1. Introduction
The genus
Vespa, natively distributed in tropical, subtropical and temperate Asia, features several invasive hornet species. The accidental introduction of
V. velutina to Europe [
1] severely affected European apiculture, leading to tens of millions of dollars in management cost [
2].
V. mandarinia is the largest species within the genus known to cause severe damage to apiculture [
3,
4] as well human deaths with multiple stings, with a mean number of only 59 stings causing death [
5]. Although its native distribution is in Asian countries and the Russian Far East [6-8], several individuals of the species have been observed in coastal regions of Washington State [
9,
10], most likely brought across the Pacific through cargo transportation. The pest has also been spotted in Nanaimo, British Columbia where three Asian giant hornets were discovered during August of 2019 [11-13]. The USA and Canadian sites where the Asian giant hornets were found have a distance of about 95 km [
9] and might have been brought to these site by the same shipping route. However, they have distinct mitochondrial genomes and might come from different source populations in Asia [
9].
These potentially harmful immigrants raised several questions. First, where is their source population in Asia? Second, could they potentially survive and reproduce in North America? The last question is often addressed with habitat niche modeling [
10,
11,
14]. However, addressing the first question would shed light on the second. If the invading Asian giant hornets were from a population in a habitat similar to that in North America, then the invaders likely will survive and reproduce. Observations in British Columbia suggested strongly that
V. mandarinia could survive and reproduce in North America. An underground nest of Asian giant hornets was found around Robin’s Park on Sept. 18th, 2019 [11-13]. All of the hornets from the nest including the queen were transferred out of the nest and the nest was destroyed by utilizing carbon dioxide [11-13]. These observations are consistent with the observation of
V. mandarinia in Blaine WA [
9], reinforcing the inference that the pest has the potential to survive and reproduce in the coastal regions of British Columbia and Washington State [
10].
In this report, we aim to address the first questions, i.e., the identification of the source population of those
V. mandarinia individuals invading Canada and USA. This can be achieved by using DNA barcoding data that include not only DNA barcodes, but also geographic coordinates or sampling locations from which geographic coordinates can be approximately derived. Such DNA barcoding data for
V. mandarinia are available for multiple populations in China, South Korea, Japan and the Russian Far East. The rationale and the operational protocol are simple. A source population needs to fulfill two criteria. First, the mitochondrial COX1 sequences from the source population should be most closely related (ideally identical) to those found in Canada and USA. Second, the COX1 sequences from other
V. mandarinia populations are more different than the source population from those found in Canada and USA. This approach is similar to identifying illegally hunted whale species in the whale meat market [
15]. If DNA in a piece of whale meat is identical to that of the protected humpback whale but different from all other known whale species, then the whale meat is from illegally hunted humpback whale. However, identifying a source population is more difficult than species identification.
V. mandarinia is a widely distributed temperate species [
3,
4,
8,
16]. If the species is genetically homogeneous so that populations in different regions all share the same genotype or the same mixture of genotypes, then the above approach for identifying the source population of the
V. mandarinia individuals invading Canada and USA would not work. However, there are indications of genetic differentiation among
V. mandarinia populations revealed in previous studies [
9,
17]. Some of
V. mandarinia lineages were even given subspecies status [
3]. This suggests the possibility of identifying the source population by using genetic markers such as mitochondrial COX1 sequences. A preliminary identification has already been made [
9] in which a phylogenetic tree including five
V. mandarinia specimens (the Canada and USA specimens plus one specimen each from China, Japan and South Korea). Based on the comparison of mitochondrial genes, the Canadian specimen was found most similar to the Japanese specimen with an evolutionary distance of 0.0012, and the USA specimen most similar to the South Korea specimen with a distance of 0.0004 [
9]. However, it is possible that
V. mandarinia populations in other Chinese populations may have a distance even smaller than those reported between the Canadian and the Japanese specimens, or between the USA and the South Korean specimens.
We compiled and analyzed mitochondrial COX1 sequences of specimens sampled from multiple populations in China, South Korea, Japan and the Russian Far East. V. mandarinia populations exhibit genetic differentiation among different geographic regions, with specimens from China, South Korea and Japan forming distinct clades. The two mitochondrial COX1 sequences from Nanaimo, British Columbia are identical to the two sequences from Japan (from Fukuoka and Yamaguchi, respectively). The COX1 sequence from Blaine, Washington State, is identical to one of six sequence from South Korea.
