Submitted:
20 January 2024
Posted:
22 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
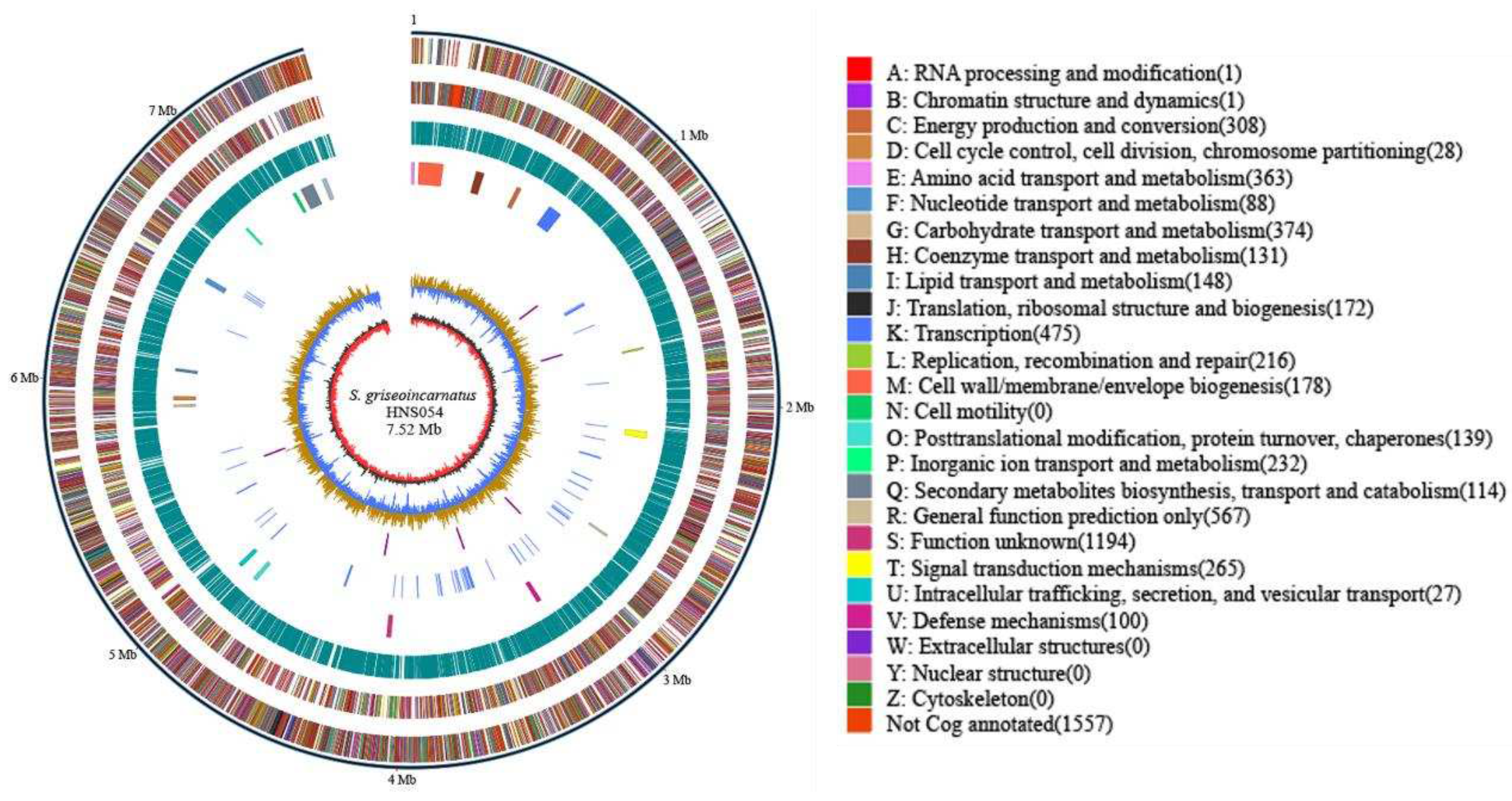
2.1. Genomic Features and Annotation of HNS054
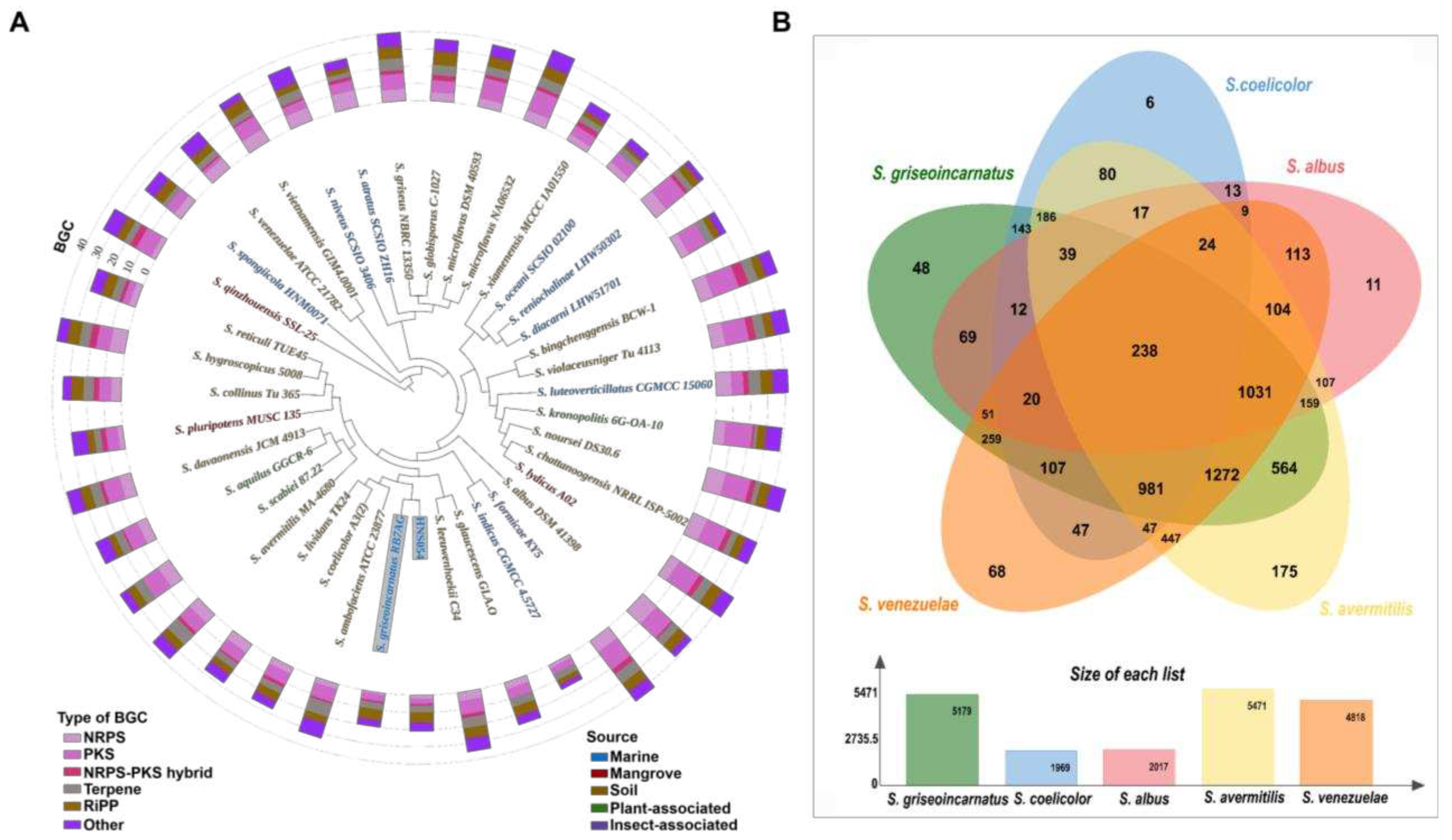
2.2. Comparative Genome and Pan-Genome Analysis
2.3. Analysis of Secondary Metabolite BGCs
2.4. Analysis of SSR Systems in Strain HNS054
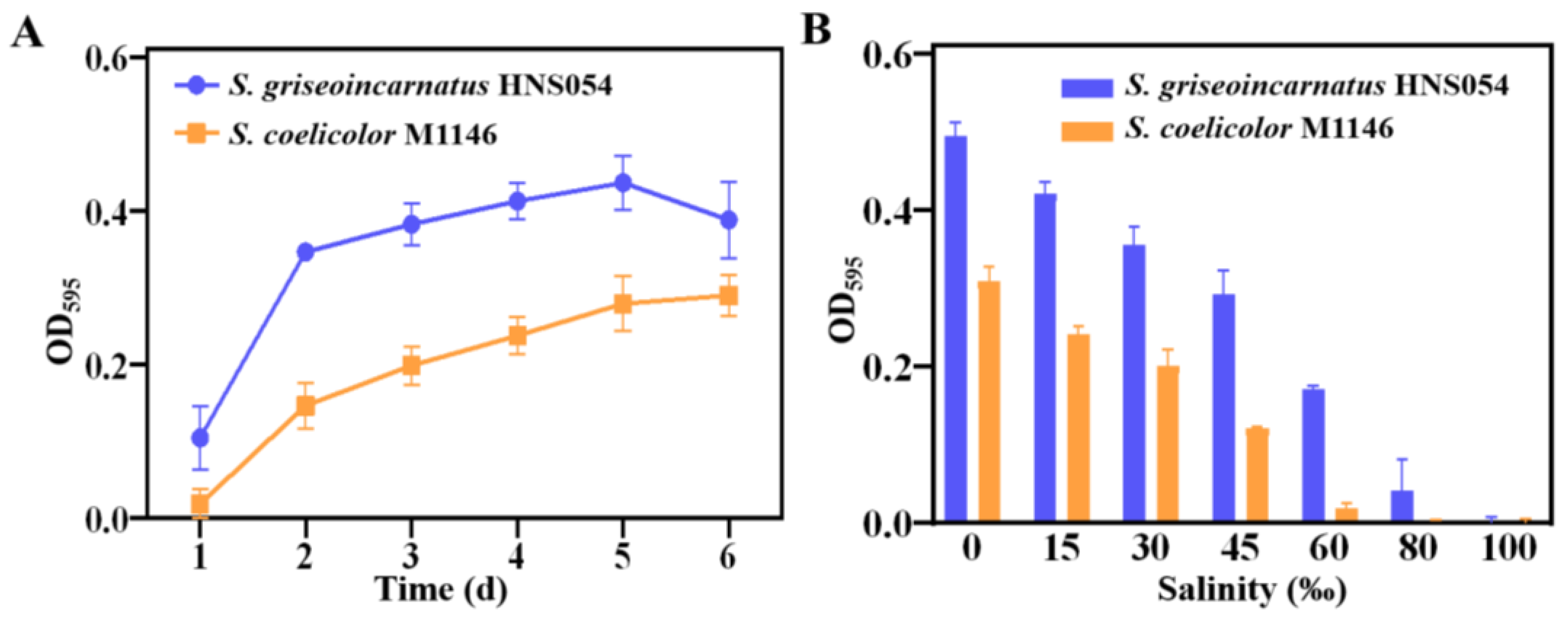
