Submitted:
24 January 2024
Posted:
25 January 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Overview of main CRISPR/Cas systems that are exploitable for both editing and non-editing uses.
CRISPR–Cas9
CRISPR–Cas12a
CRISPR/Cas13
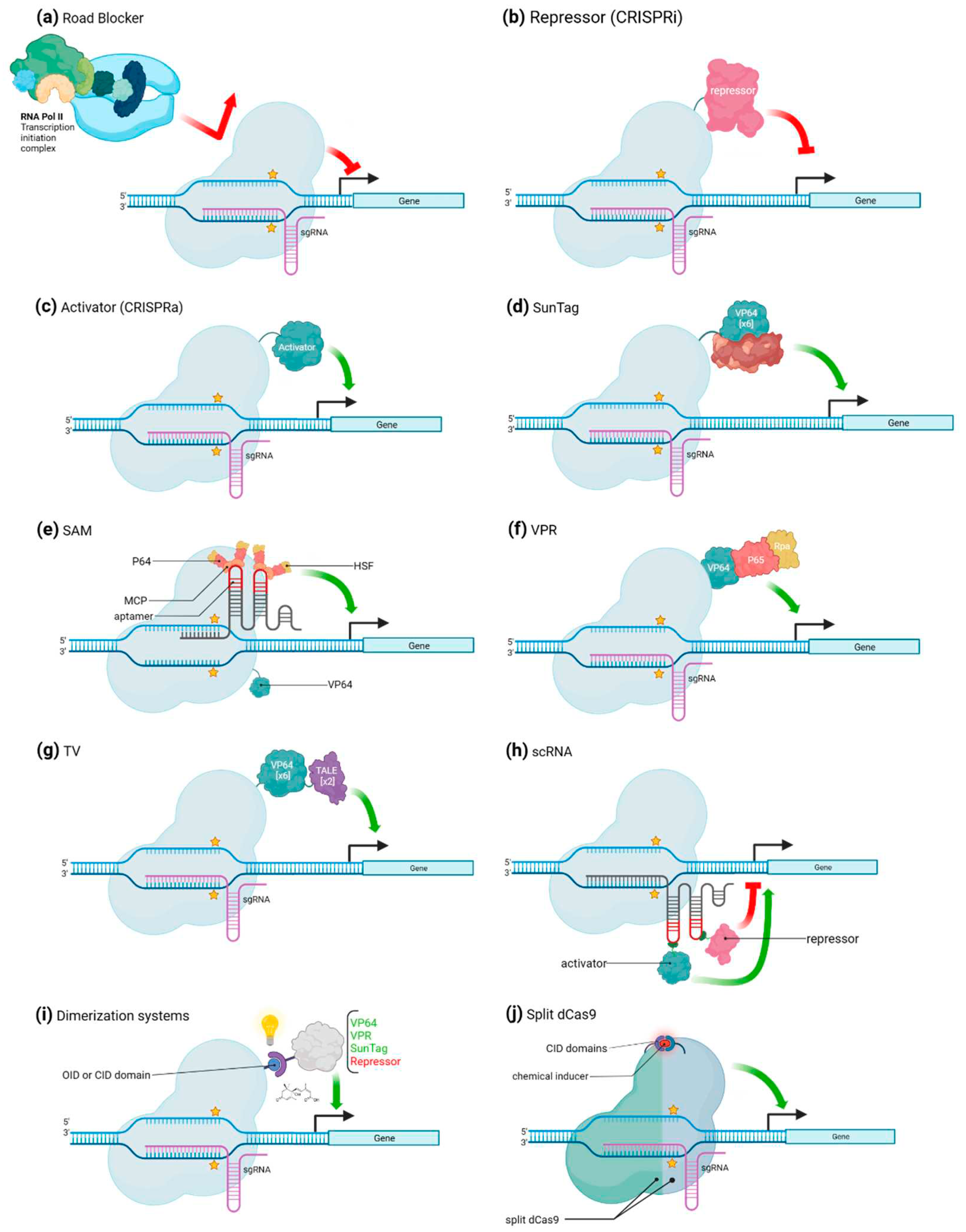
CRISPR/Cas mediated transcriptional regulation.
CRISPR/Cas mediated gene activation
CRISPR/Cas mediated gene repression
Alternative uses of CRISPR/Cas for transcriptional regulation
CRISPR/Cas in depth study of gene regulation
Study of gene regulation at the transcriptional level
Study of gene regulation at the post-transcriptional level.
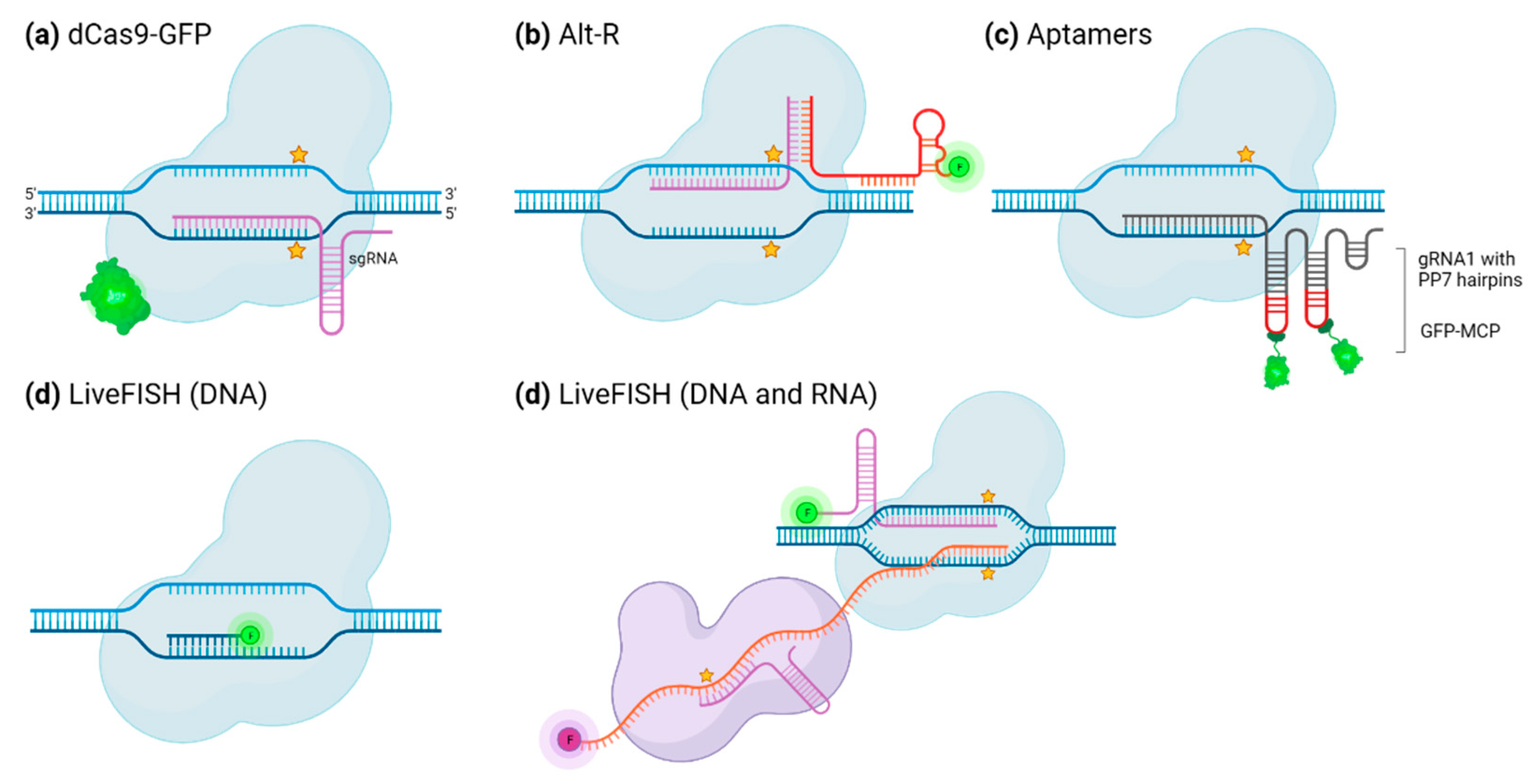
CRISPR/Cas system to image specific portions of nucleic acids in plants and animals.
