Submitted:
12 January 2024
Posted:
12 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- FAOSTAT. 2015. Available online: URL http://www.faostat.fao.org (accessed on 15 December 2023).
- Glance, H. Horticulture statistics division department of agriculture. Cooperation & Farmers ‘Welfare Ministry of Agriculture and Farmers’ Welfare Government of India. 2018. Available online: https://www.nhb.gov.in (accessed on 15 December 2023).
- Rao G.P., Chaturvedi Y., Priya M., Mall S. Association of a 16SrII group phytoplasma with dieback disease of papaya in India. Bull. Insect. 2011, 64(Suppl.), S105-S106.
- Contaldo N., Satta E., Zambon Y., Paltrinieri S., Bertaccini A. Development and evaluation of different complex media for phytoplasma isolation and growth. J. Microbiol. Meth. 2016, 127, 105–110. [CrossRef] [PubMed]
- Lee I-M., Gundersen-Rindal D.E., Davis R.E., Bartoszyk I.M. (1998). Revised classification scheme of phytoplasmas based on RFLP analyses of 16S rRNA and ribosomal protein gene sequences. Int. J. Syst. Evol. Microbiol. 1998, 48(4), 1153–1169. [CrossRef]
- Bertaccini A., Arocha-Rosete Y., Contaldo N., Duduk B., Fiore N., Guglielmi Montano H., Kube M., Kuo C-H., Martini M., Oshima K., Quaglino F., Schneider B., Wei W., Zamorano, A. Revision of the ‘Candidatus Phytoplasma’ species description guidelines. Int. J. Syst. Evol. Microbiol. 2022, 72, 005353. [CrossRef]
- Bertaccini, A. Plants and phytoplasmas: when bacteria modify plants. Plants 2022, 11(11), 1425. [Google Scholar] [CrossRef] [PubMed]
- Majumdar S., Das B.K. Studies on little leaf of brinjal and morphotaxonomy of the leafhopper species associated from Bengal. J. Entomol. Zool. 2020, 8, 514–521.
- Kumar M., Rao G.P. Molecular characterization, vector identification and sources of phytoplasmas associated with brinjal little leaf disease in India. 3 Biotech 2017, 7(1), 1-11. [CrossRef]
- Satta E. Studies on phytoplasma seed transmission in different biological systems. PhD thesis, 2017, University of Bologna, Italy.
- Khan A.J., Botti S., Paltrinieri S., Al-Subhi A.M., Bertaccini A. Phytoplasmas in alfalfa seedlings: infected or contaminated seedling?. 14th Int. Org. Mycoplasmol. Conf., Wien (Austria) 7-12, July 2002, 148.
- Calari A., Paltrinieri S., Contaldo N., Sakalieva D., Mori N., Duduk B., Bertaccini A. Molecular evidence of phytoplasmas in winter oilseed rape, tomato and corn seedlings. Bull. Insect. 2011, 64(Suppl.), S157-S158.
- Chung B.N., Jeong M.I. Identification of “stolbur” phytoplasmas in Petunia hybrida seedlings. Phytopath. Moll. 2014, 4(1), 5-8. [CrossRef]
- Mateeti S.T., Checchi G., Messina N.A., Feduzi G., Bertaccini A., Contaldo N. (2022). Presence and seed transmission of phytoplasmas in tomato fields in Italy. Phytopath. Moll. 2022, 12(1), 1–6. [CrossRef]
- Satta E., Carminati G., Bertaccini A. Phytoplasma presence in carrot seedlings. Austral. Plant Dis. Notes 2020, 15, 11. [CrossRef]
- Okuda S., Prince J.P., Davis R.E., Dally E.L., Lee I-M., Mogen B., Kato S. Two groups of phytoplasmas from Japan distinguished on the basis of amplification and restriction analysis of 16S rDNA. Plant Dis. 1997, 81(3), 301–305. [CrossRef]
- Kelly P.L., Arocha-Rosete Y., Dider S.Z. First report of a 16SrI ‘Candidatus Phytoplasma asteris’ isolate affecting eggplant and Mikania sp. in Bangladesh. Plant Pathol. 2009, 58(4), 789.
