Submitted:
10 January 2024
Posted:
10 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- F. Lebreton, R.J. Willems, and M.S. Gilmore, Enterococcus diversity, origins in nature, and gut colonization. in: M.S. Gilmore, D.B. Clewell, Y. Ike, and N. Shankar, (Eds.), Enterococci: from commensals to leading causes of drug resistant infection [Internet], Massachusetts Eye and Ear Infirmary, Boston, MA, 2014, pp. 1-46.
- Z. Zhong, W. Zhang, Y. Song, W. Liu, H. Xu, X. Xi, B. Menghe, H. Zhang, and Z. Sun, Comparative genomic analysis of the genus Enterococcus. Microbiol. Res. 196 (2017) 95-105. [CrossRef]
- S.M. Naser, M. Vancanneyt, E. De Graef, L.A. Devriese, C. Snauwaert, K. Lefebvre, B. Hoste, P. Švec, A. Decostere, and F. Haesebrouck, Enterococcus canintestini sp. nov., from faecal samples of healthy dogs. Int. J. Syst. Evol. Microbiol. 55 (2005) 2177-2182. [CrossRef]
- G.K. Walker, M.M. Suyemoto, S. Gall, L. Chen, S. Thakur, and L.B. Borst, The role of Enterococcus faecalis during co-infection with avian pathogenic Escherichia coli in avian colibacillosis. Avian Pathol. 49 (2020) 589-599. [CrossRef]
- V. Sistek, A.F. Maheux, M. Boissinot, K.A. Bernard, P. Cantin, I. Cleenwerck, P. De Vos, and M.G. Bergeron, Enterococcus ureasiticus sp. nov. and Enterococcus quebecensis sp. nov., isolated from water. Int. J. Syst. Evol. Microbiol. 62 (2012) 1314-1320. [CrossRef]
- M.F. Moreno, P. Sarantinopoulos, E. Tsakalidou, and L. De Vuyst, The role and application of enterococci in food and health. Int. J. Food. Microbiol. 106 (2006) 1-24. [CrossRef]
- T. O’Driscoll, and C.W. Crank, Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infection and drug resistance (2015) 217-230. [CrossRef]
- R.C. Moellering Jr, Emergence of Enterococcus as a significant pathogen. Clin. Infect. Dis. (1992) 1173-1176. [CrossRef]
- C.R. Jackson, P.J. Fedorka-Cray, and J.B. Barrett, Use of a Genus- and Species-Specific Multiplex PCR for Identification of Enterococci. J. Clin. Microbiol. 42 (2004) 3558-3565. [CrossRef]
- L.B. Borst, M.M. Suyemoto, A.H. Sarsour, M.C. Harris, M.P. Martin, J.D. Strickland, E.O. Oviedo, and H.J. Barnes, Pathogenesis of Enterococcal Spondylitis Caused by Enterococcus cecorum in Broiler Chickens. Vet. Pathol. 54 (2017) 61-73. [CrossRef]
- A. Jung, L.R. Chen, M.M. Suyemoto, H.J. Barnes, and L.B. Borst, A Review of Enterococcus cecorum Infection in Poultry. Avian Dis. 62 (2018) 261-271. [CrossRef]
- C.M. Logue, C.B. Andreasen, L.B. Borst, H. Eriksson, D.J. Hampson, S. Sanchez, and R.M. Fulton, Other Bacterial Diseases. in: D.E. Swayne, M. Boulianne, C.M. Logue, L.R. McDougald, V. Nair, D.L. Suarez, S. Wit, T. Grimes, D. Johnson, M. Kromm, T.Y. Prajitno, I. Rubinoff, and G. Zavala, (Eds.), Diseases of Poultry, John Wiley & Sons, 2020, pp. 995-1085.