| Name | Description | Organism | Type(s) of Cas protein | Advantages | Disadvantages | Performances | References |
|---|---|---|---|---|---|---|---|
| dCas9∷eGFP fusion protein | Imaging of DNA loci with a GFP-dCas9, expressed in situ along with the gRNA from transfected vectors. | Human | dCas9 | The use of an sgRNA guide with a custom scaffold reduces non-specific binding of Cas9. Possibility to label heterochromatin regions. |
Labelling of repetitive sequences as well as single loci. Tracking of telomere Dynamics in Live Cells Labelling of different positions of the same gene. Gene copy-number identification |
[62] | |
| Cas9-mediated fluorescence in situ hybridization (CASFISH) | dCas9 harbors a HaloTag flag and can be bound by fluorophores that are linked to HaloTag ligand. | Human | dCas9 | Highly stable sgRNA-dCas9-fluorophore complex High specificity Several loci can be stained at the same time: multiplexed imaging. Very quick protocol (15 minutes) Performed at room temperature |
Imaging of repetitive sequences in Detection of the allele of a certain sequence in the genome of cells at a tissue scale. Dual color Genetic diagnosis |
[63] | |
| LiveFISH | One Cas9 harbors a labeled sgRNA with a short protospacer to disable cutting and one other Cas9 harbors a normal unlabeled sgRNA | Human | Cas9, dCas9, Cas13, dCas13 | One type of Cas9 is used because the RNP complex is preassembled. Live imaging is possible. Combinable with other CRISPR/Cas system-based techniques. |
- Used to visualize and quantify the recruitment of a protein to a specific locus and study the kinetic of such recruitment. - Used to visualize translocations. - Dual labeling of DNA and RNA. |
[61] | |
| Labelling sgRNA scaffolds in animals | sgRNA carries a long 3’ scaffold that harbors aptamers, to which fluorescently labeled proteins bind. | Mouse | dCas9 | Fewer background compared to labeling with GFP-fused dCas9 because non-specific binding of dCas9 is not visible. A single vector encodes every component of the system. Live imaging is possible. Multiplex labelling |
Non-specific binding is not visible i.e., off target cannot be characterized. | Labelling of nuclear structures, repetitive sequences, and single loci. Study of chromatin dynamics during cell division. Labelling of two loci in different colors. |
[64] |
| Labelling sgRNA scaffolds in plants | sgRNA carries a long 3’ scaffold that harbors aptamers, to which fluorescently labeled proteins bind. | N. Benthamiana | dCas9 | Fewer background compared to labeling with GFP-fused dCas9 because non-specific binding of dCas9 is not visible. A single construct encodes every component of the system. This construct is inserted with A. tumefaciens-mediated transformation. Up to 2 simultaneous labeling. - Labelling efficiency is not dependent on dCas9 gene expression level |
Non-specific binding is not visible i.e., off target cannot be characterized. Lack of telomeric foci compared to FISH because of the working temperature in plants. Cannot be improved by modification of the RNA scaffold. Only repetitive sequences have yet been targeted. - Transient transformation of the construct is required. - Labelling efficiency is heavily dependent on the copy number of aptamers in the construct. |
Live imaging of telomeric repeats in plant cells. | [65] |
| RGEN-ISL/CRISPR-FISH | Imaging of loci in purified fixed nuclei using a preassembled ribonucleoprotein that contains the dCas9 and its sgRNA, for which the tracrRNA part is fused to a fluorophore for labelling. | Soybean, mouse, wheat, rye, maize, and Nicotiana benthamiana | dCas9 | No plasmid construct. No in vitro RNA synthesis. Theoretically available in any species Non disruptive technique Simple and fast Usable for repetitive sequences and single loci. |
Fixation of nuclei is required. ATT550-labeled tracrRNA and the crRNA that can bind to it must be ordered and are costly. |
Labeling of centromeric and telomeric repeats in diverse species. Optimization of sample fixation to increase labeling yield. Time-lapse-mediated study of the binding dynamics of dCas9-sgRNA-complex to DNA. |
[67,68,71] |
| mRNA imaging | dLwaCas9 is fused to GFP expressed along with the specific sgRNA using a transient vector. | Rice, mammals | dCas13 | Specific targeting of mRNA Applicable to live or fixed samples. |
Imaging of a specific gene’s to track its localization at stress-granules. |
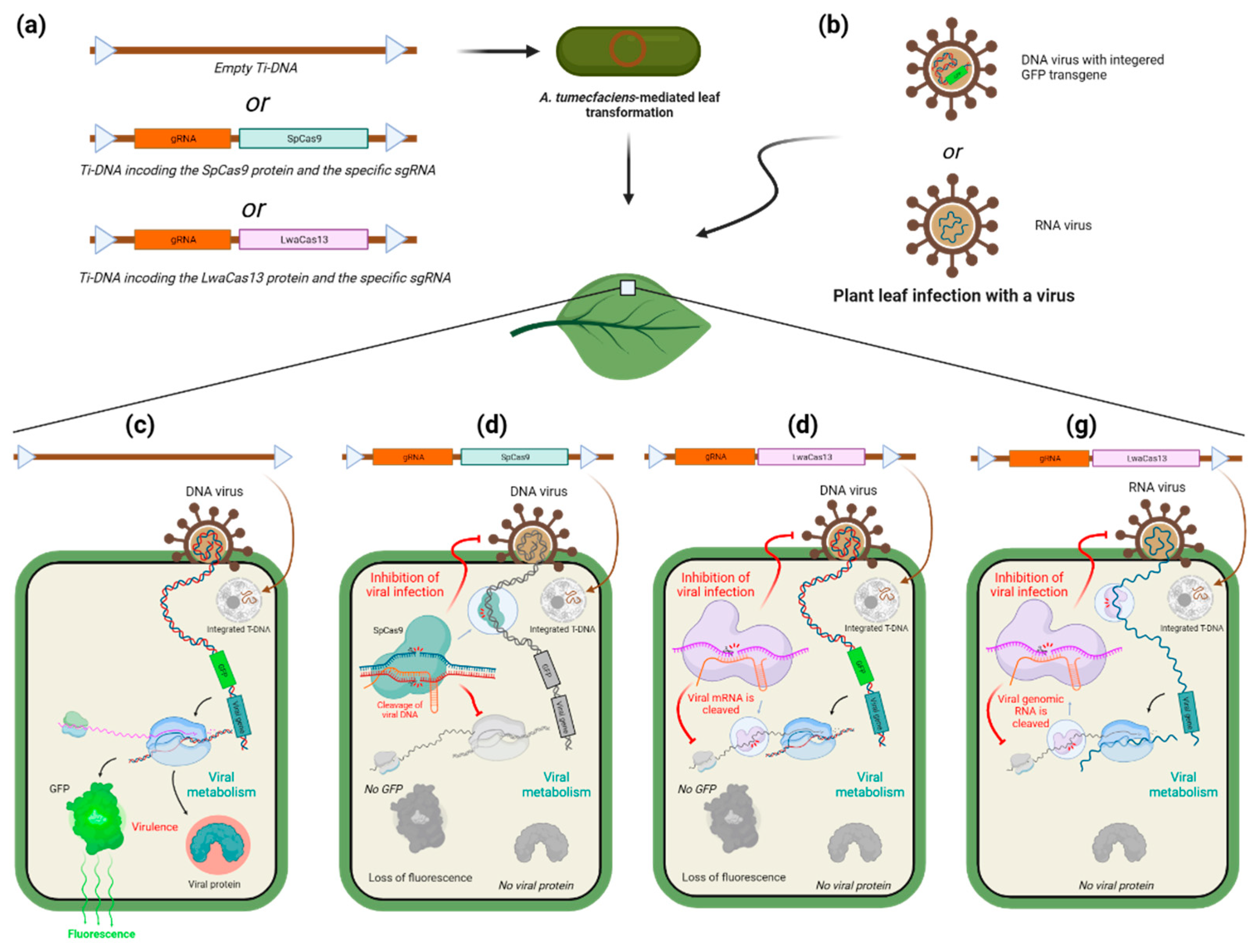
CRISPR/Cas-system as a tool to target viruses.