- Kumar J., Gunapati S., Singh S.P., Lalit A., Sharma N.C., Tuli R. First report of a ‘Candidatus Phytoplasma asteris’(16SrI group) associated with little leaf disease of Solanum melongena (brinjal) in India. New Dis. Rep. 2012, 26(21). [CrossRef]
- Al-Subhi A.M., Al-Saady N.A., Khan A.J., Deadman M.L. First report of a group 16SrII phytoplasma associated with witches’ broom of eggplant in Oman. Plant Dis. 2011, 95, 360. [CrossRef]
- Omar A.F., Foissac X. Occurrence and incidence of phytoplasmas of the 16SrII-D subgroup on solanaceous and cucurbit crops in Egypt. Eur. J. Pl. Pathol. 2012, 133(2), 353-360. [CrossRef]
- Kumar M. (2015). Genetic diversity and natural spread sources of brinjal little leaf phytoplasma. Doctoral dissertation, MSc thesis. IARI, New Delhi.
- Yadav V., Mahadevakumar S., Tejaswini G.S., Shilpa N., Sreenivasa M.Y., Amruthavalli C., Janardhana G.R. First report of 16SrII-D phytoplasma associated with eggplant big bud (Solanum melongena L.) in India. Plant Dis. 2016, 100(2), 517. [CrossRef]
- Barros T.S.L., Kitajima E.W., Resende R.O. (1998). Diversidade de isolados brasileiros de fitoplasmas através da análise do 16S rDNA. Fitopat. Brasil. 1998, 23(4), 459-465.
- Mello A., Eckstein B., Flores D., Kreyci P.F., Bedendo I.P. (2011). Identification by computer-simulated RFLP of phytoplasmas associated with eggplant giant calyx representative of two subgroups, a lineage of 16SrIII-J and the new subgroup 16SrIII-U. Int. J. Syst. Evol. Microbiol. 2011, 61(6), 1454-1461. [CrossRef]
- Sertkaya G., Martini M., Musetti R., Osler R. Detection and molecular characterization of phytoplasmas infecting sesame and solanaceous crops in Turkey. Bull. Insectol. 2007, 60(2), 141–142.
- Azadvar M., Baranwal V.K. Multilocus sequence analysis of phytoplasma associated with brinjal little leaf disease and its detection in Hishimonus phycitis in India. Phytopath. Moll. 2012, 2(1), 15–21. [CrossRef]
- Tohidi Z., Salehi M., Ghasemi S., Khanchezar A., Shahamiri S.M. Association of a 16SrIX-C phytoplasma with eggplant phyllody in Iran. J. Crop Prot. 2015, 4(2), 247-256.
- Ember I., Acs Z., Munyaneza J.E., Crosslin J.M., Kolber M. Survey and molecular detection of phytoplasmas associated with potato in Romania and southern Russia. Eur. J. Pl. Pathol. 2011, 130(3), 367–377. [CrossRef]
- Usta M., Güller A., Sipahioğlu H.M. Detection, in silico analysis and molecular diversity of phytoplasmas from solanaceous crops in Turkey. Plant Protect. Sci. 2022, 58, 31-39. [CrossRef]
- Naik D.V.K., Reddy B.B., Rani J.S., Jayalakshmi Devi R.S., Prasad K.H. Molecular characterization of phytoplasma associated with crops, weeds and forest trees in Andhra Pradesh, India. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 781-791.
- +Gawande P.Y., Karthikeyan M., Johnson I., Swarnapriya R., Boopathi N.M. Overview of little leaf disease in eggplant in Tamil Nadu. The Pharma Innov. J. 2022, SP-11(9), 2178-2182.
- Kalaria R.K., Ghanghas S., Patel A. I. Molecular detection of ‘Candidatus Phytoplasma’ associated with little leaf disease in Brinjal from Southern Gujarat region of India. J. Entomol. Zool. Stud. 2019, 7(4), 794-797.
- Maheshwari M., Kumar M., Rao G.P. Identification and characterization of phytoplasma associated with brinjal little leaf, phyllody and witches’ broom disease in India. Ind. Phytopath. 2017, 70(2), 258-261.