- L.B. Borst, M.M. Suyemoto, E.H. Scholl, F.J. Fuller, and H.J. Barnes, Comparative Genomic Analysis Identifies Divergent Genomic Features of Pathogenic Enterococcus cecorum Including a Type IC CRISPR-Cas System, a Capsule Locus, an epa-Like Locus, and Putative Host Tissue Binding Proteins. PLoS One 10 (2015) e0121294. [CrossRef]
- I.T. Paulsen, L. Banerjei, G.S. Myers, K.E. Nelson, R. Seshadri, T.D. Read, D.E. Fouts, J.A. Eisen, S.R. Gill, J.F. Heidelberg, H. Tettelin, R.J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R.T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K.A. Ketchum, B.A. Dougherty, and C.M. Fraser, Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299 (2003) 2071-4. [CrossRef]
- K.L. Palmer, P. Godfrey, A. Griggs, V.N. Kos, J. Zucker, C. Desjardins, G. Cerqueira, D. Gevers, S. Walker, J. Wortman, M. Feldgarden, B. Haas, B. Birren, and M.S. Gilmore, Comparative Genomics of Enterococci: Variation in Enterococcus faecalis, Clade Structure in E. faecium, and Defining Characteristics of E. gallinarum and E. casseliflavus. mBio 3 (2012) 10.1128/mbio.00318-11. [CrossRef]
- J. Laurentie, V. Loux, C. Hennequet-Antier, E. Chambellon, J. Deschamps, A. Trotereau, S. Furlan, C. Darrigo, F. Kempf, J. Lao, M. Milhes, C. Roques, B. Quinquis, C. Vandecasteele, R. Boyer, O. Bouchez, F. Repoila, J.L. Guennec, H. Chiapello, R. Briandet, E. Helloin, C. Schouler, I. Kempf, and P. Serror, Comparative Genome Analysis of Enterococcus cecorum Reveals Intercontinental Spread of a Lineage of Clinical Poultry Isolates. mSphere 8 (2023) e00495-22. [CrossRef]
- M. Arango, A. Forga, J. Liu, G. Zhang, L. Gray, R. Moore, M. Coles, A. Atencio, C. Trujillo, J.D. Latorre, G. Tellez-Isaias, B. Hargis, and D. Graham, Characterizing the impact of Enterococcus cecorum infection during late embryogenesis on disease progression, cecal microbiome composition, and early performance in broiler chickens. Poult. Sci. 102 (2023) 103059. [CrossRef]
- M.J. Kense, and W.J.M. Landman, Enterococcus cecorum infections in broiler breeders and their offspring: molecular epidemiology. Avian Pathol. 40 (2011) 603-612.
- G. Dunnam, J. Thornton, and M. Pulido-Landinez, An Emerging Enterococcus cecorum Outbreak in a Broiler Integrator in the Southern US: Analysis of Antimicrobial resistance Trends. Proceeedings of the 71st Western Poultry Disease Conference 2022 (2022) 55-61.
- C.R. Hodak, D.M. Bescucci, K. Shamash, L.C. Kelly, T. Montina, P.B. Savage, and G.D. Inglis, Antimicrobial Growth Promoters Altered the Function but Not the Structure of Enteric Bacterial Communities in Broiler Chicks ± Microbiota Transplantation. Animals 13 (2023) 997. [CrossRef]
- J. Sambrook, E.F. Fritsch, and T. Maniatis, Molecular cloning: a laboratory manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA, 1989.
- N.S. Ekesi, A. Hasan, A. Alrubaye, and D. Rhoads, Analysis of Genomes of Bacterial Isolates from Lameness Outbreaks in Broilers. Poult. Sci. 100 (2021) 101148. [CrossRef]
- R.D. Olson, R. Assaf, T. Brettin, N. Conrad, C. Cucinell, James J. Davis, Donald M. Dempsey, A. Dickerman, Emily M. Dietrich, Ronald W. Kenyon, M. Kuscuoglu, Elliot J. Lefkowitz, J. Lu, D. Machi, C. Macken, C. Mao, A. Niewiadomska, M. Nguyen, Gary J. Olsen, Jamie C. Overbeek, B. Parrello, V. Parrello, Jacob S. Porter, Gordon D. Pusch, M. Shukla, I. Singh, L. Stewart, G. Tan, C. Thomas, M. VanOeffelen, V. Vonstein, Zachary S. Wallace, Andrew S. Warren, Alice R. Wattam, F. Xia, H. Yoo, Y. Zhang, Christian M. Zmasek, Richard H. Scheuermann, and Rick L. Stevens, Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acid Res. 51 (2022) D678-D689. [CrossRef]
- J.R. Grant, and P. Stothard, The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acid Res. 36 (2008) W181-W184. [CrossRef]
- C.L. Brown, J. Mullet, F. Hindi, J.E. Stoll, S. Gupta, M. Choi, I. Keenum, P. Vikesland, A. Pruden, and L. Zhang, mobileOG-db: a Manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements. Appl. Environ. Microbiol. 88 (2022) e0099122.
