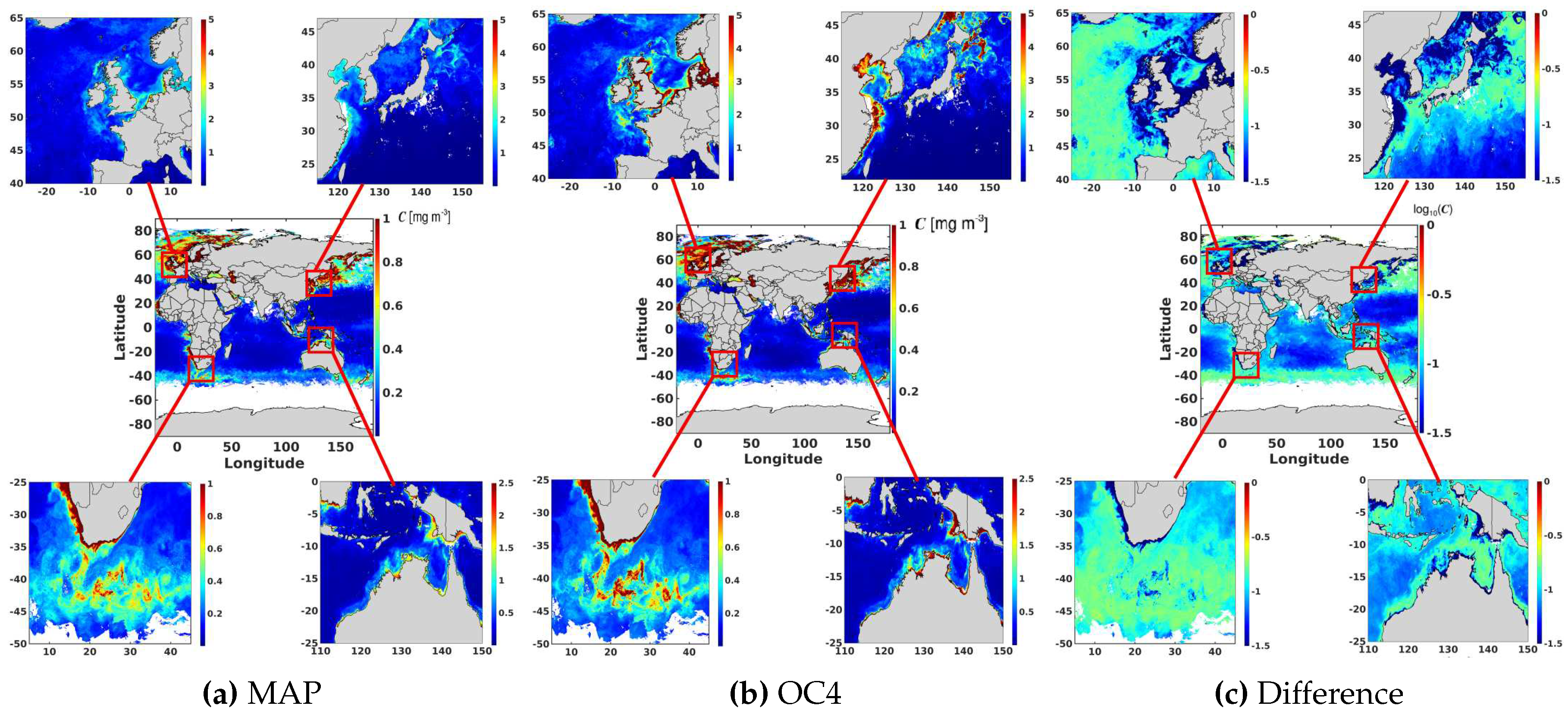
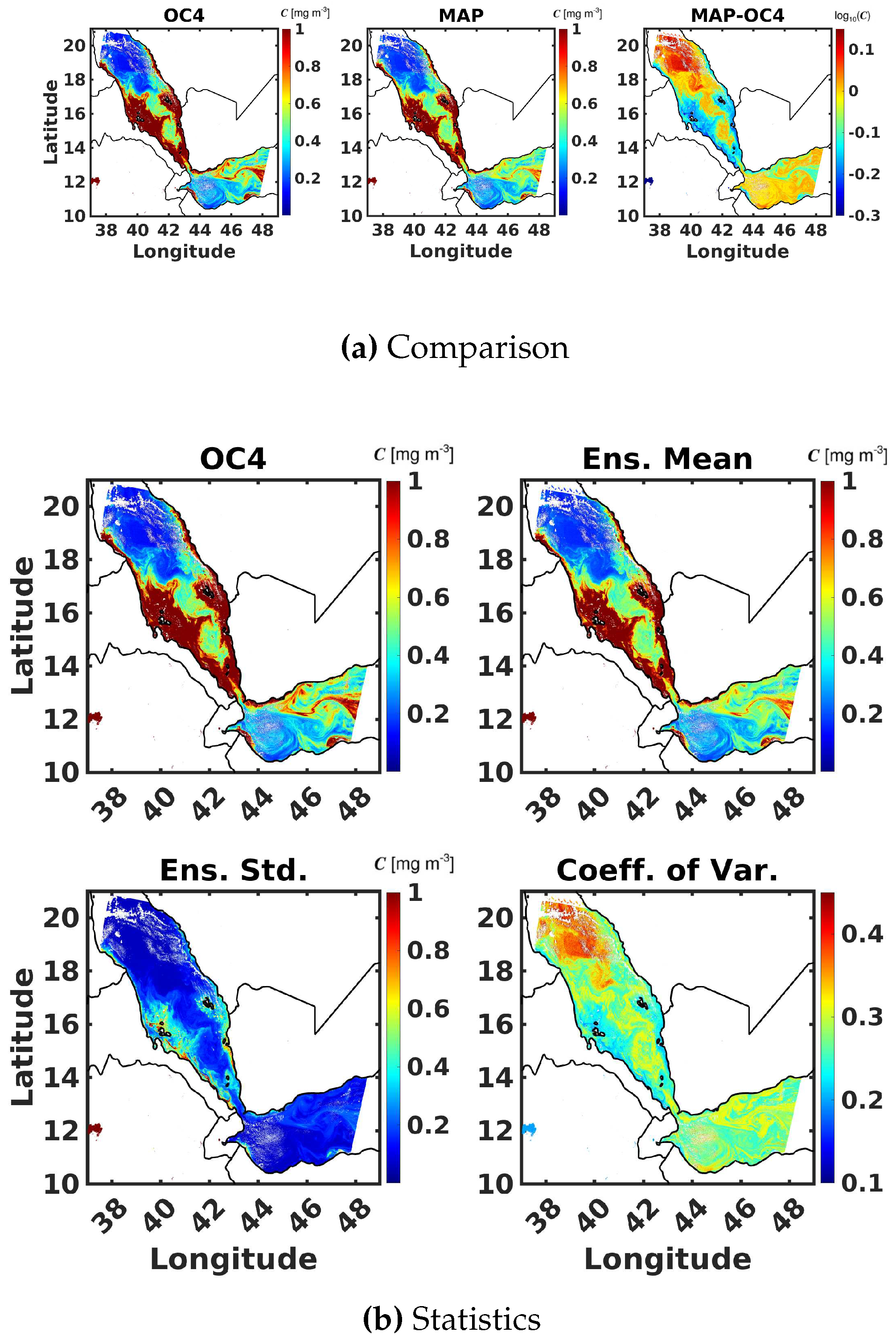
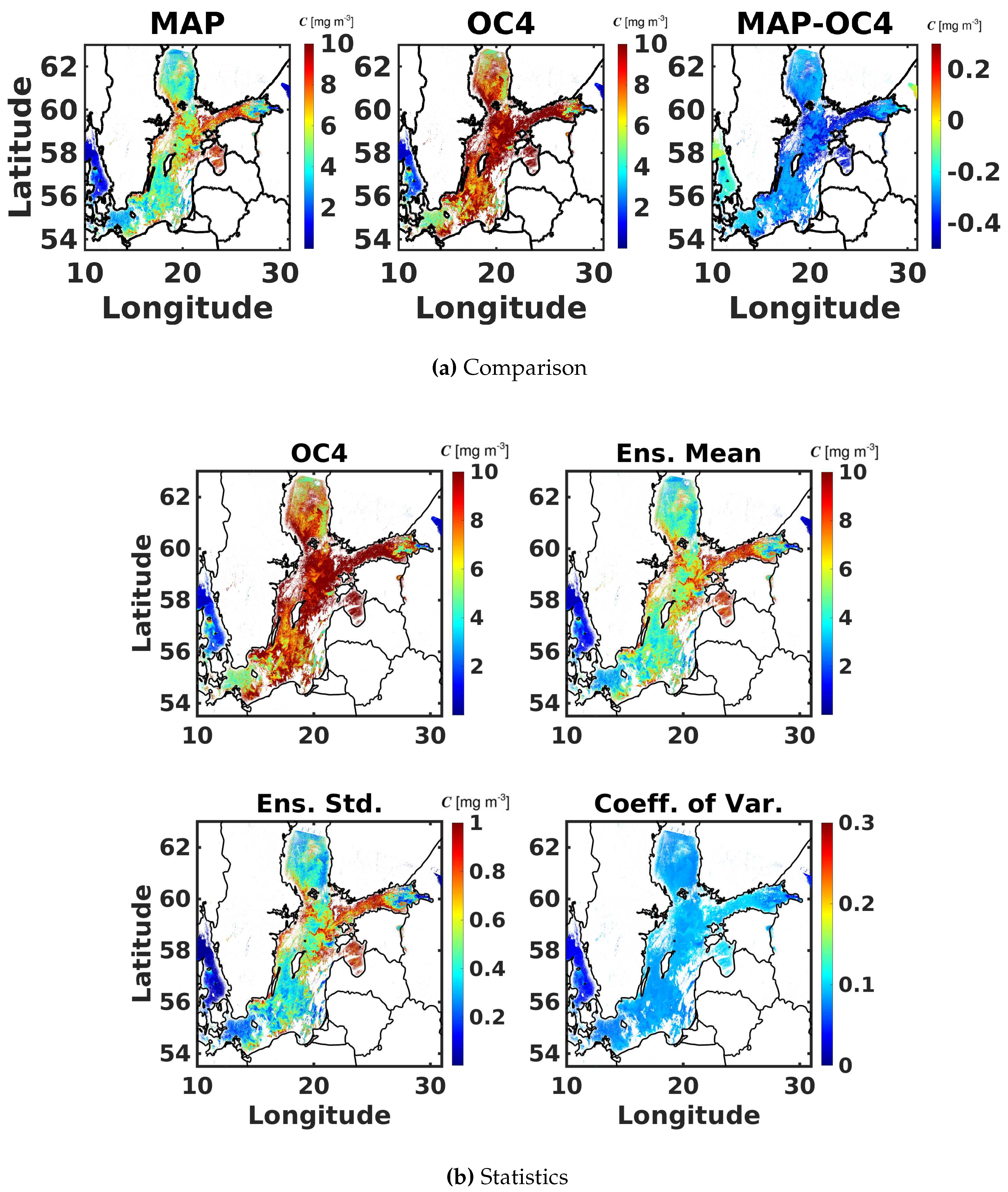
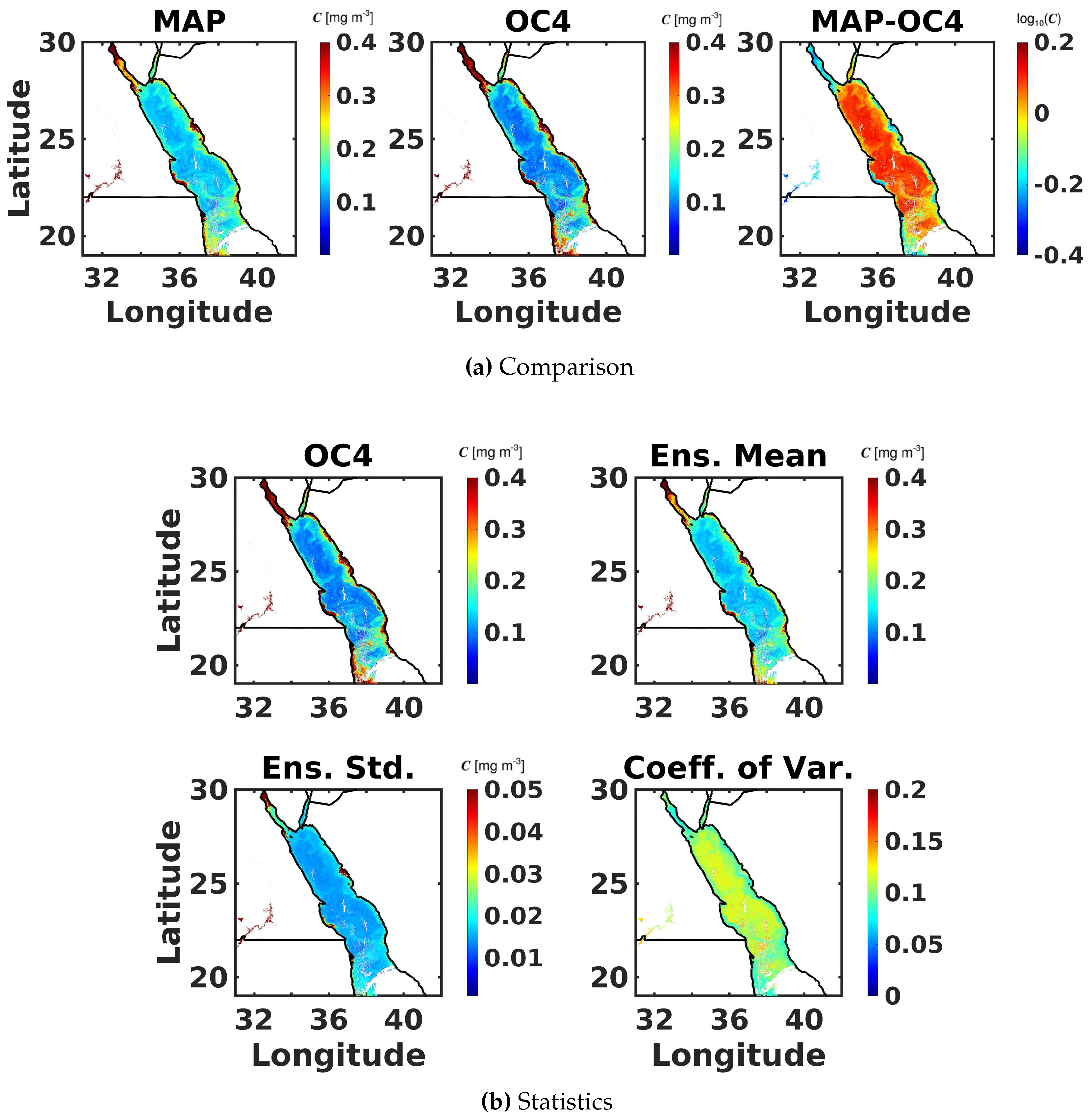
6.3. Maximum Band Ratio Bayesian Ocean Model: Generalizability
6.4. Reflectances Bayesian Ocean Model
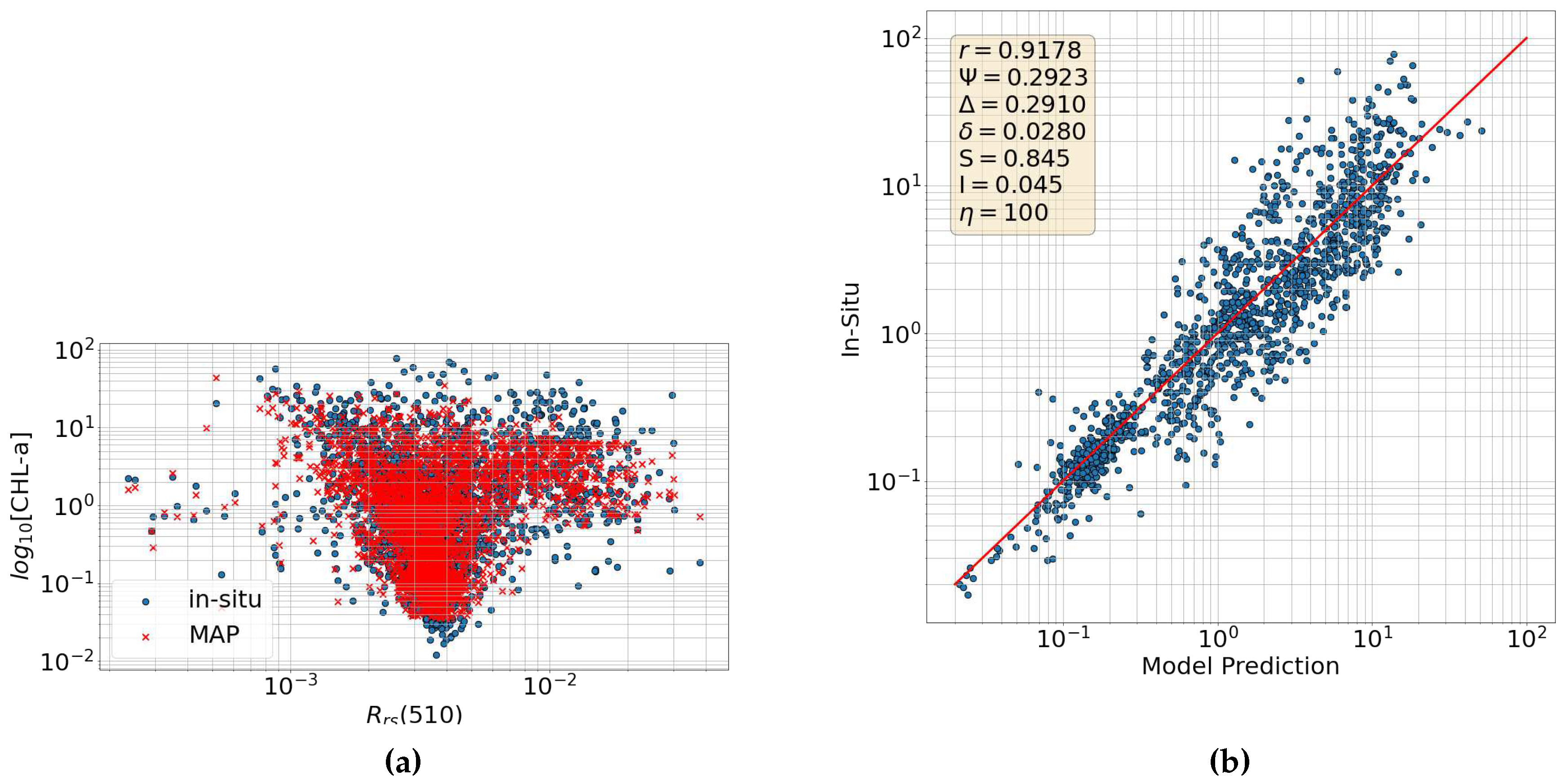
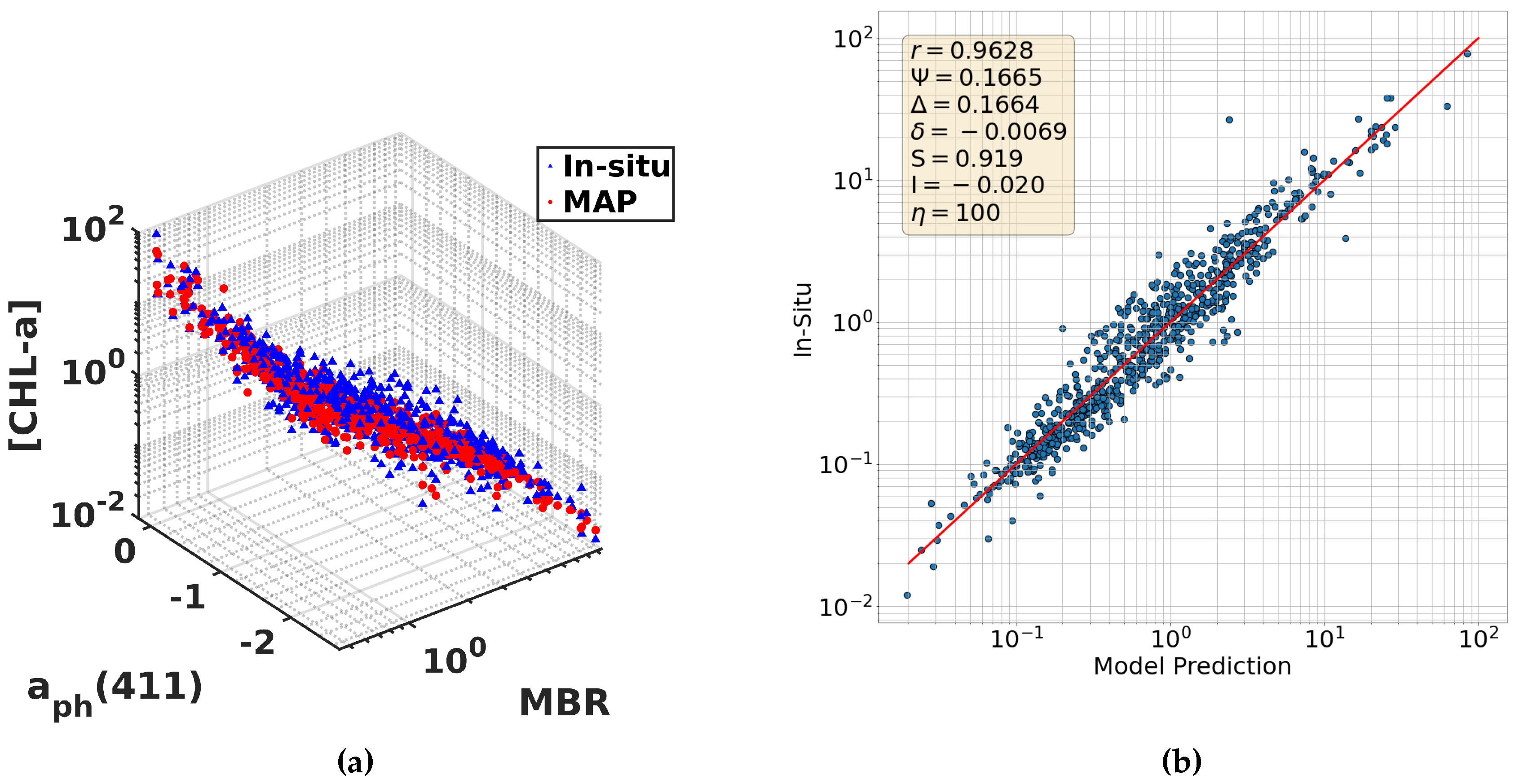
6.5. Incorporating IOPs
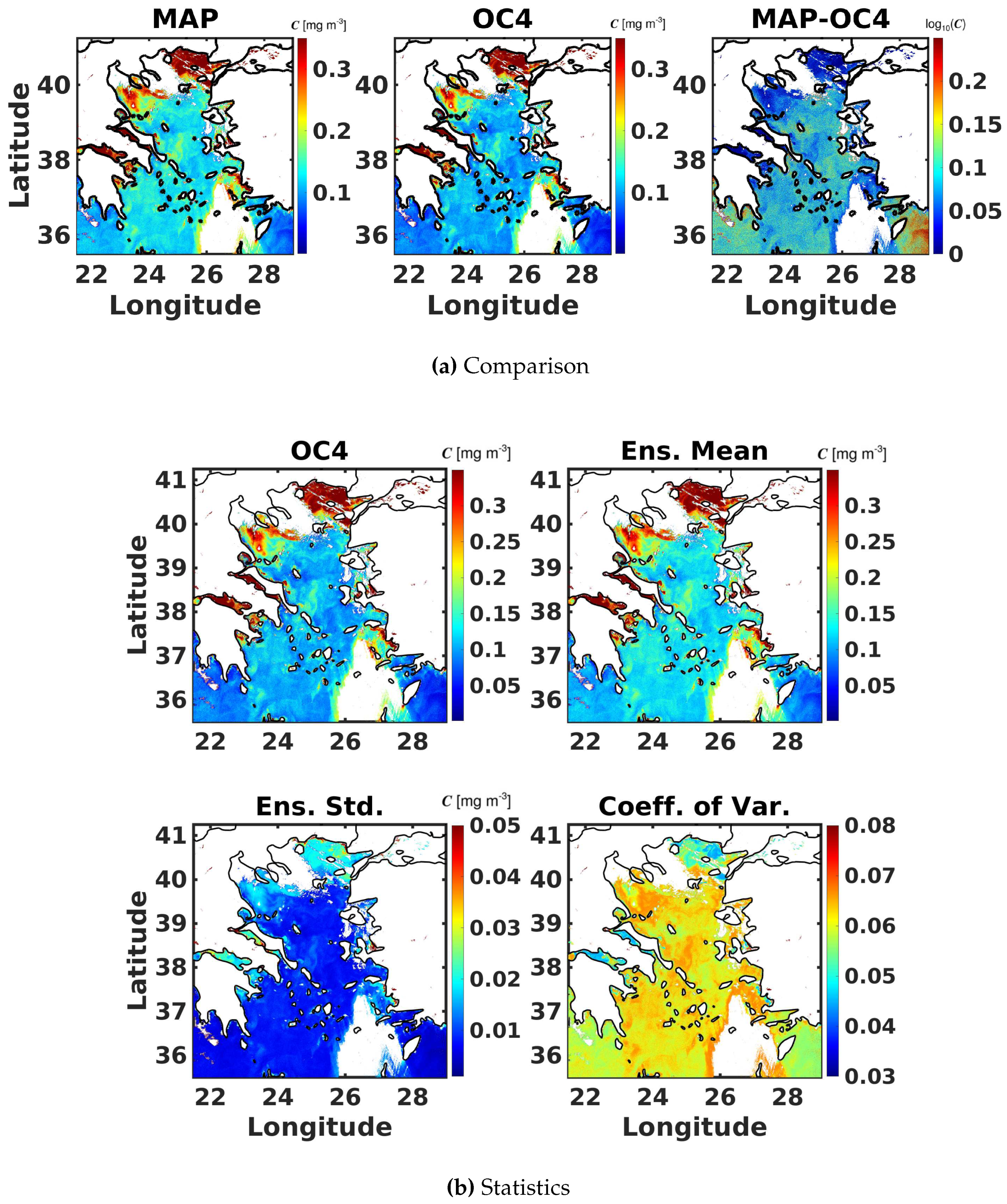
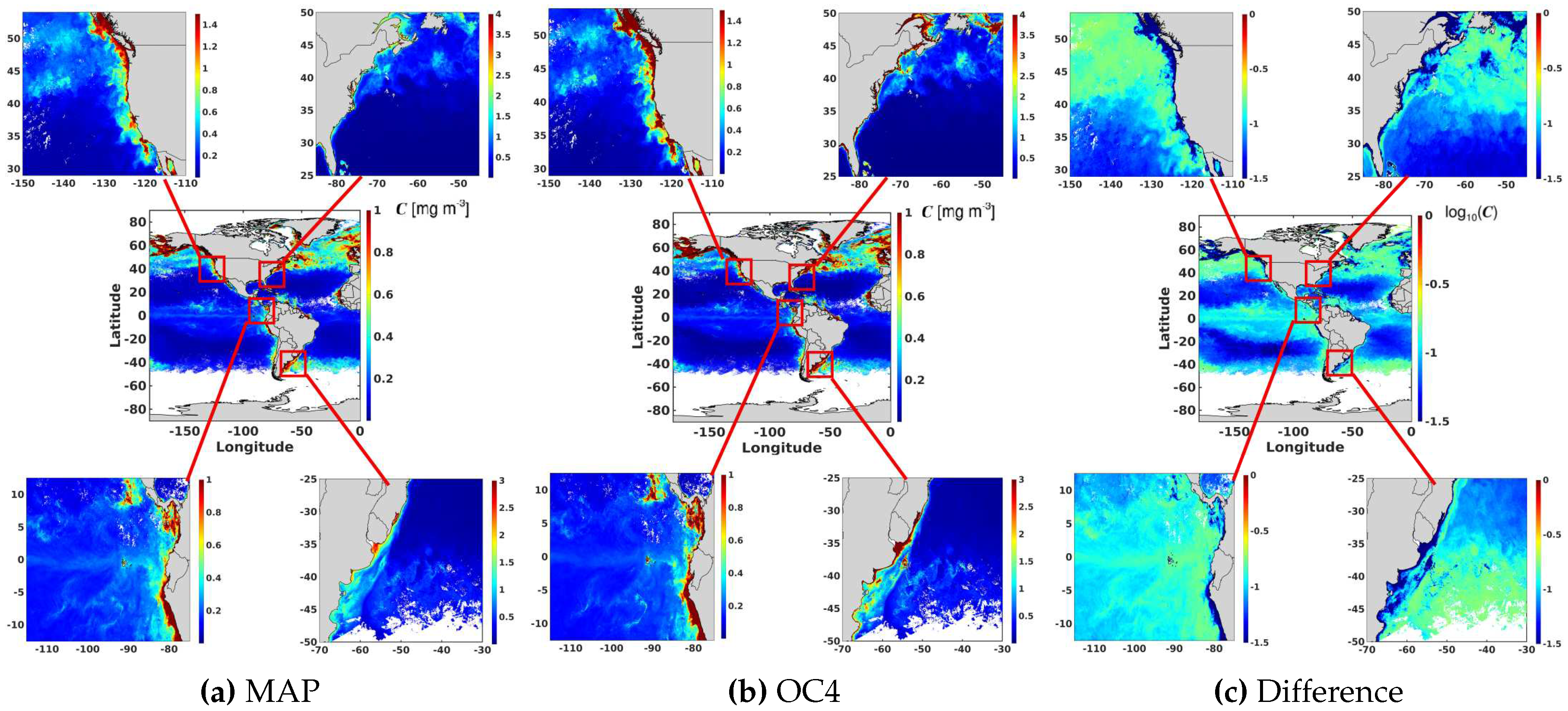
6.6. Evaluation of the Probabilistic Ocean Colour Model Using Satellite Observations
6.6.1. MODIS
6.6.2. Sentinel-3
7. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A Stochastic Variational Inference
Appendix A.1. Bayesian Statistics
Appendix A.2. Variational Inference
References
- Harris, G. Phytoplankton ecology: structure, function and fluctuation; Springer Science & Business Media, 2012.
- Jones, R.I. Phytoplankton, primary production and nutrient cycling. Aquatic humic substances: ecology and biogeochemistry 1998, pp. 145–175. [CrossRef]