- Vandana Y., Mahadevakumar S., Janardhana G.R., Amruthavalli C., Sreenivasa M.Y. Molecular detection of ‘Candidatus Phytoplasma trifolii’ associated with little leaf of brinjal from Kerala state of Southern India. Int. J. Life Sci. 2015, 9(6), 109-112. [CrossRef]
- Srilatha, V., Reddy, P.K., Priya, P.R., Reddy, M.G. Molecular identification of phytoplasmas associated with little leaf disease of brinjal in Andhra Pradesh and Telangana states of India. Phytopath. Moll. 2021, 11(1), 31–35. [CrossRef]
- Snehi S.K., Parihar S.S., Jain B. (2021). First report of a jujube witches’ broom phytoplasma (16SrV) strain associated with witches’ broom and little leaf disease of Solanum melongena in India. New Dis. Rep. 2021, 43, e120.
- Dutta D.S., Kalita M.K., Nath P.D. Detection, characterization and management of brinjal little leaf disease in Assam. J. Environ. Biol. 2022, 43(3), 460-467.
- Venkataravanappa V., Kodandaram M.H., Manjunath M., Chauhan N.S., Nagendran K., Tiwari S.K., Sarkar B., Rao G.P. (2021). Molecular characterization of phytoplasma strains associated with brinjal little leaf and screening of cultivated and wild relatives of eggplant cultivars for disease resistance. Eur. J. Pl. Pathol. 2021, 162(2), 433–453.
- Bogoutdinov D.Z., Valyunas D., Navalinskene M., Samuitene M. (2008). About specific identification of phytoplasmas in Solanaceae crops. Agricult. Biol. 2008, 1, 77-80.
- Rao G.P., Kumar M. (2017). World status of phytoplasma diseases associated with eggplant. Crop Protect. 2017, 96, 22–29. [CrossRef]
- Mateeti S., Darabakula M., Contaldo N., Pacini F., Bertaccini A. Seed transmission of phytoplasmas infecting eggplants in India. Phytopath. Moll. 2023, 13(1), 57–58. [CrossRef]
- Menon K.P.V., Pandalai K.M. The Coconut Palm, A monograph 133. 1960, Indian Central Coconut Committee, Ernakulam, India.
- Bertaccini A., Marani F. Electron microscopy of two viruses and mycoplasma-like organisms in lilies with deformed flowers. Phytopath. Medit. 1982, 21, 8–14.
- Cordova I., Jones P., Harrison N.A., Oropeza C. (2003). In situ PCR detection of phytoplasma DNA in embryos from coconut palms with lethal yellowing disease. Mol. Pl. Pathol. 2003, 4(2), 99–108. [CrossRef]
- Nipah J.O., Jones P., Hodgetts J., Dickinson M. Detection of phytoplasma DNA in embryos from coconut palms in Ghana, and kernels from maize in Peru. Bull. Insect. 2007, 60(2), 385-386.
- Nečas T., Mašková V., Krška B. The possibility of ESFY phytoplasma transmission through flowers and seeds. Acta Hortic. 2005, 781, 443–448. [CrossRef]
- Seruga-Music M., Vrek I., Skoric D. (2004). Dianthus croaticus Borb _a new host for phytoplasma from ribosomal groups 16SrI and 16SrIII. Proc. 15th Int. Congr. Mycoplasmol. IOM, Athens, USA, 2004, 122-123.
- Botti, S., Bertaccini, A. (2006). Phytoplasma infection through seed transmission: further observations. Proc. 16th Int. Int. Congr. Mycoplasmol. IOM, Cambridge, UK, 2006, 76.
- Zwolinska A., Krawczyk K., Pospieszny H. First report of “stolbur” phytoplasma infecting pea plants. Proc. 18th Int. Int. Congr. Mycoplasmol. IOM, Chianciano Terme, Italy, 2010, 11, 16.