- C. Bertelli, M.R. Laird, K.P. Williams, Simon Fraser University Research Computing Group, B.Y. Lau, G. Hoad, G.L. Winsor, and F.S. Brinkman, IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acid Res. 45 (2017) W30-W35. [CrossRef]
- A. Pon, A. Marcu, D. Arndt, J.R. Grant, T. Sajed, Y. Liang, and D.S. Wishart, PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acid Res. 44 (2016) W16-W21. [CrossRef]
- J.A. Lees, S.R. Harris, G. Tonkin-Hill, R.A. Gladstone, S.W. Lo, J.N. Weiser, J. Corander, S.D. Bentley, and N.J. Croucher, Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 29 (2019) 304-316. [CrossRef]
- T.J. Treangen, B.D. Ondov, S. Koren, and A.M. Phillippy, The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15 (2014) 524. [CrossRef]
- M.V. Han, and C.M. Zmasek, phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics 10 (2009) 356. [CrossRef]
- S. Argimón, K. Abudahab, R.J.E. Goater, A. Fedosejev, J. Bhai, C. Glasner, E.J. Feil, M.T.G. Holden, C.A. Yeats, H. Grundmann, B.G. Spratt, and D.M. Aanensen, Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom. 2 (2016). [CrossRef]
- T. Seemann, Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 (2014) 2068-2069. [CrossRef]
- A.J. Page, C.A. Cummins, M. Hunt, V.K. Wong, S. Reuter, M.T.G. Holden, M. Fookes, D. Falush, J.A. Keane, and J. Parkhill, Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31 (2015) 3691-3693. [CrossRef]
- O. Brynildsrud, J. Bohlin, L. Scheffer, and V. Eldholm, Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 17 (2016) 238. [CrossRef]
- Z.J. Shi, S. Nayfach, and K.S. Pollard, Maast: genotyping thousands of microbial strains efficiently. Genome Biol. 24 (2023) 186. [CrossRef]
- F. Teufel, J.J. Almagro Armenteros, A.R. Johansen, M.H. Gíslason, S.I. Pihl, K.D. Tsirigos, O. Winther, S. Brunak, G. von Heijne, and H. Nielsen, SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotech. (2022). [CrossRef]
- Y. Huang, V. Eeckhaut, E. Goossens, G. Rasschaert, J. Van Erum, G. Roovers, R. Ducatelle, G. Antonissen, and F. Van Immerseel, Bacterial chondronecrosis with osteomyelitis related Enterococcus cecorum isolates are genetically distinct from the commensal population and are more virulent in an embryo mortality model. Vet. Res. 54 (2023) 13. [CrossRef]
- S.R. Fiddaman, E.A. Dimopoulos, O. Lebrasseur, L. du Plessis, B. Vrancken, S. Charlton, A.F. Haruda, K. Tabbada, P.G. Flammer, S. Dascalu, N. Marković, H. Li, G. Franklin, R. Symmons, H. Baron, L. Daróczi-Szabó, D.N. Shaymuratova, I.V. Askeyev, O. Putelat, M. Sana, H. Davoudi, H. Fathi, A.S. Mucheshi, A.A. Vahdati, L. Zhang, A. Foster, N. Sykes, G.C. Baumberg, J. Bulatović, A.O. Askeyev, O.V. Askeyev, M. Mashkour, O.G. Pybus, V. Nair, G. Larson, A.L. Smith, and L.A.F. Frantz, Ancient chicken remains reveal the origins of virulence in Marek’s disease virus. Science 382 (2023) 1276-1281. [CrossRef]