- Falkowski, P.G.; Raven, J.A. Aquatic photosynthesis; Princeton University Press, 2013.
- Hays, G.C.; Richardson, A.J.; Robinson, C. Climate change and marine plankton. Trends in ecology & evolution 2005, 20, 337–344. [CrossRef]
- Haeder, D.P.; Villafane, V.E.; Helbling, E.W. Productivity of aquatic primary producers under global climate change. Photochemical & Photobiological Sciences 2014, 13, 1370–1392. [CrossRef]
- Basu, S.; Mackey, K.R. Phytoplankton as key mediators of the biological carbon pump: Their responses to a changing climate. Sustainability 2018, 10, 869. [CrossRef]
- Falkowski, P.G. The role of phytoplankton photosynthesis in global biogeochemical cycles. Photosynthesis research 1994, 39, 235–258. [CrossRef]
- Platt, T.; Fuentes-Yaco, C.; Frank, K.T. Spring algal bloom and larval fish survival. Nature 2003, 423, 398–399. [CrossRef]
- Anderson, D.M.; Glibert, P.M.; Burkholder, J.M. Harmful algal blooms and eutrophication: nutrient sources, composition, and consequences. Estuaries 2002, 25, 704–726. [CrossRef]
- Klemas, V. Remote sensing of algal blooms: an overview with case studies. Journal of coastal research 2012, 28, 34–43. [CrossRef]
- Racault, M.F.; Platt, T.; Sathyendranath, S.; Ağirbaş, E.; Martinez Vicente, V.; Brewin, R. Plankton indicators and ocean observing systems: support to the marine ecosystem state assessment. Journal of Plankton Research 2014, 36, 621–629. [CrossRef]
- Platt, T.; White III, G.N.; Zhai, L.; Sathyendranath, S.; Roy, S. The phenology of phytoplankton blooms: Ecosystem indicators from remote sensing. Ecological Modelling 2009, 220, 3057–3069. [CrossRef]
- Hazen, E.L.; Suryan, R.M.; Santora, J.A.; Bograd, S.J.; Watanuki, Y.; Wilson, R.P. Scales and mechanisms of marine hotspot formation. Marine Ecology Progress Series 2013, 487, 177–183. [CrossRef]