- Çağlar B.K., Satar S., Bertaccini A., Elbeaino T. Detection and seed transmission of Bermudagrass phytoplasma in maize in Turkey. J. Phytopath. 2019, 167(4), 248–255. [CrossRef]
- Satta E., Paltrinieri S., Bertaccini A. Phytoplasma transmission by seed. In Phytoplasmas: Plant Pathogenic Bacteria-II Transmission and Management of Phytoplasma-Associated Diseases. Eds. Bertaccini A., Weintraub P.G, Rao G.P, Mori N. Springer, Singapore, 2019; Volume 2, pp. 131-147. [CrossRef]
- Vijay Kumar N.D., Bhaskara R.B.V., Sailaja R.J., Sarada Jayalakshmi D.R., Hari Prasa, K.V. ‘Candidatus Phytoplasma trifolii’ associated with little leaf disease of Solanum melongena (brinjal) in Andhra Pradesh, India. J. Pharmacogn. and Phytochem. 2018, 7(3), 3695–3697.
- Salehi M., Esmaeilzadeh-Hosseini S.A., Salehi E., Bertaccini A. Molecular diversity of phytoplasmas associated with eggplant phyllody disease in Iran. Eur. J. Pl. Pathol. 2021, 161, 195–205. [CrossRef]
- Angelini E., Clair D., Borgo M., Bertaccini A., Boudon-Padieu E. “Flavescence dorée” in France and Italy-occurrence of closely related phytoplasma isolates and their near relationships to Palatinate grapevine yellows and an alder yellows phytoplasma. Vitis 2001, 40(2), 79-86.
- Lee I-M., Bertaccini A., Vibio M., Gundersen D.E. Detection of multiple phytoplasmas in perennial fruit trees with decline symptoms in Italy. Phytopathology 1995, 85(6), 728–735. [CrossRef]
- Lee I-M., Gundersen D.E., Hammond R.W., Davis R.E. Use of mycoplasmalike organism (MLO) group-specific oligonucleotide primers for nested-PCR assays to detect mixed-MLO infections in a single host plant. Phytopathology 1994, 84(6), 559-566. [CrossRef]
- Lorenz K H., Schneider B., Ahrens U., Seemüller E. Detection of the apple proliferation and pear decline phytoplasmas by PCR amplification of ribosomal and nonribosomal DNA. Phytopathology 1995, 85(7), 771-776. [CrossRef]