Use of CRISPR/Cas system to target viruses in animals.
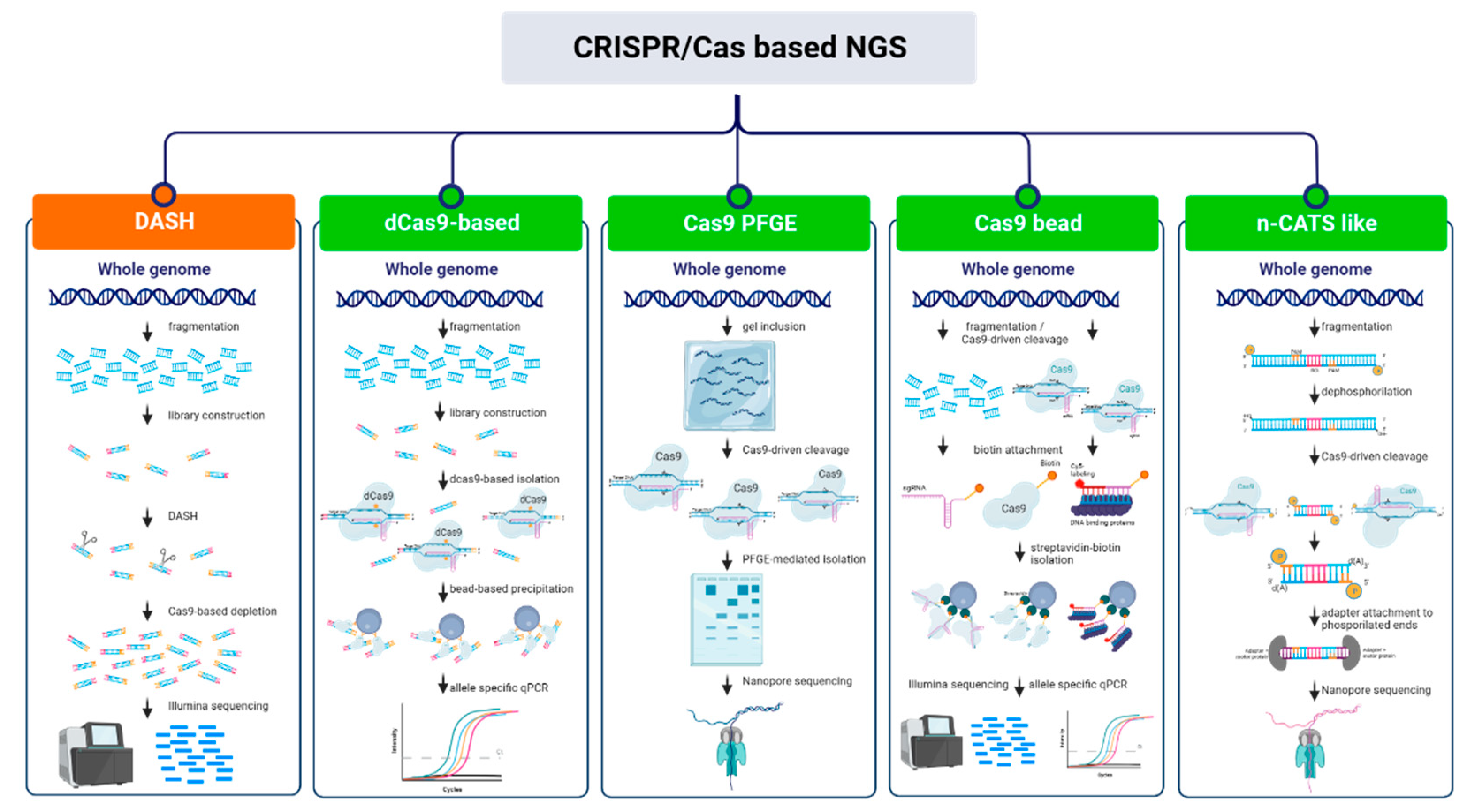
CRISPR/Cas as enrichment tool for next generation sequencing
CRISPR/Cas NGS approaches based on depletion of undesired region.
CRISPR/Cas NGS approaches based on enrichment of regions of interest (ROI)
Conclusions and future perspectives
Funding
Abbreviations
| 53BP1 | p53-binding protein 1 |
| ABA | Abscisic acid |
| CAPTURE | CRISPR affinity purification in-situ of regulatory elements |
| CASFISH | Cas9-mediated fluorescence in situ hybridization |
| CATCH | Cas9-assisted targeting of chromosome segments |
| CATE-seq | CRISPR-assisted targeted enrichment-sequencing |
| nCATS | Nanopore Cas9 targeted sequencing |
| CIDs | Chemically inducible dimerizing domains |
| CISMR | CRISPR mediated isolation of specific megabase-sized regions of the genome |
| CRISDA | CRISPR–Cas9-triggered nicking endonuclease mediated strand displacement amplification |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| CRISPR-DS | CRISPR-duplex sequencing |
| CRISPRa | CRISPR activation |
| CRISPRi | CRISPR inhibition |
| CS | Chromoshadow domain |
| CUT-PCR | CRISPR-mediated, Ultrasensitive detection of Target DNA by PCR |
| Cas | CRISPR associated protein |
| dCas | Dead Cas |
| LwaCas13 | Cas13a from Leptotrichia wadei |
| SpCas9 | Cas9 from Streptococcus pyogenes |
| dCas9VPR | dCas9 fused to VPC domain |
| DASH | Depletion of abundant sequences by hybridization |
| dsDNA | Double-stranded DNA |
| ssDNA | Single-stranded DNA |
| DSB | Double stranded breaks |
| ERT | Ligand-binding domain ERT from estrogen receptor |
| FISH | Fluorescence in-situ hybridization |
| LiveFISH | CRISPR live-cell fluorescent in situ hybridization |
| FLASH | Finding low abundance sequences by hybridization |
| GA | Gibberellin |
| GFP | Green fluorescent protein |
| eGFP | Enhanced green fluorescent protein |
| HEPN | Higher eukaryotes and prokaryotes nucleotide-binding |
| HNH | Histidine-asparagine-histidine endonuclease domain |
| HSV | Herpes simplex virus 1 |
| OIDs | Optogenetically inducible dimerizing domains |
| PAM | Protospacer associated motif |
| RGEN-ISL | RNA-guided endonuclease - in situ labeling |
| crRNA | CRISPR RNA |
| gRNA | Guide RNA |
| scRNA | Scaffold RNA |
| sgRNA | Short guide RNA |
| ssRNA | Single-stranded RNA |
| tracrRNA | Trans-activating crRNA |
| RNAP | RNA polymerase |
| RNP | Ribonucleoprotein |
| RNase | Ribonuclease |
| ROIs | Regions of interest |
| RuvC | Recombination UV C |
| SAM | Synergistic Activation Mediator |
| SHERLOCK | High-sensitivity enzymatic reporter unlocking |
| SID4X | mSin3 interaction domain |
| SMRT | Single molecule - real time |
| SRDX | SUPERMAN Repression Domain X |
| SVs | Structural variants |
| TAD | TALE transcription activation domain |
| TEIs | Transposable element insertions |
| TRV | Tobacco rattle virus |
| VPR | VP64, p65 and Rta |
References
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Pickar-Oliver, A.; Gersbach, C.A. The next Generation of CRISPR-Cas Technologies and Applications. Nat Rev Mol Cell Biol 2019, 20, 490–507. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary Classification of CRISPR-Cas Systems: A Burst of Class 2 and Derived Variants. Nat Rev Microbiol 2020, 18, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Evolutionary Classification of CRISPR-Cas Systems. In Crispr; John Wiley & Sons, Ltd, 2022; pp. 13–38 ISBN 978-1-68367-379-8. [CrossRef]
- Hryhorowicz, M.; Lipiński, D.; Zeyland, J.; Słomski, R. CRISPR/Cas9 Immune System as a Tool for Genome Engineering. Arch. Immunol. Ther. Exp. 2017, 65, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Kozovska, Z.; Rajcaniova, S.; Munteanu, P.; Dzacovska, S.; Demkova, L. CRISPR: History and Perspectives to the Future. Biomedicine & Pharmacotherapy 2021, 141, 111917. [Google Scholar] [CrossRef]
- Chen, W.; Ma, J.; Wu, Z.; Wang, Z.; Zhang, H.; Fu, W.; Pan, D.; Shi, J.; Ji, Q. Cas12n Nucleases, Early Evolutionary Intermediates of Type V CRISPR, Comprise a Distinct Family of Miniature Genome Editors. Molecular Cell 2023, 83, 2768–2780.e6. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 Structures and Mechanisms. Annual Review of Biophysics 2017, 46, 505–529. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.R.; Oakes, B.L.; Sternberg, S.H.; East-Seletsky, A.; Kaplan, M.; Doudna, J.A. Programmable RNA Recognition and Cleavage by CRISPR/Cas9. Nature 2014, 516, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.W.; Wang, H.; Yang, H.; Shi, L.; Katz, Y.; Theunissen, T.W.; Rangarajan, S.; Shivalila, C.S.; Dadon, D.B.; Jaenisch, R. Multiplexed Activation of Endogenous Genes by CRISPR-on, an RNA-Guided Transcriptional Activator System. Cell Res 2013, 23, 1163–1171. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA Editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA Targeting with CRISPR–Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic Acid Detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Gasiunas, G.; Young, J.K.; Karvelis, T.; Kazlauskas, D.; Urbaitis, T.; Jasnauskaite, M.; Grusyte, M.M.; Paulraj, S.; Wang, P.-H.; Hou, Z.; et al. A Catalogue of Biochemically Diverse CRISPR-Cas9 Orthologs. Nat Commun 2020, 11, 5512. [Google Scholar] [CrossRef]