- Racault, M.F.; Le Quéré, C.; Buitenhuis, E.; Sathyendranath, S.; Platt, T. Phytoplankton phenology in the global ocean. Ecological Indicators 2012, 14, 152–163. [CrossRef]
- Gokul, E.A.; Raitsos, D.E.; Gittings, J.A.; Alkawri, A.; Hoteit, I. Remotely sensing harmful algal blooms in the Red Sea. PLoS One 2019, 14, e0215463. [CrossRef]
- Lin, J.; Miller, P.I.; Jönsson, B.F.; Bedington, M. Early Warning of Harmful Algal Bloom Risk Using Satellite Ocean Color and Lagrangian Particle Trajectories. Frontiers in Marine Science 2021, 8. [CrossRef]
- Shu, C.; Xiu, P.; Xing, X.; Qiu, G.; Ma, W.; Brewin, R.J.; Ciavatta, S. Biogeochemical Model Optimization by Using Satellite-Derived Phytoplankton Functional Type Data and BGC-Argo Observations in the Northern South China Sea. Remote Sensing 2022, 14, 1297. [CrossRef]
- Joint, I.; Groom, S.B. Estimation of phytoplankton production from space: current status and future potential of satellite remote sensing. Journal of experimental marine Biology and Ecology 2000, 250, 233–255. [CrossRef]
- Morel, A.; Prieur, L. Analysis of variations in ocean color1. Limnology and Oceanography 1977, 22, 709–722, [https://aslopubs.onlinelibrary.wiley.com/doi/pdf/10.4319/lo.1977.22.4.0709]. [CrossRef]
- Bricaud, A.; Babin, M.; Morel, A.; Claustre, H. Variability in the chlorophyll-specific absorption coefficients of natural phytoplankton: Analysis and parameterization. Journal of Geophysical Research: Oceans 1995, 100, 13321–13332. [CrossRef]
- Groom, S.; Sathyendranath, S.; Ban, Y.; Bernard, S.; Brewin, R.; Brotas, V.; Brockmann, C.; Chauhan, P.; Choi, J.k.; Chuprin, A.; others. Satellite ocean colour: current status and future perspective. Frontiers in Marine Science 2019, 6, 485. [CrossRef]
- O’Reilly, J.E.; Werdell, P.J. Chlorophyll algorithms for ocean color sensors-OC4, OC5 & OC6. Remote sensing of environment 2019, 229, 32–47. [CrossRef]
- Mélin, F.; others. Uncertainties in ocean colour remote sensing 2019.