- R. Madan-Lala, J.K. Sia, R. King, T. Adekambi, L. Monin, S.A. Khader, B. Pulendran, and J. Rengarajan, Mycobacterium tuberculosis Impairs Dendritic Cell Functions through the Serine Hydrolase Hip1. J Immunol 192 (2014) 4263-4272. [CrossRef]
- Y. Liu, B.B. Yoo, C.-A. Hwang, Y. Suo, S. Sheen, P. Khosravi, and L. Huang, LMOf2365_0442 Encoding for a Fructose Specific PTS Permease IIA May Be Required for Virulence in L. monocytogenes Strain F2365. Front. Microbiol. 8 (2017). [CrossRef]
- F.L. Paganelli, J. Huebner, K.V. Singh, X. Zhang, W. van Schaik, D. Wobser, J.C. Braat, B.E. Murray, M.J.M. Bonten, R.J.L. Willems, and H.L. Leavis, Genome-wide Screening Identifies Phosphotransferase System Permease BepA to Be Involved in Enterococcus faecium Endocarditis and Biofilm Formation. J. Infect. Dis. 214 (2016) 189-195. [CrossRef]
- J.P. Coleman, L.L. Hudson, S.L. McKnight, J.M. Farrow, M.W. Calfee, C.A. Lindsey, and E.C. Pesci, Pseudomonas aeruginosa PqsA Is an Anthranilate-Coenzyme A Ligase. J. Bact. 190 (2008) 1247-1255.
- A. Dash, and R. Modak, Protein Acetyltransferases Mediate Bacterial Adaptation to a Diverse Environment. J. Bact. 203 (2021) 10.1128/jb.00231-21. [CrossRef]
- G. Boël, P.C. Smith, W. Ning, M.T. Englander, B. Chen, Y. Hashem, A.J. Testa, J.J. Fischer, H.-J. Wieden, J. Frank, R.L. Gonzalez, and J.F. Hunt, The ABC-F protein EttA gates ribosome entry into the translation elongation cycle. Na.t Struct. Mol. Biol. 21 (2014) 143-151. [CrossRef]
- M.J. Kazmierczak, M. Wiedmann, and K.J. Boor, Alternative Sigma Factors and Their Roles in Bacterial Virulence. Microbiology and Molecular Biology Reviews 69 (2005) 527-543. [CrossRef]
- S. Fukushima, M. Yoshimura, T. Chibazakura, T. Sato, and H. Yoshikawa, The putative ABC transporter YheH/YheI is involved in the signalling pathway that activates KinA during sporulation initiation. FEMS Microbiol. Lett. 256 (2006) 90-97. [CrossRef]
- A. Shwani, B. Zuo, A. Alrubaye, J. Zhao, and D.D. Rhoads, A Simple, Inexpensive Alkaline Method for Bacterial DNA Extraction from Environmental Samples for PCR Surveillance and Microbiome Analyses. Appl. Sci. 14 (2024) 141. [CrossRef]
- L.B. Borst, M.M. Suyemoto, S. Keelara, S.E. Dunningan, J.S. Guy, and H.J. Barnes, A Chicken Embryo Lethality Assay for Pathogenic Enterococcus cecorum. Avian Dis. 58 (2014) 244-248. [CrossRef]
- B. Dolka, D. Chrobak-Chmiel, M. Czopowicz, and P. Szeleszczuk, Characterization of pathogenic Enterococcus cecorum from different poultry groups: Broiler chickens, layers, turkeys, and waterfowl. PLoS One 12 (2017) e0185199. [CrossRef]
- N. Ekesi, A. Hasan, A. Parveen, A. Shwani, and D. Rhoads, Embryo Lethality Assay for Evaluating Virulence of Isolates from Bacterial Chondronecrosis with Osteomyelitis in Broilers. Poult. Sci. (in review) (2021).