2.5. Growth, Salt Tolerance, and Antibiotic Susceptibility of Strain HNS054
2.6. Development of MSGE Hosts from Strain HNS054 Using CRISPR/Cas9 Methodology
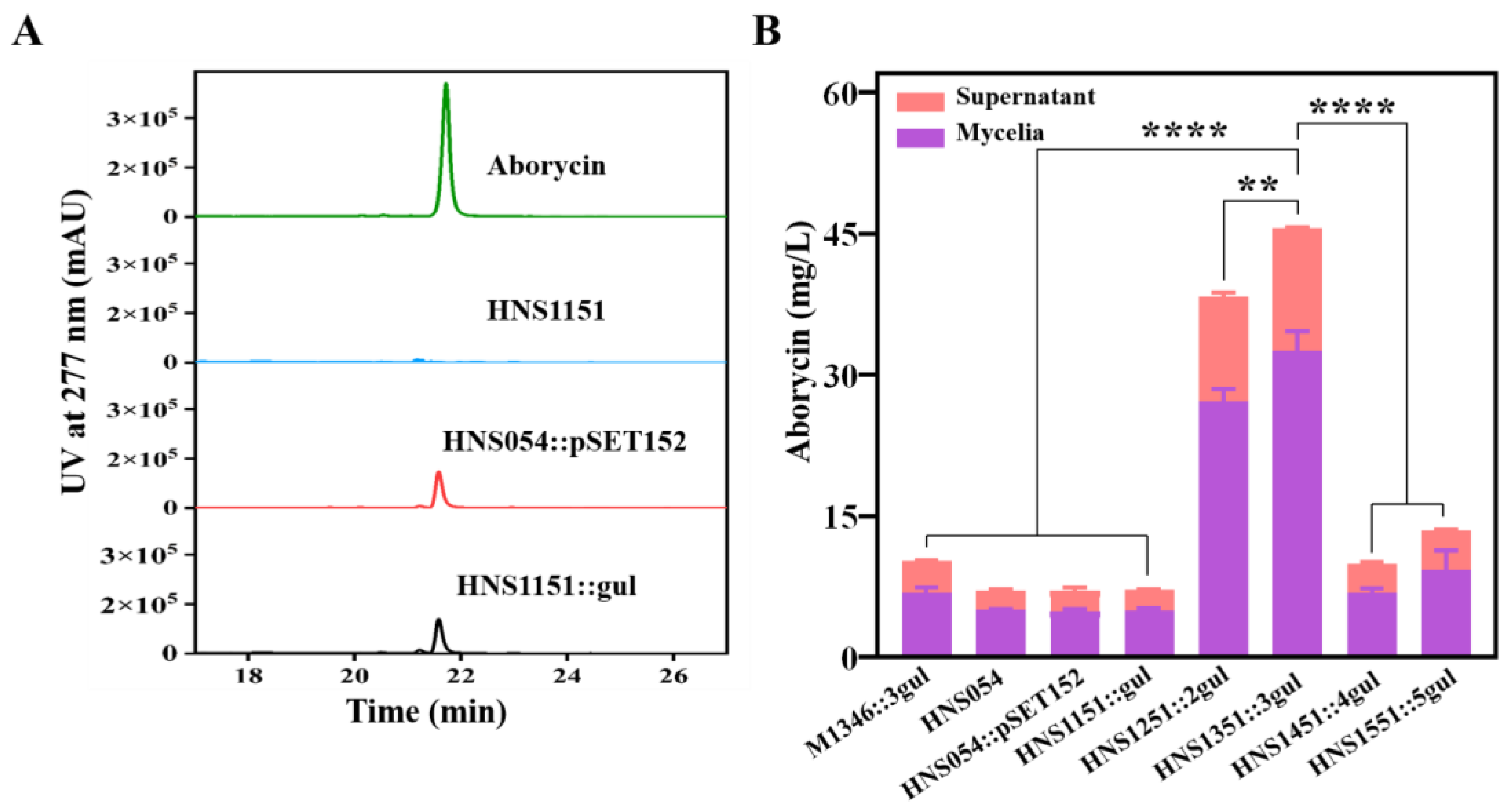
2.7. Improvement of Aborycin Production
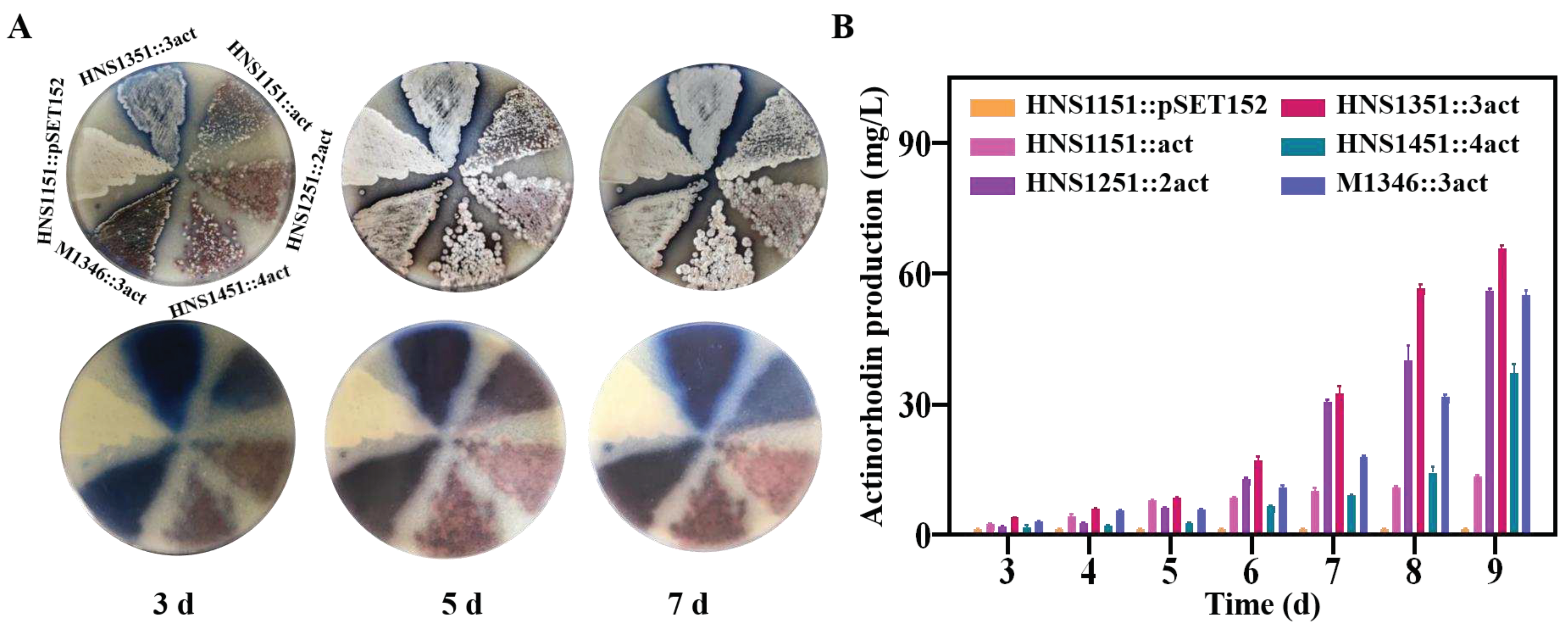
2.8. Improvement of Actinorhodin Production
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, Primers, and Culture Conditions
4.2. Genome Sequencing, Assembly and Annotation
4.3. Comparative Genomic Analysis
4.4. Assessment of Growth, Salt Tolerance, and Antibiotic Sensitivity
4.5. Construction of the HNS054-derived Hosts
- Amplification of Upstream and Downstream Regions: PCR amplification of the upstream and downstream regions of the targeted BGCs (e.g., BGC11, BGC14, BGC17, BGC2) using specific primer pairs (e.g., Del-BGC11-up-B-fwd/rev, Del-BGC11-down-B-fwd/rev).
- Amplification of sgRNA Expression Cassette: PCR amplification of the sgRNA expression cassette using a plasmid template (e.g., pKCcas9dO) and specific primers (e.g., BGC11-sgRNA-B-fwd, sgRNA-rev).
- Gibson Assembly: assembly of the three DNA fragments (upstream region, downstream region, sgRNA expression cassette) using the Gibson assembly method to generate plasmids (e.g., pKY01dB11::attB).
- Plasmid Introduction: introduction of the constructed plasmids into the HNS054-derived hosts through conjugal transfer on MS solid media.
- Verification of Double Crossovers: verification of correct double crossovers in the transformed strains by PCR using specific primers (e.g., ID-BGC11-B-fwd/rev).