- Nakagawa, R.; Ishiguro, S.; Okazaki, S.; Mori, H.; Tanaka, M.; Aburatani, H.; Yachie, N.; Nishimasu, H.; Nureki, O. Engineered Campylobacter Jejuni Cas9 Variant with Enhanced Activity and Broader Targeting Range. Commun Biol 2022, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a Target Binding Unleashes Indiscriminate Single-Stranded DNase Activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 Is a Single-Component Programmable RNA-Guided RNA-Targeting CRISPR Effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Zhang, N.; Xia, S.; Tian, P.; Lu, L.; Du, J.; Du, Y. Progress of CRISPR-Cas13 Mediated Live-Cell RNA Imaging and Detection of RNA-Protein Interactions. Frontiers in Cell and Developmental Biology 2022, 10. [Google Scholar] [CrossRef]
- Yu, Y.; Pan, Z.; Wang, X.; Bian, X.; Wang, W.; Liang, Q.; Kou, M.; Ji, H.; Li, Y.; Ma, D.; et al. Targeting of SPCSV-RNase3 via CRISPR-Cas13 Confers Resistance against Sweet Potato Virus Disease. Molecular Plant Pathology 2022, 23, 104–117. [Google Scholar] [CrossRef]
- Schmitz, R.J.; Grotewold, E.; Stam, M. Cis-Regulatory Sequences in Plants: Their Importance, Discovery, and Future Challenges. The Plant Cell 2022, 34, 718–741. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and Validation of Promoters and Cis-Acting Regulatory Elements. Plant Science 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable Repression and Activation of Bacterial Gene Expression Using an Engineered CRISPR-Cas System. Nucleic Acids Research 2013, 41, 7429–7437. [Google Scholar] [CrossRef]
- La Russa, M.F.; Qi, L.S. The New State of the Art: Cas9 for Gene Activation and Repression. Molecular and Cellular Biology 2015, 35, 3800–3809. [Google Scholar] [CrossRef] [PubMed]
- Morita, S.; Horii, T.; Hatada, I. Regulation of Gene Expression Using dCas9-SunTag Platforms. In Epigenomics: Methods and Protocols; Hatada, I., Horii, T., Eds.; Methods in Molecular Biology; Springer US: New York, NY, 2023; pp. 189–195. ISBN 978-1-07-162724-2. [Google Scholar] [CrossRef]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef]
- Zalatan, J.G.; Lee, M.E.; Almeida, R.; Gilbert, L.A.; Whitehead, E.H.; La Russa, M.; Tsai, J.C.; Weissman, J.S.; Dueber, J.E.; Qi, L.S.; et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell 2015, 160, 339–350. [Google Scholar] [CrossRef]
- Papikian, A.; Liu, W.; Gallego-Bartolomé, J.; Jacobsen, S.E. Site-Specific Manipulation of Arabidopsis Loci Using CRISPR-Cas9 SunTag Systems. Nature Communications 2019, 10. [Google Scholar] [CrossRef]
- Morita, S.; Horii, T.; Kimura, M.; Hatada, I. Synergistic Upregulation of Target Genes by TET1 and VP64 in the dCas9–SunTag Platform. International Journal of Molecular Sciences 2020, 21, 1574. [Google Scholar] [CrossRef]
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; P R Iyer, E.; Lin, S.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly Efficient Cas9-Mediated Transcriptional Programming. Nat Methods 2015, 12, 326–328. [Google Scholar] [CrossRef]
- Selma, S.; Bernabé-Orts, J.M.; Vazquez-Vilar, M.; Diego-Martin, B.; Ajenjo, M.; Garcia-Carpintero, V.; Granell, A.; Orzaez, D. Strong Gene Activation in Plants with Genome-Wide Specificity Using a New Orthogonal CRISPR/Cas9-Based Programmable Transcriptional Activator. Plant Biotechnology Journal 2019, 17, 1703–1705. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-Scale Transcriptional Activation by an Engineered CRISPR-Cas9 Complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, D.; Xiong, X.; Yan, B.; Xie, W.; Sheen, J.; Li, J.-F. A Potent Cas9-Derived Gene Activator for Plant and Mammalian Cells. Nature Plants 2017, 3, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W., III; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.-F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant Physiology 2015, 169, 971–985. [Google Scholar] [CrossRef]
- Piatek, A.; Ali, Z.; Baazim, H.; Li, L.; Abulfaraj, A.; Al-Shareef, S.; Aouida, M.; Mahfouz, M.M. RNA-Guided Transcriptional Regulation in Planta via Synthetic dCas9-Based Transcription Factors. Plant Biotechnology Journal 2015, 13, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Sretenovic, S.; Qi, Y. CRISPR/dCas-Mediated Transcriptional and Epigenetic Regulation in Plants. Current Opinion in Plant Biology 2021, 60, 101980. [Google Scholar] [CrossRef]
- Xu, X.; Qi, L.S. A CRISPR–dCas Toolbox for Genetic Engineering and Synthetic Biology. Journal of Molecular Biology 2019, 431, 34–47. [Google Scholar] [CrossRef]
- Bao, Z.; Jain, S.; Jaroenpuntaruk, V.; Zhao, H. Orthogonal Genetic Regulation in Human Cells Using Chemically Induced CRISPR/Cas9 Activators. ACS Synth. Biol. 2017, 6, 686–693. [Google Scholar] [CrossRef]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY Transcription Factor Family in Model Plants and Crops. Critical Reviews in Plant Sciences 2017, 36, 311–335. [Google Scholar] [CrossRef]