- Gibb K.S., Padovan A.C., Mogen B.D. Studies on sweet potato little-leaf phytoplasma detected in sweet potato and other plant species growing in Northern Australia. Phytopathology 1995, 85(2), 169-174. [CrossRef]
- Bertaccini A., Paltrinieri S., Contaldo N. Standard detection protocol: PCR and RFLP analyses based on 16S rRNA gene. In Phytoplasmas. Humana Press, New York, NY, USA, 2019; pp. 83-95. [CrossRef]
- Jukes T.H., Cantor C.R. Evolution of protein molecules. In: Mammalian Protein Metabolism, Munro HN, editor, Academic Press, New York USA, 1969; pp. 21-132. [CrossRef]
- Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [CrossRef] [PubMed]
- Hodgetts J., Boonham N., Mumford R., Harrison N., Dickinson M. 2008. Phytoplasma phylogenetics based on analysis of secA and 23S rRNA gene sequences for improved resolution of candidate species of ‘Candidatus Phytoplasma’. Int. J. Syst. Evol. Microbiol. 2008, 58, 1826-183. [CrossRef]
- Abeysinghe S., Abeysinghe P., Kanatiwela de Silva C., Udagama P.V., Warawichanee K., Aljafar N., Kawicha P., Dickinson M. Refinement of the taxonomic structure of 16SrXI and 16SrXIV phytoplasmas of gramineous plants using multilocus sequence typing. Plant Dis. 2016, 100(10), 2001-2010. [CrossRef]

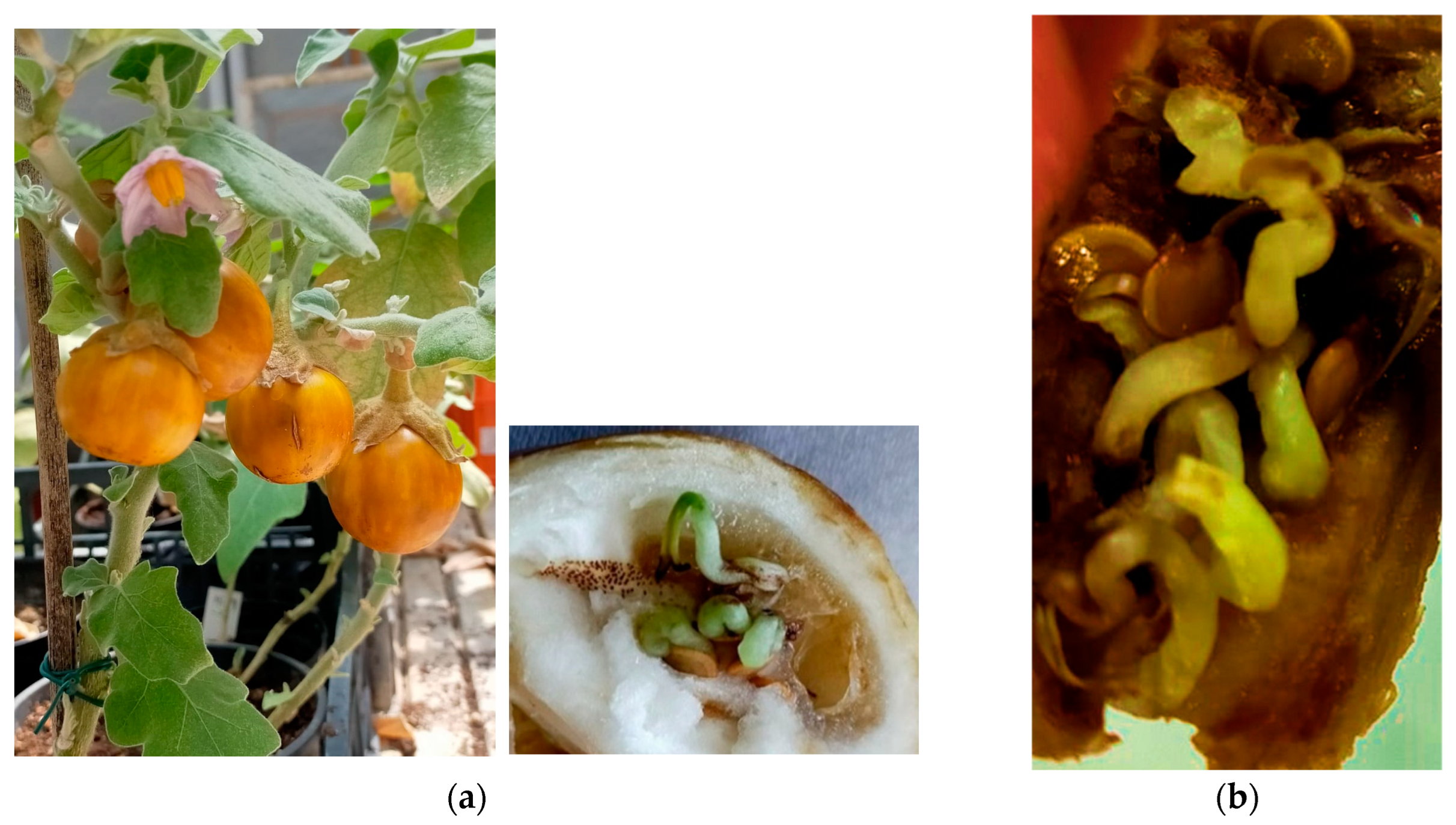
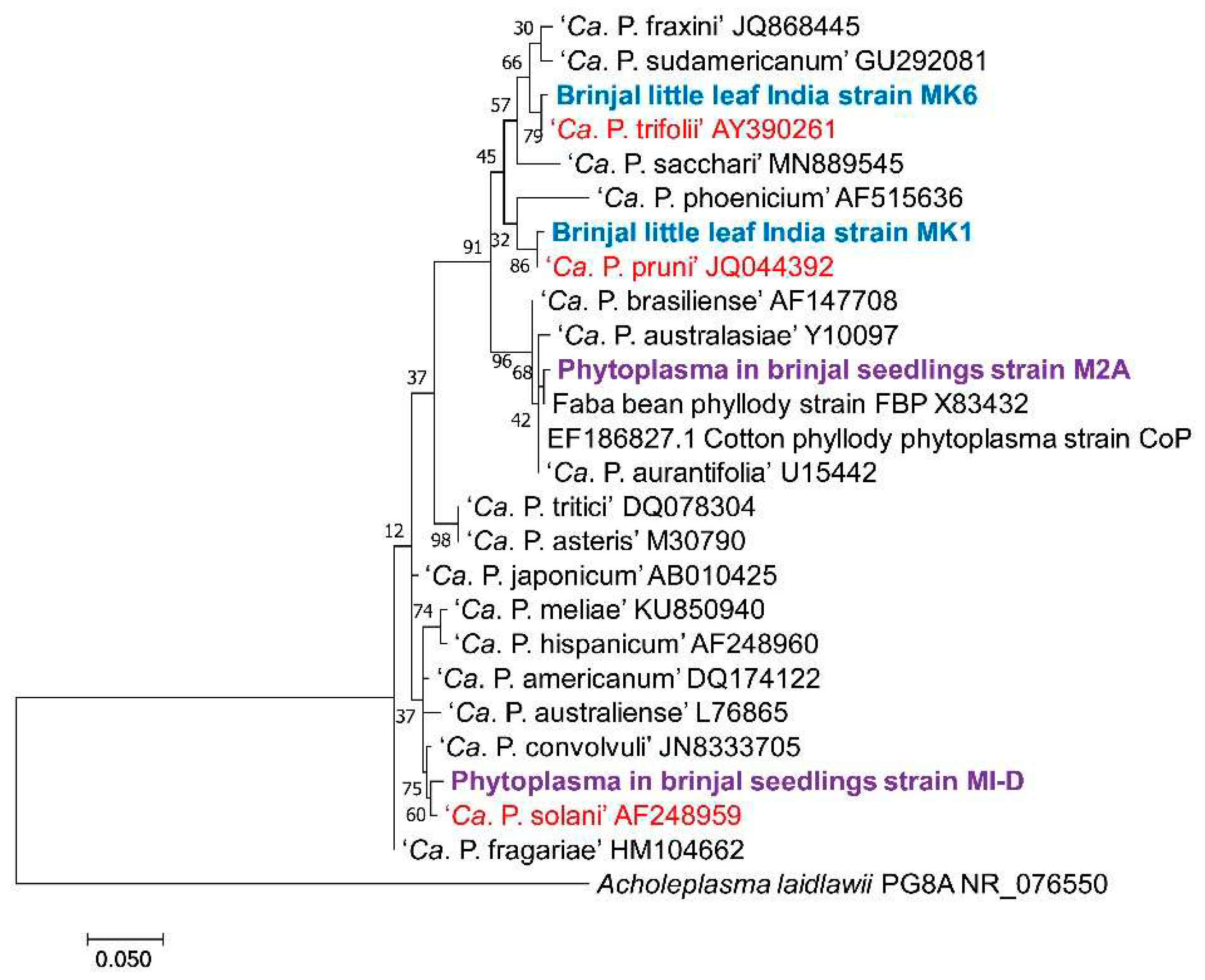
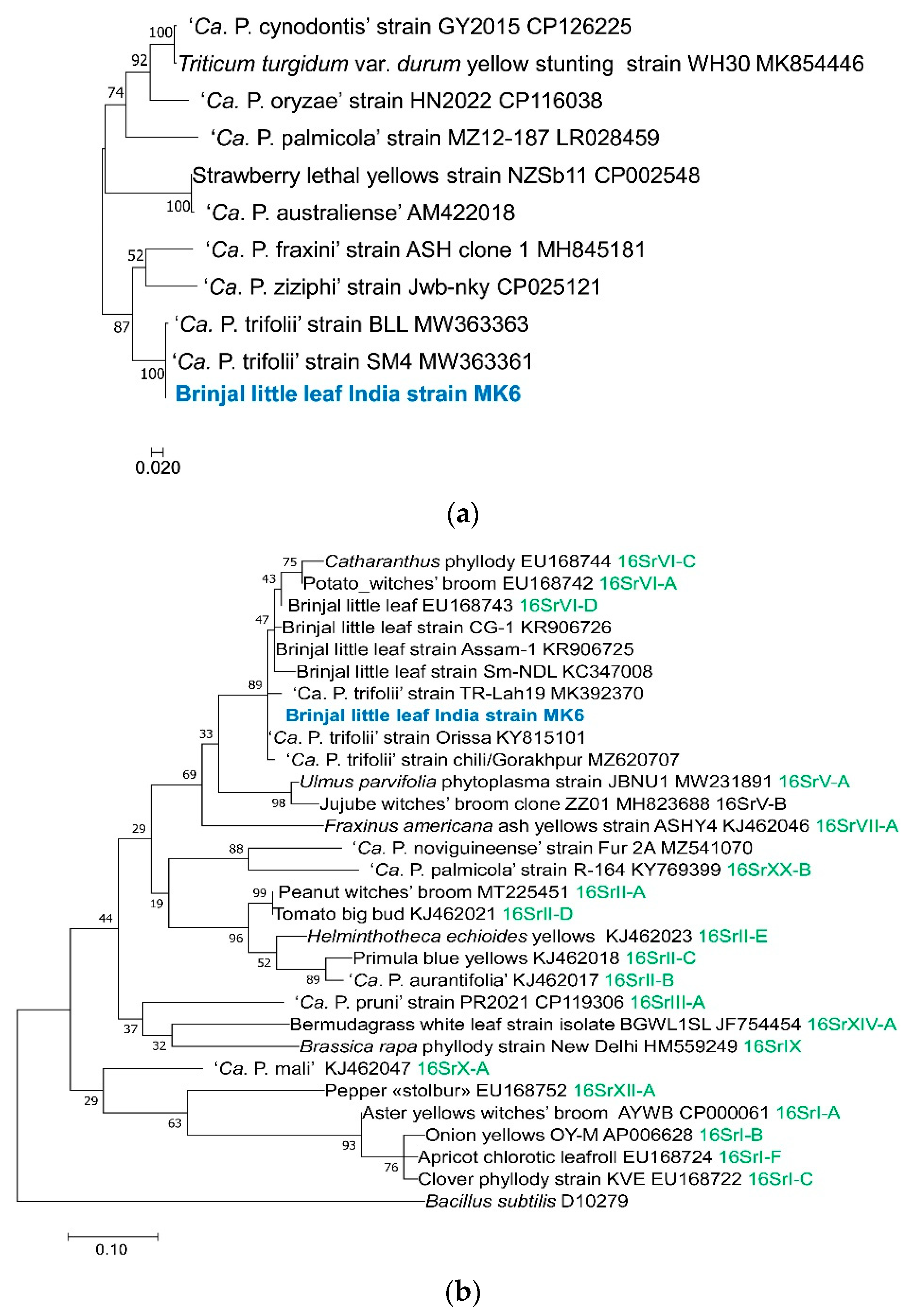
| Seed batches | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Average |
|---|---|---|---|---|---|---|---|---|---|
| Germinated seeds (%) | 92 | 92 | 93 | 89 | 93 | 81 | 95 | 98 | 91.63% |
| Transplanted seedlings (%) | 97 | 90 | 93 | 90 | 97 | 90 | 87 | 80 | 91.00% |
| Seedlings | Phytoplasma | Second generation seedlings | Phytoplasma |
|---|---|---|---|
| M1.B | 16SrI | M1.B(N/G), M1.B(SM), M1.B(SM) | 16SrI |
| M3.D, M2.5, M8.18 | 16SrII | M1.A (N/G) | 16SrI |
| M2-M20 | 16SrV | M1.C(G) | 16SrI |
| M3.B | 16SrVI | M3.D(G) | 16SrXII |
| M1.D, M2.A | 16SrXII | M5.A(G) | 16SrXII |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