- Neil, C.; Spyrakos, E.; Hunter, P.D.; Tyler, A.N. A global approach for chlorophyll-a retrieval across optically complex inland waters based on optical water types. Remote Sensing of Environment 2019, 229, 159–178. [CrossRef]
- Doerffer, R.; Schiller, H. The MERIS Case 2 water algorithm. International Journal of Remote Sensing 2007, 28, 517–535. [CrossRef]
- Yu, B.; Xu, L.; Peng, J.; Hu, Z.; Wong, A. Global chlorophyll-a concentration estimation from moderate resolution imaging spectroradiometer using convolutional neural networks. Journal of Applied Remote Sensing 2020, 14, 034520–034520. [CrossRef]
- Ye, H.; Tang, S.; Yang, C. Deep learning for Chlorophyll-a concentration retrieval: A case study for the Pearl River Estuary. Remote Sensing 2021, 13, 3717. [CrossRef]
- Fan, D.; He, H.; Wang, R.; Zeng, Y.; Fu, B.; Xiong, Y.; Liu, L.; Xu, Y.; Gao, E. CHLNET: A novel hybrid 1D CNN-SVR algorithm for estimating ocean surface chlorophyll-a. Frontiers in Marine Science 2022, 9, 934536. [CrossRef]
- Hadjal, M.; Medina-Lopez, E.; Ren, J.; Gallego, A.; McKee, D. An artificial neural network algorithm to retrieve chlorophyll a for Northwest European shelf seas from top of atmosphere ocean colour reflectance. Remote sensing 2022, 14, 3353. [CrossRef]
- MacKay, D.J. Bayesian neural networks and density networks. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 1995, 354, 73–80. Proceedings of the Third Workshop on Neutron Scattering Data Analysis. [CrossRef]
- Jospin, L.V.; Laga, H.; Boussaid, F.; Buntine, W.; Bennamoun, M. Hands-On Bayesian Neural Networks—A Tutorial for Deep Learning Users. IEEE Computational Intelligence Magazine 2022, 17, 29–48. [CrossRef]
- Shen, G.; Chen, X.; Deng, Z. Variational Learning of Bayesian Neural Networks via Bayesian Dark Knowledge. Proceedings of the Twenty-Ninth International Joint Conference on Artificial Intelligence, IJCAI-20; Bessiere, C., Ed. International Joint Conferences on Artificial Intelligence Organization, 2020, pp. 2037–2043. Main track, . [CrossRef]
- Izmailov, P.; Vikram, S.; Hoffman, M.D.; Wilson, A.G.G. What Are Bayesian Neural Network Posteriors Really Like? Proceedings of the 38th International Conference on Machine Learning; Meila, M.; Zhang, T., Eds. PMLR, 2021, Vol. 139, Proceedings of Machine Learning Research, pp. 4629–4640.
- Magris, M.; Iosifidis, A. Bayesian learning for neural networks: an algorithmic survey. Artificial Intelligence Review 2023, 56, 11773–11823. [CrossRef]
- Kendall, A.; Gal, Y. What Uncertainties Do We Need in Bayesian Deep Learning for Computer Vision? Advances in Neural Information Processing Systems; Guyon, I.; Luxburg, U.V.; Bengio, S.; Wallach, H.; Fergus, R.; Vishwanathan, S.; Garnett, R., Eds. Curran Associates, Inc., 2017, Vol. 30.