- Gao, Y.; Xiong, X.; Wong, S.; Charles, E.J.; Lim, W.A.; Qi, L.S. Complex Transcriptional Modulation with Orthogonal and Inducible dCas9 Regulators. Nat Methods 2016, 13, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Levskaya, A.; Weiner, O.D.; Lim, W.A.; Voigt, C.A. Spatiotemporal Control of Cell Signalling Using a Light-Switchable Protein Interaction. Nature 2009, 461, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Nihongaki, Y.; Yamamoto, S.; Kawano, F.; Suzuki, H.; Sato, M. CRISPR-Cas9-Based Photoactivatable Transcription System. Chemistry & Biology 2015, 22, 169–174. [Google Scholar] [CrossRef]
- Polstein, L.R.; Gersbach, C.A. A Light-Inducible CRISPR-Cas9 System for Control of Endogenous Gene Activation. Nat Chem Biol 2015, 11, 198–200. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.P.; Miyaoka, Y.; Gilbert, L.A.; Mayerl, S.J.; Lee, B.H.; Weissman, J.S.; Conklin, B.R.; Wells, J.A. Ligand-Binding Domains of Nuclear Receptors Facilitate Tight Control of Split CRISPR Activity. Nature Communications 2016, 7. [Google Scholar] [CrossRef]
- Tarasava, K.; Oh, E.J.; Eckert, C.A.; Gill, R.T. CRISPR-Enabled Tools for Engineering Microbial Genomes and Phenotypes. Biotechnol J 2018, 13, e1700586. [Google Scholar] [CrossRef]
- Jing, X.; Xie, B.; Chen, L.; Zhang, N.; Jiang, Y.; Qin, H.; Wang, H.; Hao, P.; Yang, S.; Li, X. Implementation of the CRISPR-Cas13a System in Fission Yeast and Its Repurposing for Precise RNA Editing. Nucleic Acids Research 2018, 46, e90. [Google Scholar] [CrossRef] [PubMed]
- Kushawah, G.; Hernandez-Huertas, L.; Abugattas-Nuñez del Prado, J.; Martinez-Morales, J.R.; DeVore, M.L.; Hassan, H.; Moreno-Sanchez, I.; Tomas-Gallardo, L.; Diaz-Moscoso, A.; Monges, D.E.; et al. CRISPR-Cas13d Induces Efficient mRNA Knockdown in Animal Embryos. Developmental Cell 2020, 54, 805–817.e7. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Qi, Y. Plant Gene Knockout and Knockdown by CRISPR-Cpf1 (Cas12a) Systems. In Plant Genome Editing with CRISPR Systems: Methods and Protocols; Qi, Y., Ed.; Methods in Molecular Biology; Springer: New York, NY, 2019; pp. 245–256. ISBN 978-1-4939-8991-1. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Chen, Y.; Li, M.; Zhou, F.; Li, K.; Cao, H.; Ni, M.; Liu, Y.; Gu, Z.; et al. In Situ Capture of Chromatin Interactions by Biotinylated dCas9. Cell 2017, 170, 1028–1043.e19. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Chen, Y.; Li, M.; Shao, Z.; Zhang, M.Q.; Xu, J. CAPTURE: In Situ Analysis of Chromatin Composition of Endogenous Genomic Loci by Biotinylated dCas9. Current Protocols in Molecular Biology 2018, 123, e64. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; He, Z.; Liu, Z.; Qu, M.; Gao, C.; Wang, C.; Wang, Y. A Reverse Chromatin Immunoprecipitation Technique Based on the CRISPR–dCas9 System. Plant Physiology 2023, 191, 1505–1519. [Google Scholar] [CrossRef]
- Orphanides, G.; Reinberg, D. A Unified Theory of Gene Expression. Cell 2002, 108, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chen, S.; Jia, G. Detection, Regulation, and Functions of RNA N6-Methyladenosine Modification in Plants. Plant Communications 2023, 4, 100546. [Google Scholar] [CrossRef] [PubMed]
- Maslova, A.; Krasikova, A. FISH Going Meso-Scale: A Microscopic Search for Chromatin Domains. Front Cell Dev Biol 2021, 9, 753097. [Google Scholar] [CrossRef]
- Khosravi, S.; Ishii, T.; Dreissig, S.; Houben, A. Application and Prospects of CRISPR/Cas9-Based Methods to Trace Defined Genomic Sequences in Living and Fixed Plant Cells. Chromosome Res 2020, 28, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, S.; Dreissig, S.; Schindele, P.; Wolter, F.; Rutten, T.; Puchta, H.; Houben, A. Live-Cell CRISPR Imaging in Plant Cells with a Telomere-Specific Guide RNA. In RNA Tagging: Methods and Protocols; Heinlein, M., Ed.; Methods in Molecular Biology; Springer US: New York, NY, 2020; pp. 343–356. ISBN 978-1-07-160712-1. [Google Scholar] [CrossRef]
- Wu, X.; Mao, S.; Ying, Y.; Krueger, C.J.; Chen, A.K. Progress and Challenges for Live-Cell Imaging of Genomic Loci Using CRISPR-Based Platforms. Genomics, Proteomics & Bioinformatics 2019, 17, 119–128. [Google Scholar] [CrossRef]
- Wang, H.; Nakamura, M.; Abbott, T.R.; Zhao, D.; Luo, K.; Yu, C.; Nguyen, C.M.; Lo, A.; Daley, T.P.; La Russa, M.; et al. CRISPR-Mediated Live Imaging of Genome Editing and Transcription. Science 2019, 365, 1301–1305. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Gilbert, L.A.; Cimini, B.A.; Schnitzbauer, J.; Zhang, W.; Li, G.-W.; Park, J.; Blackburn, E.H.; Weissman, J.S.; Qi, L.S.; et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 2013, 155, 1479–1491. [Google Scholar] [CrossRef]
- Deng, W.; Shi, X.; Tjian, R.; Lionnet, T.; Singer, R.H. CASFISH: CRISPR/Cas9-Mediated in Situ Labeling of Genomic Loci in Fixed Cells. Proceedings of the National Academy of Sciences 2015, 112, 11870–11875. [Google Scholar] [CrossRef]