- D.A. Garsin, K.L. Frank, J. Silanpää, F.M. Ausubel, A. Hartke, N. Shankar, and B.E. Murray, Pathogenesis and Models of Enterococcal Infection. in: M.S. Gilmore, D.B. Clewell, Y. Ike, and N. Shankar, (Eds.), Enterococci: from commensals to leading causes of drug resistant infection [Internet], Massachusetts Eye and Ear Infirmary, Boston, MA, 2014, pp. 139-192.
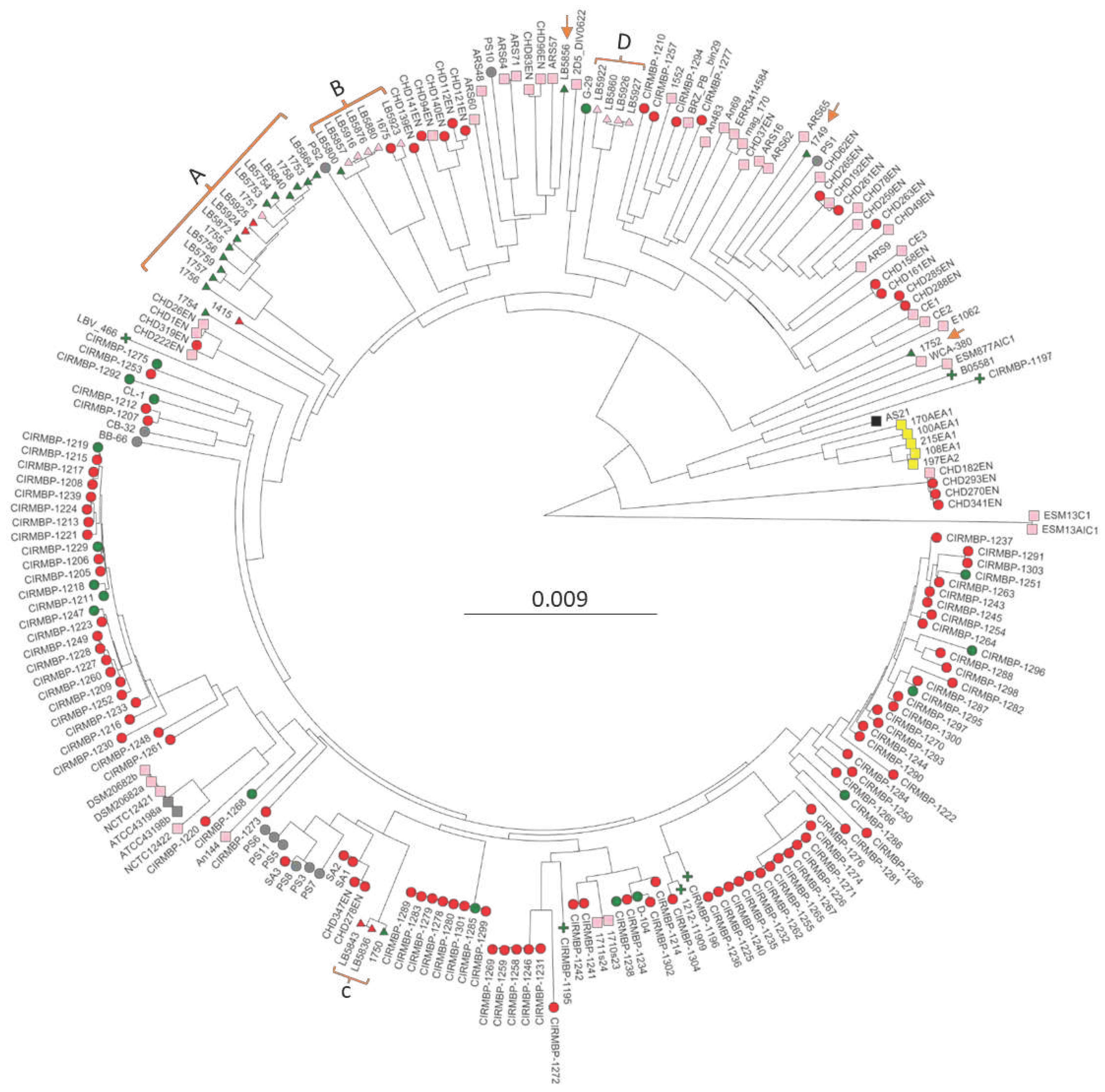
| Strain | Source | Date Collected | Collect Location | Bird Age | Assembly contigs | Total bp | BioSample | BLASTp | PopPUNK cluster |
| 1415 | Tibial pus | 6/23/2016 | farm16 | 4.3 | 65 | 2411963 | SAMN38750234 | 100 | A |
| 1675 | Femoral head necrosis | 11/26/2020 | UAPRF | 8 | 56 | 2188186 | SAMN38750235 | 100 | B |
| 1749 | Liver | n/a | farm15 | 0.5 | 93 | 2494030 | SAMN38750236 | 15 | solo |
| 1750 | Heart | n/a | farm22 | 3 | 49 | 2281603 | SAMN38750237 | 100 | C |
| 1751 | n/a | n/a | farm19 | 2.5 | 49 | 2300397 | SAMN38750238 | 100 | A |
| 1752 | Liver | n/a | farm8 | 3.6 | 32 | 2279238 | SAMN38750239 | 17 | solo |
| 1753 | Heart | n/a | farm15 | 3.1 | 51 | 2240969 | SAMN38750240 | 100 | A |
| 1754 | n/a | n/a | farm2 | n/a | 47 | 2180738 | SAMN38750241 | 100 | A |
| 1755 | Heart | n/a | farm2 | n/a | 58 | 2366287 | SAMN38750242 | 100 | A |
| 1756 | Heart | n/a | farm8 | 3.2 | 62 | 2288864 | SAMN38750243 | 100 | A |
| 1757 | Heart | n/a | farm4 | 3.3 | 53 | 2297190 | SAMN38750244 | 100 | A |
| 1758 | Heart | n/a | farm22 | 3 | 53 | 2249959 | SAMN38750245 | 100 | A |
| LB5753 | Heart | 2/5/2021 | farm17 | 3 | 56 | 2226311 | SAMN38750246 | 100 | A |
| LB5754 | Heart | 2/12/2021 | farm9 | 5 | 62 | 2234879 | SAMN38750247 | 100 | A |