2. Material and Methods
All sequences were downloaded from GenBank except for four sequences downloaded from the DNA barcoding Bold System [
18] with sample IDs BIOUG26171-B05, BIOUG24885-F10, NIBGE HYM-01572 and NIBGE HYM-01001). The criteria for inclusion are 1) that the original publication contains a description of sampling locations from which approximate latitude and longitude values can be derived, and 2) that the sequence is not a fragment of a longer sequence sampled from the same location. For example, sequence with GenBank accession MZ165595 was included, but MZ165596 was not because the latter is identical to a segment within the former and the two were sampled at the same location, i.e., Ruili, China, near the border between China and Myanmar.
For specimens with complete mitochondrial genomes, the COX1 sequences were extracted from the GenBank file with DAMBE [
19]. Four COX1 sequences from
V. tropica and three COX1 sequences from
V. soror were used as outgroups. Previous studies [
16,
17,
20,
21] have shown
V. soror to be the closest relative of
V. mandarinia within the genus. A total of 36 COX1 sequences were included.
One included sequence (Accession KR059904) requires some clarification. It is the first
V. mandarinia mitochondrial genome sequenced in China [
22] and has often been used as a reference genome to assemble mitochondrial genomes [
9]. However, its sampling location has not been recorded in publications. We emailed Dr. Shu-Jun Wei and learned that the specimen was collected in Chengde City in Hebei Province in Northern China, which yields an approximate latitude and longitude values 40.94 and 117.99, respectively. The GenBank accession number, species name, geographic locations in latitude and longitude, COX1 length and GC% are listed in
Table 1.
The resulting
COX1 sequences were aligned using MAFFT [
23] with the most accurate LINSI option (‘–localpair’ and ‘–maxiterate = 1000’). For phylogenetic reconstruction, the GTR+Γ model was used with four discrete rate categories for approximating a continuous gamma distribution [
24]. This model was chosen based on information-theoretic index AIC (
Table 2) and the likelihood ratio tests (
Table 3) [
25,
26] among the nested HKY [
27], TN93 [
28] and GTR [
29,
30] models with or without the discrete gamma distribution to accommodate rate heterogeneity in substitution rate among sites.
The tree log-likelihood (lnL) was obtained with PhyML v3.3 [
31]. The tree improvement option (‘-s’) was set to ‘BEST’ (best of NNI and SPR search). The ‘-o’ option was set to ‘tlr’ which optimizes the topology, the branch lengths and rate parameters. The phylogenetic trees reported in the Results section was from PhyML reconstruction with GTR+Γ which has the smallest AIC (
Table 2) and fits the data significantly better than the alternative models (
Table 3). MAFFT and PhyML are included in DAMBE and called to analyze sequences with a consistent user interface.