- Fu, Y.; Rocha, P.P.; Luo, V.M.; Raviram, R.; Deng, Y.; Mazzoni, E.O.; Skok, J.A. CRISPR-dCas9 and sgRNA Scaffolds Enable Dual-Colour Live Imaging of Satellite Sequences and Repeat-Enriched Individual Loci. Nat Commun 2016, 7, 11707. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, S.; Schindele, P.; Gladilin, E.; Dunemann, F.; Rutten, T.; Puchta, H.; Houben, A. Application of Aptamers Improves CRISPR-Based Live Imaging of Plant Telomeres. Front Plant Sci 2020, 11, 1254. [Google Scholar] [CrossRef] [PubMed]
- Dreissig, S.; Schiml, S.; Schindele, P.; Weiss, O.; Rutten, T.; Schubert, V.; Gladilin, E.; Mette, M.F.; Puchta, H.; Houben, A. Live-Cell CRISPR Imaging in Plants Reveals Dynamic Telomere Movements. The Plant Journal 2017, 91, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Schubert, V.; Khosravi, S.; Dreissig, S.; Metje-Sprink, J.; Sprink, T.; Fuchs, J.; Meister, A.; Houben, A. RNA-Guided Endonuclease – in Situ Labelling (RGEN-ISL): A Fast CRISPR/Cas9-Based Method to Label Genomic Sequences in Various Species. New Phytologist 2019, 222, 1652–1661. [Google Scholar] [CrossRef] [PubMed]
- Potlapalli, B.P.; Schubert, V.; Metje-Sprink, J.; Liehr, T.; Houben, A. Application of Tris-HCl Allows the Specific Labeling of Regularly Prepared Chromosomes by CRISPR-FISH. Cytogenet Genome Res 2020, 160, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Jacobi, A.M.; Rettig, G.R.; Turk, R.; Collingwood, M.A.; Zeiner, S.A.; Quadros, R.M.; Harms, D.W.; Bonthuis, P.J.; Gregg, C.; Ohtsuka, M.; et al. Simplified CRISPR Tools for Efficient Genome Editing and Streamlined Protocols for Their Delivery into Mammalian Cells and Mouse Zygotes. Methods 2017, 121–122, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Nagaki, K.; Yamaji, N. Decrosslinking Enables Visualization of RNA-Guided Endonuclease–in Situ Labeling Signals for DNA Sequences in Plant Tissues. Journal of Experimental Botany 2020, 71, 1792–1800. [Google Scholar] [CrossRef] [PubMed]
- Potlapalli, B.P.; Ishii, T.; Nagaki, K.; Somasundaram, S.; Houben, A. CRISPR-FISH: A CRISPR/Cas9-Based In Situ Labeling Method. In Plant Cytogenetics and Cytogenomics: Methods and Protocols; Heitkam, T., Garcia, S., Eds.; Methods in Molecular Biology; Springer US: New York, NY, 2023; pp. 315–335. ISBN 978-1-07-163226-0. [Google Scholar] [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and Classification of the CRISPR–Cas Systems. Nat Rev Microbiol 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Manjunatha, L.; Rajashekara, H.; Uppala, L.S.; Ambika, D.S.; Patil, B.; Shankarappa, K.S.; Nath, V.S.; Kavitha, T.R.; Mishra, A.K. Mechanisms of Microbial Plant Protection and Control of Plant Viruses. Plants 2022, 11, 3449. [Google Scholar] [CrossRef]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-Mediated Viral Interference in Plants. Genome Biology 2015, 16, 238. [Google Scholar] [CrossRef]
- Kalinina, N.O.; Khromov, A.; Love, A.J.; Taliansky, M.E. CRISPR Applications in Plant Virology: Virus Resistance and Beyond. Phytopathology® 2020, 110, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Taliansky, M.; Samarskaya, V.; Zavriev, S.K.; Fesenko, I.; Kalinina, N.O.; Love, A.J. RNA-Based Technologies for Engineering Plant Virus Resistance. Plants 2021, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Karpov, D.S.; Demidova, N.A.; Kulagin, K.A.; Shuvalova, A.I.; Kovalev, M.A.; Simonov, R.A.; Karpov, V.L.; Snezhkina, A.V.; Kudryavtseva, A.V.; Klimova, R.R.; et al. Complete and Prolonged Inhibition of Herpes Simplex Virus Type 1 Infection In Vitro by CRISPR/Cas9 and CRISPR/CasX Systems. Int J Mol Sci 2022, 23, 14847. [Google Scholar] [CrossRef] [PubMed]
- Bayoumi, M.; Munir, M. Potential Use of CRISPR/Cas13 Machinery in Understanding Virus-Host Interaction. Front Microbiol 2021, 12, 743580. [Google Scholar] [CrossRef] [PubMed]
- Schultzhaus, Z.; Wang, Z.; Stenger, D. CRISPR-Based Enrichment Strategies for Targeted Sequencing. Biotechnology Advances 2021, 46, 107672. [Google Scholar] [CrossRef] [PubMed]
- Stevens, R.C.; Steele, J.L.; Glover, W.R.; Sanchez-Garcia, J.F.; Simpson, S.D.; O’Rourke, D.; Ramsdell, J.S.; MacManes, M.D.; Thomas, W.K.; Shuber, A.P. A Novel CRISPR/Cas9 Associated Technology for Sequence-Specific Nucleic Acid Enrichment. PLoS ONE 2019, 14, e0215441. [Google Scholar] [CrossRef] [PubMed]
- Ramani, V.; Shendure, J. Smash and DASH with Cas9. Genome Biol 2016, 17, 42. [Google Scholar] [CrossRef]
- Gu, W.; Crawford, E.D.; O’Donovan, B.D.; Wilson, M.R.; Chow, E.D.; Retallack, H.; DeRisi, J.L. Depletion of Abundant Sequences by Hybridization (DASH): Using Cas9 to Remove Unwanted High-Abundance Species in Sequencing Libraries and Molecular Counting Applications. Genome Biol 2016, 17, 41. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Yu, J.; Hwang, G.-H.; Kim, S.; Kim, H.S.; Ye, S.; Kim, K.; Park, J.; Park, D.Y.; Cho, Y.-K.; et al. CUT-PCR: CRISPR-Mediated, Ultrasensitive Detection of Target DNA Using PCR. Oncogene 2017, 36, 6823–6829. [Google Scholar] [CrossRef]