| LB5756 | Heart | 3/8/2021 | farm13 | 2.3 | 62 | 2239264 | SAMN38750248 | 100 | A |
| LB5759 | Heart | 3/8/2021 | farm11 | 2.6 | 75 | 2392627 | SAMN38750249 | 100 | A |
| LB5800 | Heart | 3/11/2021 | farm3 | 3 | 42 | 2212442 | SAMN38750250 | 100 | B |
| LB5836 | Vertebral osteomyelitis | 9/29/2020 | farm6 | 12 | 69 | 2320142 | SAMN38750251 | 100 | C |
| LB5840 | Heart | 10/13/2020 | farm5 | n/a | 57 | 2258228 | SAMN38750252 | 100 | A |
| LB5843 | Vertebral osteomyelitis | 10/20/2020 | farm7 | 8 | 70 | 2319059 | SAMN38750253 | 100 | C |
| LB5856 | Heart | 12/21/2020 | farm14 | n/a | 81 | 2378014 | SAMN38750254 | 40 | solo |
| LB5857 | Egg transfer residue | 3/26/2021 | hatchery 1 | Eggs | 47 | 2180187 | SAMN38750255 | 100 | B |
| LB5860 | Cull eggs | 3/24/2021 | hatchery 1 | Eggs | 102 | 2230151 | SAMN38750256 | 100 | D |
| LB5864 | Heart | 12/9/2020 | farm20 | 2.3 | 61 | 2239173 | SAMN38750257 | 100 | A |
| LB5872 | Heart | 4/21/2021 | farm18 | n/a | 65 | 2365322 | SAMN38750258 | 100 | A |
| LB5876 | Egg transfer residue | 5/10/2021 | hatchery 1 | Eggs | 37 | 2183317 | SAMN38750259 | 100 | B |
| LB5880 | Air sacculitis | 4/29/2021 | farm21 | 2 | 38 | 2146928 | SAMN38750260 | 100 | B |
| LB5916 | Egg transfer residue | 6/4/2021 | hatchery 1 | Eggs | 40 | 2193104 | SAMN38750261 | 100 | B |
| LB5922 | n/a | 6/16/2021 | farm12 | 1 | 88 | 2169308 | SAMN38750262 | 99 | D |
| LB5923 | intestinal tract | 6/21/2021 | farm 1 | 1.3 | 58 | 2239406 | SAMN38750263 | 100 | B |
| LB5924 | Air sacculitis | 4/2/2021 | farm10 | 2.4 | 61 | 2296440 | SAMN38750264 | 100 | A |
| LB5925 | Air sacculitis | 4/2/2021 | farm10 | 2.4 | 61 | 2297136 | SAMN38750265 | 94 | A |
| LB5926 | Egg transfer residue | 4/16/2021 | hatchery 2 | Eggs | 103 | 2277367 | SAMN38750266 | 100 | D |