- Goan, E.; Fookes, C. Bayesian Neural Networks: An Introduction and Survey. In Case Studies in Applied Bayesian Data Science: CIRM Jean-Morlet Chair, Fall 2018; Mengersen, K.L.; Pudlo, P.; Robert, C.P., Eds.; Springer International Publishing: Cham, 2020; pp. 45–87. [CrossRef]
- Chen, B.; Zhang, A.; Cao, L. Autonomous intelligent decision-making system based on Bayesian SOM neural network for robot soccer. Neurocomputing 2014, 128, 447–458. [CrossRef]
- Abdullah, A.A.; Hassan, M.M.; Mustafa, Y.T. A Review on Bayesian Deep Learning in Healthcare: Applications and Challenges. IEEE Access 2022, 10, 36538–36562. [CrossRef]
- Werther, M.; Odermatt, D.; Simis, S.G.; Gurlin, D.; Lehmann, M.K.; Kutser, T.; Gupana, R.; Varley, A.; Hunter, P.D.; Tyler, A.N.; Spyrakos, E. A Bayesian approach for remote sensing of chlorophyll-a and associated retrieval uncertainty in oligotrophic and mesotrophic lakes. Remote Sensing of Environment 2022, 283, 113295. [CrossRef]
- Valente, A.; Sathyendranath, S.; Brotas, V.; Groom, S.; Grant, M.; Jackson, T.; Chuprin, A.; Taberner, M.; Airs, R.; Antoine, D.; Arnone, R.; Balch, W.M.; Barker, K.; Barlow, R.; Bélanger, S.; Berthon, J.F.; Beşiktepe, c.; Borsheim, Y.; Bracher, A.; Brando, V.; Brewin, R.J.W.; Canuti, E.; Chavez, F.P.; Cianca, A.; Claustre, H.; Clementson, L.; Crout, R.; Ferreira, A.; Freeman, S.; Frouin, R.; García-Soto, C.; Gibb, S.W.; Goericke, R.; Gould, R.; Guillocheau, N.; Hooker, S.B.; Hu, C.; Kahru, M.; Kampel, M.; Klein, H.; Kratzer, S.; Kudela, R.; Ledesma, J.; Lohrenz, S.; Loisel, H.; Mannino, A.; Martinez-Vicente, V.; Matrai, P.; McKee, D.; Mitchell, B.G.; Moisan, T.; Montes, E.; Muller-Karger, F.; Neeley, A.; Novak, M.; O’Dowd, L.; Ondrusek, M.; Platt, T.; Poulton, A.J.; Repecaud, M.; Röttgers, R.; Schroeder, T.; Smyth, T.; Smythe-Wright, D.; Sosik, H.M.; Thomas, C.; Thomas, R.; Tilstone, G.; Tracana, A.; Twardowski, M.; Vellucci, V.; Voss, K.; Werdell, J.; Wernand, M.; Wojtasiewicz, B.; Wright, S.; Zibordi, G. A compilation of global bio-optical in situ data for ocean colour satellite applications – version three. Earth System Science Data 2022, 14, 5737–5770. [CrossRef]
- Werdell, P.; Bailey, S. The SeaWiFS Bio-optical Archive and Storage System (SeaBASS): Current Architecture and Implementation. technical memorandum 2002-211617, NASA Goddard Space Flight Center, Greenbelt, Maryland, 2002.
- Werdell, P.J.; Bailey, S.W. An improved in-situ bio-optical data set for ocean color algorithm development and satellite data product validation. Remote Sensing of Environment 2005, 98, 122–140. [CrossRef]
- Barker, K.; Mazeran, C.; Lerebourg, C.; Bouvet, M.; Antoine, D.; Ondrusek, M.; Zibordi, G.; Lavender, S. Mermaid: The MERIS matchup in-situ database. Proceedings of the 2nd (A) ATSR and MERIS Workshop, Frascati, Italy, 2008, pp. 22–26.
- Matrai, P.; Olson, E.; Suttles, S.; Hill, V.; Codispoti, L.; Light, B.; Steele, M. Synthesis of primary production in the Arctic Ocean: I. Surface waters, 1954–2007. Progress in Oceanography 2013, 110, 93–106. [CrossRef]
- Devine, L.; Galbraith, P.S.; Joly, P.; Plourde, S.; Saint-Amand.; Pierre, J.S.; Starr, M. Chemical and biological oceanographic conditions in the estuary and Gulf of St. Lawrence during 2015; Fisheries and Oceans Canada, Ecosystems and Oceans Science, 2015.
- Nechad, B.; Ruddick, K.; Schroeder, T.; Oubelkheir, K.; Blondeau-Patissier, D.; Cherukuru, N.; Brando, V.; Dekker, A.; Clementson, L.; Banks, A.C.; Maritorena, S.; Werdell, P.J.; Sá, C.; Brotas, V.; Caballero de Frutos, I.; Ahn, Y.H.; Salama, S.; Tilstone, G.; Martinez-Vicente, V.; Foley, D.; McKibben, M.; Nahorniak, J.; Peterson, T.; Siliò-Calzada, A.; Röttgers, R.; Lee, Z.; Peters, M.; Brockmann, C. CoastColour Round Robin data sets: a database to evaluate the performance of algorithms for the retrieval of water quality parameters in coastal waters. Earth System Science Data 2015, 7, 319–348. [CrossRef]
- Peloquin, J.M.; Swan, C.; Gruber, N.; Vogt, M.; Claustre, H.; Ras, J.; Uitz, J.; Barlow, R.G.; Behrenfeld, M.J.; Bidigare, R.R.; Dierssen, H.M.; Ditullio, G.; Fernández, E.; Gallienne, C.; Gibb, S.W.; Goericke, R.; Harding, L.; Head, E.J.H.; Holligan, P.M.; Hooker, S.B.; Karl, D.; Landry, M.R.; Letelier, R.; Llewellyn, C.; Lomas, M.W.; Lucas, M.; Mannino, A.; Marty, J.C.; Mitchell, B.G.; Muller-Karger, F.E.; Nelson, N.; O’Brien, C.J.; Prezelin, B.; Repeta, D.J.; Smith, W.O.J.; Smythe-Wright, D.; Stumpf, R.; Subramaniam, A.; Suzuki, K.; Trees, C.; Vernet, M.; Wasmund, N.; Wright, S. The MAREDAT global database of high performance liquid chromatography marine pigment measurements - Gridded data product (NetCDF) - Contribution to the MAREDAT World Ocean Atlas of Plankton Functional Types, 2013. [CrossRef]
- Clark, D.; Murphy, M.; Yarbrough, M.; Feinholz, M.; Flora, S.; Broenkow, W.; Johnson, B.; Brown, S.; Kim, Y.; Mueller, J. MOBY, A Radiometric Buoy for Performance Monitoring and Vicarious Calibration of Satellite Ocean Color Sensors: Measurement and Data Analysis Protocols, 2003.
- Neal, R.M. Bayesian Learning for Neural Networks; Springer New York, 1996. [CrossRef]
- Samaniego, F.J. A comparison of the Bayesian and frequentist approaches to estimation; Vol. 24, Springer, 2010.
- Chen, T.; Chen, H. Universal approximation to nonlinear operators by neural networks with arbitrary activation functions and its application to dynamical systems. IEEE Transactions on Neural Networks 1995, 6, 911–917. [CrossRef]
- Moore, T.S.; Campbell, J.W.; Dowell, M.D. A class-based approach to characterizing and mapping the uncertainty of the MODIS ocean chlorophyll product. Remote Sensing of Environment 2009, 113, 2424–2430. [CrossRef]
- IOCCG. Uncertainties in Ocean Colour Remote Sensing. International Ocean Colour Coordinating Group, Dartmouth, Canada 2019. [CrossRef]
- Amini, A.; Schwarting, W.; Soleimany, A.; Rus, D. Deep evidential regression. Advances in Neural Information Processing Systems 2020, 33, 14927–14937.
- Bingham, E.; Chen, J.P.; Jankowiak, M.; Obermeyer, F.; Pradhan, N.; Karaletsos, T.; Singh, R.; Szerlip, P.; Horsfall, P.; Goodman, N.D. Pyro: Deep Universal Probabilistic Programming. J. Mach. Learn. Res. 2019, 20, 973–978. [CrossRef]
- Hoffman, M.D.; Blei, D.M.; Wang, C.; Paisley, J. Stochastic variational inference. Journal of Machine Learning Research 2013.