- Song, L.; Xie, K. Engineering CRISPR/Cas9 to Mitigate Abundant Host Contamination for 16S rRNA Gene-Based Amplicon Sequencing. Microbiome 2020, 8, 80. [Google Scholar] [CrossRef]
- Rossato, M.; Marcolungo, L.; De Antoni, L.; Lopatriello, G.; Bellucci, E.; Cortinovis, G.; Frascarelli, G.; Nanni, L.; Bitocchi, E.; Di Vittori, V.; et al. CRISPR/Cas9-Based Repeat Depletion for the High-Throughput Genotyping of Complex Plant Genomes; Genomics, 2022; 2022. [CrossRef]
- Aalipour, A.; Dudley, J.C.; Park, S.; Murty, S.; Chabon, J.J.; Boyle, E.A.; Diehn, M.; Gambhir, S.S. Deactivated CRISPR Associated Protein 9 for Minor-Allele Enrichment in Cell-Free DNA. Clinical Chemistry 2018, 64, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xia, Q.; Zhang, S.; Gao, J.; Dai, W.; Wu, J.; Wang, J. CRISPR-Assisted Targeted Enrichment-Sequencing (CATE-Seq). 2019. [CrossRef]
- Zhou, W.; Hu, L.; Ying, L.; Zhao, Z.; Chu, P.K.; Yu, X.-F. A CRISPR–Cas9-Triggered Strand Displacement Amplification Method for Ultrasensitive DNA Detection. Nat Commun 2018, 9, 5012. [Google Scholar] [CrossRef]
- Lee, J.; Lim, H.; Jang, H.; Hwang, B.; Lee, J.H.; Cho, J.; Lee, J.H.; Bang, D. CRISPR-Cap: Multiplexed Double-Stranded DNA Enrichment Based on the CRISPR System. Nucleic Acids Research 2019, 47, e1–e1. [Google Scholar] [CrossRef]
- Slesarev, A.; Viswanathan, L.; Tang, Y.; Borgschulte, T.; Achtien, K.; Razafsky, D.; Onions, D.; Chang, A.; Cote, C. CRISPR/Cas9 Targeted CAPTURE of Mammalian Genomic Regions for Characterization by NGS. Sci Rep 2019, 9, 3587. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-C.; Greenberg, D.; Powell, J.; Höijer, I.; Ameur, A.; Strahl, M.; Ellis, E.; Jonasson, I.; Mouro Pinto, R.; Wheeler, V.C.; et al. Amplification-Free, CRISPR-Cas9 Targeted Enrichment and SMRT Sequencing of Repeat-Expansion Disease Causative Genomic Regions; Genomics, 2017; [CrossRef]
- Bennett-Baker, P.E.; Mueller, J.L. CRISPR-Mediated Isolation of Specific Megabase Segments of Genomic DNA. Nucleic Acids Research 2017, 45, e165–e165. [Google Scholar] [CrossRef]
- Gabrieli, T.; Sharim, H.; Fridman, D.; Arbib, N.; Michaeli, Y.; Ebenstein, Y. Selective Nanopore Sequencing of Human BRCA1 by Cas9-Assisted Targeting of Chromosome Segments (CATCH). Nucleic Acids Research 2018, 46, e87–e87. [Google Scholar] [CrossRef] [PubMed]
- Gilpatrick, T.; Lee, I.; Graham, J.E.; Raimondeau, E.; Bowen, R.; Heron, A.; Downs, B.; Sukumar, S.; Sedlazeck, F.J.; Timp, W. Targeted Nanopore Sequencing with Cas9-Guided Adapter Ligation. Nat Biotechnol 2020, 38, 433–438. [Google Scholar] [CrossRef]
- Watson, C.M.; Crinnion, L.A.; Hewitt, S.; Bates, J.; Robinson, R.; Carr, I.M.; Sheridan, E.; Adlard, J.; Bonthron, D.T. Cas9-Based Enrichment and Single-Molecule Sequencing for Precise Characterization of Genomic Duplications. Laboratory Investigation 2020, 100, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Stangl, C.; de Blank, S.; Renkens, I.; Westera, L.; Verbeek, T.; Valle-Inclan, J.E.; González, R.C.; Henssen, A.G.; van Roosmalen, M.J.; Stam, R.W.; et al. Partner Independent Fusion Gene Detection by Multiplexed CRISPR-Cas9 Enrichment and Long Read Nanopore Sequencing. Nat Commun 2020, 11, 2861. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.V.; Kramer, M.; Goodwin, S.; McCombie, W.R. ACME: An Affinity-Based Cas9 Mediated Enrichment Method for Targeted Nanopore Sequencing 2022, 2022.02.03.478550. [CrossRef]
- Kirov, I.; Merkulov, P.; Gvaramiya, S.; Komakhin, R.; Omarov, M.; Dudnikov, M.; Kocheshkova, A.; Soloviev, A.; Karlov, G.; Divashuk, M. Illuminating the Transposon Insertion Landscape in Plants Using Cas9-Targeted Nanopore Sequencing and a Novel Pipeline 2021, 2021.06.11.448052. [CrossRef]
- Steele, J.L.; Stevens, R.C.; Cabrera, O.A.; Bassill, G.J.; Cramer, S.M.; Guzman, F.; Shuber, A.P. Novel CRISPR-Based Sequence Specific Enrichment Methods for Target Loci and Single Base Mutations. PLoS ONE 2020, 15, e0243781. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Langelier, C.; Kuchta, A.; Batson, J.; Teyssier, N.; Lyden, A.; Caldera, S.; McGeever, A.; Dimitrov, B.; King, R.; et al. FLASH: A next-Generation CRISPR Diagnostic for Multiplexed Detection of Antimicrobial Resistance Sequences. Nucleic Acids Research 2019, 47, e83–e83. [Google Scholar] [CrossRef] [PubMed]
- McDonald, T.L.; Zhou, W.; Castro, C.P.; Mumm, C.; Switzenberg, J.A.; Mills, R.E.; Boyle, A.P. Cas9 Targeted Enrichment of Mobile Elements Using Nanopore Sequencing. Nat Commun 2021, 12, 3586. [Google Scholar] [CrossRef] [PubMed]
- Merkulov, P.; Gvaramiya, S.; Komakhin, R.; Omarov, M.; Dudnikov, M.; Kocheshkova, A.; Konstantinov, Z.; Soloviev, A.; Karlov, G.; Divashuk, M.; et al. Cas9-Targeted Nanopore Sequencing Rapidly Elucidates the Transposition Preferences and DNA Methylation Profiles of Mobile Elements in Plants; Genomics, 2021; [CrossRef]