| LB5927 | Egg transfer residue | 4/16/2021 | hatchery 2 | Eggs | 105 | 2276597 | SAMN38750267 | 97 | D |
| phentotype | count | Borst gene cluster protein encoded gene | average±SEM | |||||||||||
| 312 | 313 | 314 | 315 | 316 | 317 | 318 | 319 | 320 | 321 | 322 | 323 | |||
| None | 82 | 53 | 35 | 37 | 49 | 30 | 43 | 31 | 31 | 45 | 43 | 39 | 91 | 44±5 |
| CD | 145 | 85 | 77 | 79 | 83 | 75 | 79 | 74 | 75 | 84 | 85 | 82 | 98 | 81±2 |
| BCO | 113 | 84 | 76 | 77 | 82 | 73 | 77 | 72 | 73 | 81 | 83 | 79 | 97 | 80±2 |
| SS | 34 | 88 | 80 | 83 | 86 | 81 | 83 | 79 | 79 | 89 | 88 | 88 | 98 | 85±2 |
| Gene | Phenotypic Trait Group | ||||||
| Name | Function | AA Pos | Ref | Alt | BCO | Sepsis Strict | Sepsis Strict Reduced |
| PEG231 | Serine aminopeptidase S33 domain-containing protein | 261 | T | A | 35 | 66 | 50 |
| EIIABC | PTS fructose-specific EIIABC component | 555 | R | Q | 33 | 75 | 64 |
| 590 | A | V | 29 | 75 | 64 | ||
| CBCL1 | 4-chlorobenzoate--CoA ligase | 244 | G | H | 43 | 69 | 55 |
| 246 | I | L | 43 | 69 | 55 | ||
| 249 | H | Y | 43 | 69 | 55 | ||
| ElaA | Gcn5-related N-acetyltransferase (GNAT family) | 128 | N | K | 34 | 69 | 55 |
| EttA | Energy-dependent translational throttle protein | 106 | S | A | 40 | 69 | 55 |
| 534 | T | A | 39 | 66 | 50 | ||
| RpoN | RNA polymerase σ54 factor | 12-13 | TQ | -- | 25 | 38 | 50 |
| 22 | T | S | 26 | 38 | 50 | ||
| YheH | putative multidrug resistance ABC transporter ATP-binding/permease protein | 13 | I | L | 43 | 72 | 59 |
| 578 | D | N | 28 | 66 | 50 | ||
| 590 | S | I | 28 | 66 | 50 | ||
| 592-593 | EEI | GAD | 28 | 69 | 55 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