4.6. Production of Aborycin
4.7. Production of Actinorhodin
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paoli, L.; Ruscheweyh, H.-J.; Forneris, C. C.; Hubrich, F.; Kautsar, S.; Bhushan, A.; Lotti, A.; Clayssen, Q.; Salazar, G.; Milanese, A. Biosynthetic potential of the global ocean microbiome. Nature 2022, 607, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Jose, P. A.; Maharshi, A.; Jha, B. Actinobacteria in natural products research: Progress and prospects. Microbiol. Res. 2021, 246, 126708. [Google Scholar] [CrossRef] [PubMed]
- Harir, M.; Bendif, H.; Bellahcene, M.; Fortas, Z.; Pogni, R. Streptomyces Secondary Metabolites. In Basic Biology and Applications of Actinobacteria; Enany, S., Ed.; IntechOpen: London, UK, 2018; pp. 99–122. [Google Scholar]
- Gomez-Escribano, J. P.; Bibb, M. J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 2011, 4, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Y.; Rebets, Y.; Estévez, M. R.; Zapp, J.; Myronovskyi, M.; Luzhetskyy, A. Engineering of Streptomyces lividans for heterologous expression of secondary metabolite gene clusters. Microb. Cell. Fact. 2020, 19, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Gao, G.; Lu, J.; Long, Q.; Chen, X.; Zhang, F.; Xu, M.; Liu, K.; Wang, Y.; Deng, Z.; et al. Engineered Streptomyces lividans strains for optimal identification and expression of cryptic biosynthetic gene clusters. Front. Microbiol. 2018, 9, 3042. [Google Scholar] [CrossRef] [PubMed]
- Myronovskyi, M.; Rosenkränzer, B.; Nadmid, S.; Pujic, P.; Normand, P.; Luzhetskyy, A. Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metab. Eng. 2018; 49, 316–324. [Google Scholar]
- Komatsu, M.; Komatsu, K.; Koiwai, H.; Yamada, Y.; Kozone, I.; Izumikawa, M.; Hashimoto, J.; Takagi, M.; Omura, S.; Shin-ya, K.; et al. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth. Biol. 2013, 2, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. J.; Moore, B. S.; Tang, X. Engineering Salinispora tropica for heterologous expression of natural product biosynthetic gene clusters. Appl. Microbiol. Biotechnol. 2018, 102, 8437–8446. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, C.; Wang, Y.; Chen, Y.; Li, Q.; Zhang, Y.; Chen, Q.; Ju, J.; Ma, J. MGCEP 1.0: A genetic-engineered marine-derived chassis cell for a scaled heterologous expression platform of microbial bioactive metabolites. ACS Synth. Biol. 2022, 11, 3772–3784. [Google Scholar] [CrossRef] [PubMed]
- Komatsua, M.; Uchiyama, T.; Ōmura, S.; Cane, D. E.; Ikeda, H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. USA 2010, 107, 2646–2651. [Google Scholar] [CrossRef]
- Hwang, S.; Lee, Y.; Kim, J. H.; Kim, G.; Kim, H.; Kim, W.; Cho, S.; Palsson, B. O.; Cho, B.-K. Streptomyces as microbial chassis for heterologous protein expression. Front. Bioeng. Biotechnol. 2021, 9, 1–23. [Google Scholar] [CrossRef]
- Myronovskyi, M.; Luzhetskyy, A. Genome engineering in actinomycetes using site-specific recombinases. Appl. Microbiol. Biotechnol. 2013, 97, 4701–4712. [Google Scholar] [CrossRef] [PubMed]
- Nah, H.-J.; Park, J.; Choi, S.; Kim, E.-S. WblA, a global regulator of antibiotic biosynthesis in Streptomyces. J. Ind. Microbiol. Biotechnol. 2021, 48, kuab007. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liang, J.; Liu, Z.; Yi, Y.; Zhao, J.; Huang, Z.; Chen, J. Multicopy chromosome integration and deletion of negative global regulators significantly increased the heterologous production of aborycin in Streptomyces coelicolor. Mar. Drugs 2023, 21, 534. [Google Scholar] [CrossRef]