- Nachmanson, D.; Lian, S.; Schmidt, E.K.; Hipp, M.J.; Baker, K.T.; Zhang, Y.; Tretiakova, M.; Loubet-Senear, K.; Kohrn, B.F.; Salk, J.J.; et al. Targeted Genome Fragmentation with CRISPR/Cas9 Enables Fast and Efficient Enrichment of Small Genomic Regions and Ultra-Accurate Sequencing with Low DNA Input (CRISPR-DS). Genome Res. 2018, 28, 1589–1599. [Google Scholar] [CrossRef] [PubMed]
- Bruijnesteijn, J.; van der Wiel, M.; de Groot, N.G.; Bontrop, R.E. Rapid Characterization of Complex Killer Cell Immunoglobulin-Like Receptor (KIR) Regions Using Cas9 Enrichment and Nanopore Sequencing. Frontiers in Immunology 2021, 12. [CrossRef]
- Rubben, K.; Tilleman, L.; Deserranno, K.; Tytgat, O.; Deforce, D.; Nieuwerburgh, F.V. Cas9 Targeted Nanopore Sequencing with Enhanced Variant Calling Improves CYP2D6-CYP2D7 Hybrid Allele Genotyping. PLOS Genetics 2022, 18, e1010176. [Google Scholar] [CrossRef]
- Lopatriello, G.; Maestri, S.; Alfano, M.; Papa, R.; Di Vittori, V.; De Antoni, L.; Bellucci, E.; Pieri, A.; Bitocchi, E.; Delledonne, M.; et al. CRISPR/Cas9-Mediated Enrichment Coupled to Nanopore Sequencing Provides a Valuable Tool for the Precise Reconstruction of Large Genomic Target Regions. International Journal of Molecular Sciences 2023, 24, 1076. [Google Scholar] [CrossRef]
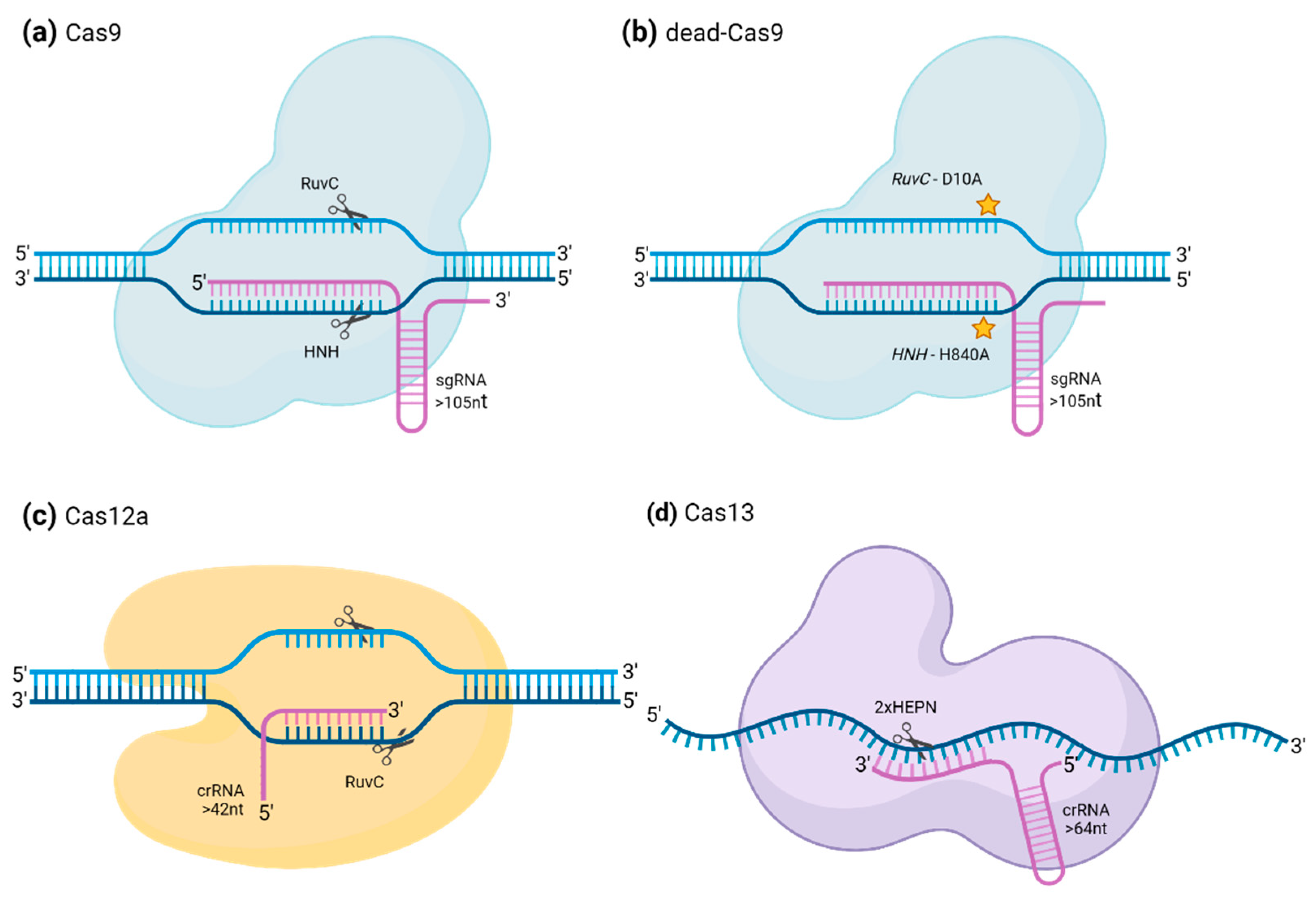
| Features | Cas9 | Cas12a | Cas13 |
|---|---|---|---|
| Other names | - | Cpf1 | C2c2 |
| Type of CRISPR/Cas system | Class 2 type II | Class 2 type V | Class 2 type VI |
| Size (aminoacids) | 1,368 (SpCas9) | 1,307 (AaCas12a) | 1389 (LshCas13a) |
| Nuclease domain | RuvC and HNH | RuvC | HEPN (×2) |
| Mutations inducing loss of function in Nuclease domain. | D10A and H840A | - | D474A and D1046A |
| sgRNA components | crRNA and tracrRNA | crRNA | crRNA |
| sgRNA crRNA processing | tracrRNA-dependent | tracrRNA-independent | - |
| sgRNA protospacer length (nucleotides) | 20 (minimum ensure DNA cleavage) | 20 | 22 - 28 |
| sgRNA total length (nucleotides) | >105 | >42 | >140 |
| Targeted nuclei acid | dsDNA (can be induced to cleave ssRNA) | dsDNA, ssDNA (not cleavable) | ssRNA |
| PAM sequence (5’-3’) | NGG (SpCas9) | T-reachTTTV (AsCas12a, LbCas12a) | None |
| Cleavage | Blunt ended double-stranded break, 3 nucleotides before PAM sequence. Each nuclease domain cleaves one strand. | PAM-distal dsDNA break with staggered 5’ and 3’ ends | Single mismatches may be tolerated. Cleavage patterns depend on features of the target sequence (like accessibility) rather than the distance from the binding site. |
| Other properties | - | Non-targeted ssDNA cleaving activity upon recognition of target sequence | Non-targeted ssRNA cleaving activity upon recognition of target sequence |
| Non-editing applications presented in this review | Modulation of gene expression and regulation Viral DNA targeting In situ DNA imaging New sequencing techniques |
Modulation of gene expression and regulation Viral DNA targeting |
In situ RNA imaging Viral RNA targeting Gene post-transcriptional regulation RNA detection techniques |
| References | [2,4,7,8] | [2,4,9] | [4,10,11,12] |
| Name | Description | Organism | CRISPR/Cas system | Type of regulation | Performance | References |
|---|---|---|---|---|---|---|
| VP64 | Single activator (VP16 or p65) | Mammalian cells and budding yeast | dCas9 | CRISPRa | Between 2- and 5-fold | [15,25] |
| SunTag | Tandem array of peptides which recruits several copies of VP64 | HEK293 and U2OS cells. Arabidopsis thaliana | dCas9 | CRISPRa | Up to 50-fold | [27,29] |
| VPR | Tripartite peptide composed by the VP64, p65 and Rta activators placed in a specific order to maximize gene activation | HEK293T and Neuro-2A cells. Nicotiana benthamiana | dCas9 | CRISPRa | Up to 300-fold | [31,32] |
| SAM | VP64 and sgRNA with two MS2 on turn fused to p65 and HSF1 | HEK293FT and Neuro-2a cells | dCas9 | CRISPRa | Variable | [33] |
| TV | Six copies of TAD motif and two copies of the VP64 activator | HEK293T cells. Arabidopsis thaliana and Oryza sativa | dCas9 | CRISPRa | Variable | [34] |
| Road blocker | Steric hamper due to simple bound of dCas9 | E. coli and mammalian | dCas9 | CRISPRi | Depends on organism | [16] |
| Transcriptional repressors | KRAB, CS, WPRW, SID4X, 3xSRDX and SRDX domains | Mammalian cells. Arabidopsis thaliana and Nicotiana benthamiana | dCas9 | CRISPRi | Between 40 and 99 % | [35,36,37,38,39] |
| scRNA | Differential regulation (both activation or repression) of a set of gene targets simultaneously | Human cells | dCas9 | Both | NA | [28] |
| Dimerization systems | Spatial and temporal control of gene function through sense input signals and generate functional outputs | HEK293T cells, mice and Avena sativa | dCas9 | Both | NA | [2,41,43] |
| Split dCas9 | Fusing ligand-binding domains of nuclear receptors to split Cas9 protein fragments can provide chemical control over split Cas9 activity | HEK293T cells. | dCas10 | Both | NA | [47] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