- Ranganath, R.; Gerrish, S.; Blei, D. Black box variational inference. Artificial intelligence and statistics. PMLR, 2014, pp. 814–822.
- Blei, D.M.; Kucukelbir, A.; McAuliffe, J.D. Variational Inference: A Review for Statisticians. Journal of the American Statistical Association 2017, 112, 859–877, [https://doi.org/10.1080/01621459.2017.1285773]. [CrossRef]
- Kingma, D.P.; Welling, M. Auto-Encoding Variational Bayes, 2022, [arXiv:stat.ML/1312.6114].
- Olivier, A.; Shields, M.D.; Graham-Brady, L. Bayesian neural networks for uncertainty quantification in data-driven materials modeling. Computer Methods in Applied Mechanics and Engineering 2021, 386, 114079. [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization, 2017, [arXiv:cs.LG/1412.6980].
- O’Reilly, J.; Maritorena, S.; Siegel, D.; O’Brien, M.; Toole, D.; Mitchell, B.; Kahru, M.; Chavez, F.; Strutton, P.; Cota, G. Ocean color chlorophyll a algorithms for SeaWiFS, OC2, and OC4: Technical report. SeaWiFS postlaunch technical report series, SeaWiFS postlaunch calibration and validation analyses part 3,Vol. 11, NASA, Goddard Space Flight Center, Greenbelt, Maryland, 2000.
- Hammoud, M.A.E.R.; Mittal, H.V.R.; Le Maître, O.; Hoteit, I.; Knio, O. Variance-based sensitivity analysis of oil spill predictions in the Red Sea region. Frontiers in Marine Science 2023, 10. [CrossRef]
- Hu, C.; Lee, Z.; Franz, B. Chlorophyll aalgorithms for oligotrophic oceans: A novel approach based on three-band reflectance difference. Journal of Geophysical Research: Oceans 2012, 117, [https://agupubs.onlinelibrary.wiley.com/doi/pdf/10.1029/2011JC007395]. [CrossRef]
- Hu, C.; Lee, Z.; Franz, B. Chlorophyll aalgorithms for oligotrophic oceans: A novel approach based on three-band reflectance difference. Journal of Geophysical Research: Oceans 2012, 117, [https://agupubs.onlinelibrary.wiley.com/doi/pdf/10.1029/2011JC007395]. [CrossRef]
- Brewin, R.J.; Sathyendranath, S.; Müller, D.; Brockmann, C.; Deschamps, P.Y.; Devred, E.; Doerffer, R.; Fomferra, N.; Franz, B.; Grant, M.; Groom, S.; Horseman, A.; Hu, C.; Krasemann, H.; Lee, Z.; Maritorena, S.; Mélin, F.; Peters, M.; Platt, T.; Regner, P.; Smyth, T.; Steinmetz, F.; Swinton, J.; Werdell, J.; White, G.N. The Ocean Colour Climate Change Initiative: III. A round-robin comparison on in-water bio-optical algorithms. Remote Sensing of Environment 2015, 162, 271–294. [CrossRef]
- O’Reilly, J.E.; Werdell, P.J. Chlorophyll algorithms for ocean color sensors - OC4, OC5 & OC6. Remote Sensing of Environment 2019, 229, 32–47. [CrossRef]
- Woźniak, S.B.; Meler, J.; Stoń-Egiert, J. Inherent optical properties of suspended particulate matter in the southern Baltic Sea in relation to the concentration, composition and characteristics of the particle size distribution; new forms of multicomponent parameterizations of optical properties. Journal of Marine Systems 2022, 229, 103720. [CrossRef]
- Lavigne, H.; Van der Zande, D.; Ruddick, K.; Cardoso Dos Santos, J.; Gohin, F.; Brotas, V.; Kratzer, S. Quality-control tests for OC4, OC5 and NIR-red satellite chlorophyll-a algorithms applied to coastal waters. Remote Sensing of Environment 2021, 255, 112237. [CrossRef]
- Raitsos, D.E.; Pradhan, Y.; Brewin, R.J.W.; Stenchikov, G.; Hoteit, I. Remote Sensing the Phytoplankton Seasonal Succession of the Red Sea. PLOS ONE 2013, 8, 1–9. [CrossRef]
- Raitsos, D.E.; Yi, X.; Platt, T.; Racault, M.F.; Brewin, R.J.W.; Pradhan, Y.; Papadopoulos, V.P.; Sathyendranath, S.; Hoteit, I. Monsoon oscillations regulate fertility of the Red Sea. Geophysical Research Letters 2015, 42, 855–862, [https://agupubs.onlinelibrary.wiley.com/doi/pdf/10.1002/2014GL062882]. [CrossRef]
- Raitsos, D.E.; Brewin, R.J.W.; Zhan, P.; Dreano, D.; Pradhan, Y.; Nanninga, G.B.; Hoteit, I. Sensing coral reef connectivity pathways from space. Scientific Reports 2017, 7, 9338. [CrossRef]
- Garcia, V.M.T.; Signorini, S.; Garcia, C.A.E.; McClain, C.R. Empirical and semi-analytical chlorophyll algorithms in the south-western Atlantic coastal region (25–40°S and 60–45°W). International Journal of Remote Sensing 2006, 27, 1539–1562, [https://doi.org/10.1080/01431160500382857]. [CrossRef]
- Brewin, R.J.; Raitsos, D.E.; Pradhan, Y.; Hoteit, I. Comparison of chlorophyll in the Red Sea derived from MODIS-Aqua and in vivo fluorescence. Remote Sensing of Environment 2013, 136, 218–224.
- Racault, M.F.; Raitsos, D.E.; Berumen, M.L.; Brewin, R.J.; Platt, T.; Sathyendranath, S.; Hoteit, I. Phytoplankton phenology indices in coral reef ecosystems: Application to ocean-color observations in the Red Sea. Remote Sensing of Environment 2015, 160, 222–234. [CrossRef]
- Darecki, M.; Stramski, D. An evaluation of MODIS and SeaWiFS bio-optical algorithms in the Baltic Sea. Remote sensing of Environment 2004, 89, 326–350.
- Kowalczuk, P. Seasonal variability of yellow substance absorption in the surface layer of the Baltic Sea. Journal of Geophysical Research: Oceans 1999, 104, 30047–30058.
- Pitarch, J.; Volpe, G.; Colella, S.; Krasemann, H.; Santoleri, R. Remote sensing of chlorophyll in the Baltic Sea at basin scale from 1997 to 2012 using merged multi-sensor data. Ocean Science 2016, 12, 379–389.
- Darecki, M.; Weeks, A.; Sagan, S.; Kowalczuk, P.; Kaczmarek, S. Optical characteristics of two contrasting Case 2 waters and their influence on remote sensing algorithms. Continental Shelf Research 2003, 23, 237–250.

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