- Su, P.; Wang, D.; Ding, S.; Zhao, J. Isolation and diversity of natural product biosynthetic genes of cultivable bacteria associated with marine sponge Mycale sp. from the coast of Fujian, China. Can. J. Microbiol. 2014; 60, 217–225. [Google Scholar]
- Liu, T.; Huang, Z.; Gui, X.; Xiang, W.; Jin, Y.; Chen, J.; Zhao, J. Multi-omics comparative analysis of Streptomyces mutants obtained by iterative atmosphere and room-temperature plasma mutagenesis. Front. Microbiol. 2021, 11, 630309. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S. D.; Chater, K. F.; Cerdeño-Tárraga, A. M.; Challis, G. L.; Thomson, N. R.; James, K. D.; Harris, D. E.; Quail, M. A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wei, K.; Liu, X.; Wu, Y.; Zheng, G.; Chen, S.; Jiang, W.; Lu, Y. aMSGE: advanced multiplex site-specific genome engineering with orthogonal modular recombinases in actinomycetes. Metab. Eng. 2019, 52, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Li, Y.; Xie, H.; Wang, J.; Li, Z.; Chen, X.; Mao, X.; Li, Y. Comprehensive dissection of dispensable genomic regions in Streptomyces based on comparative analysis approach. Microb. Cell. Fact. 2020, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, C.; Liu, C.; Ju, J.; Ma, J. Genome sequencing of Streptomyces atratus SCSIOZH16 and activation production of nocardamine via metabolic engineering. Front. Microbiol. 2018, 9, 1269. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Wang, J.; Bu, X.; Yu, H.; Li, P.; Ou, H.; He, Y.; Xu, F.; Hu, X.; Zhu, X.; et al. Deciphering the streamlined genome of Streptomyces xiamenensis 318 as the producer of the anti-fibrotic drug candidate xiamenmycin. Sci. Rep. 2016, 6, 18977. [Google Scholar] [CrossRef]
- Yano, K.; Wada, T.; Suzuki, S.; Tagami, K.; Matsumoto, T.; Shiwa, Y.; Ishige, T.; Kawaguchi, Y.; Masuda, K.; Akanuma, G.; et al. Multiple rRNA operons are essential for efficient cell growth and sporulation as well as outgrowth in Bacillus subtilis. Microbiology 2013, 159, 2225–2236. [Google Scholar] [CrossRef]
- Ye, J.; Chen, G. Halomonas as a chassis. Essays Biochem. 2021, 65, 393–403. [Google Scholar] [PubMed]
- Pikl, Š.; Carrillo Rincón, A. F.; Slemc, L.; Goranovič, D.; Avbelj, M.; Gjuračić, K.; Sucipto, H.; Stare, K.; Baebler, Š.; Šala, M.; et al. Multiple copies of the oxytetracycline gene cluster in selected Streptomyces rimosus strains can provide significantly increased titers. Microb. Cell. Fact. 2021, 20, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zheng, G.; Chen, J.; Ge, M.; Jiang, W.; Lu, Y. Multiplexed site-specific genome engineering for overproducing bioactive secondary metabolites in actinomycetes. Metab. Eng. 2017, 40, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Concepcion, G. T.; Feng, X.; Zhang, H.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Lindgreen, S. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res. Notes 2012, 5, 337. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xiang, S.; Dai, X.; Yang, K. A simplified diphenylamine colorimetric method for growth quantification. Appl. Microbiol. Biotechnol. 2013, 97, 5069–5077. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fu, Y.; Wang, M.; Niu, G. Synthetic cellobiose-inducible regulatory systems allow tight and dynamic controls of gene expression in Streptomyces. ACS Synth. Biol. 2021, 10, 1956–1965. [Google Scholar] [CrossRef]
- Demir, Z.; Bayraktar, A.; Tunca, S. One extra copy of ion gene causes a dramatic increase in actinorhodin production by Streptomyces coelicolor A3(2). Curr. Microbiol. 2019, 76, 1045–1054. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