Phylogeographic analysis aims to visualize genetic variation over time and space. A geophylogeny combines a phylogenetic tree together with the geographic coordinates of sampling locations. We used PGT software [
32] for generating geophylogenies for visualization. PGT makes use of both Google Maps and Microsoft Bing Maps with regular map view and satellite terrain view.
3. Results
3.1. Identification of the source population for the Canadian and USA specimens
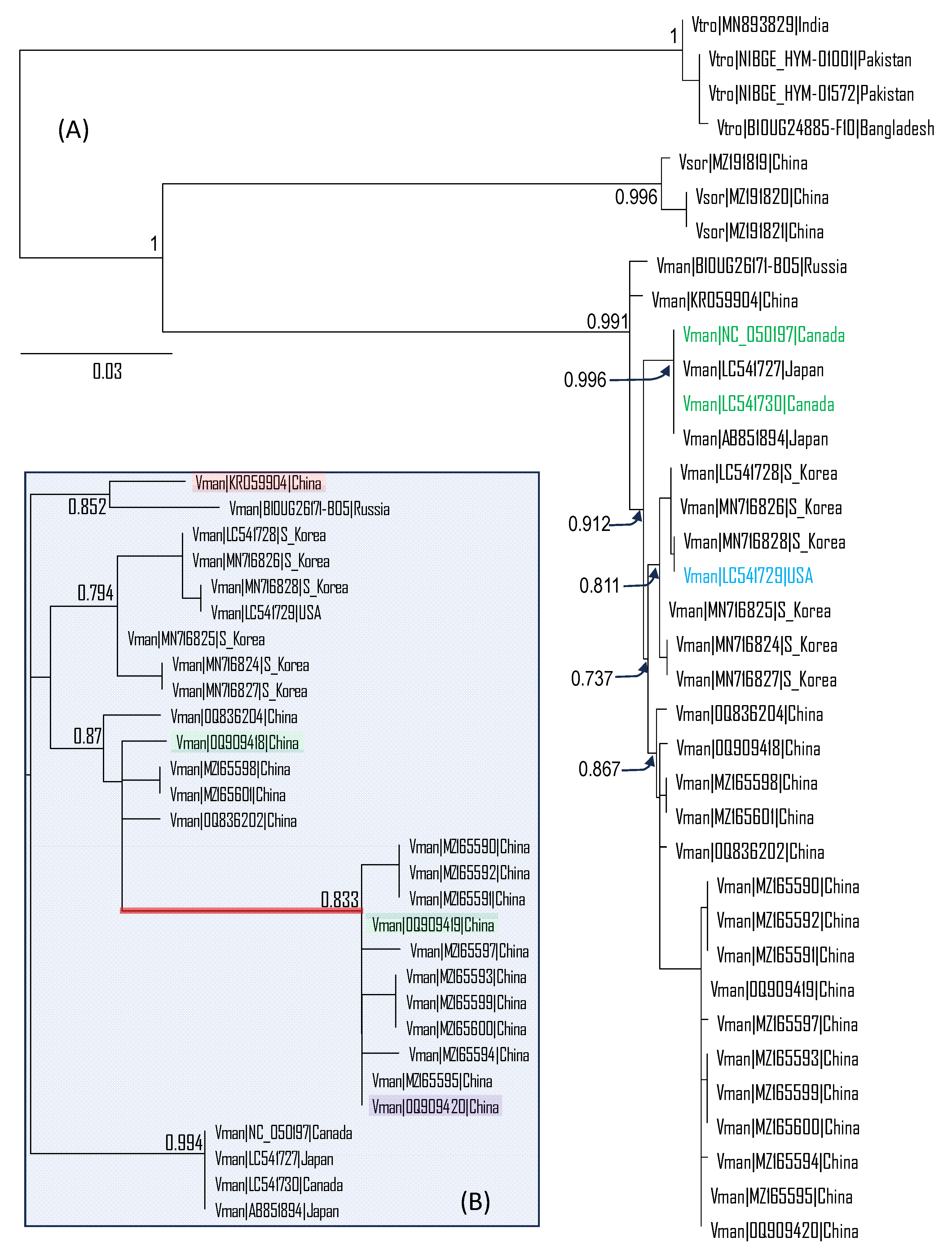
The three
Vespa species,
V. tropica, V. soror and
V. mandarinia, are well differentiated genetically from each other (
Figure 1A). There is also clear but less pronounced geographic differentiation within
V. mandarinia, forming monophyletic clades for 16 specimens from China (Yunnan, Guangdong and Sichuan provinces), six specimens from South Korea and two specimens from Japan (
Figure 1A). This genetic differentiation over space facilitates the identification of the source populations to which the invading individuals found in Canada and USA belong. If all
V. mandarinia populations are genetically homogeneous, then there would no chance of identifying a specific source population for any invading individual.
The two mitochondrial COX1 sequences of the
V. mandarinia specimens from Nanaimo, British Columbia are identical to each other and to the two sequences from Japan (which are also identical to each other). The collection sites of the two in Japan has a straight-line distance of 120 km, between Fukuoka and Yamaguchi. The lack of genetic variation between the two
V. mandarinia specimens suggests that they were recent descendants of a common ancestor. In contrast, the South Korean specimens exhibited substantial genetic variation as exemplified by the branch lengths in
Figure 1. The COX1 sequence from Blaine, Washington State, clustered with those from South Korea, and is identical to one sequence from South Korea (GenBank accession MN716828,
Figure 1A). Thus, one may tentatively infer that the source populations for the Canada and USA specimens of
V. mandarinia were most likely in Japan and South Korea, respectively. However, this inference is not strong because there is no specimen sampled along the coastal regions of East Chian opposite Japan and South Korea. It is possible that specimens from such coastal regions may also have COX1 sequences identical to the Canada and USA specimens, respectively. Thus, identifying a specimen to its source population is much more difficult than identifying a piece of whale meat to whale species.
The unrooted phylogeny (
Figure 1B) shows the genetic variation within
V. mandarinia. In addition to the clades in Japan and South Korea, the phylogeny also suggests 1) a northern clade represented by the two specimens from the Russian Far East and Northern China (pink-shaded), and 2) a relatively diverse clade in mainland China separated into two subgroups by the long red branch (
Figure 1B). What is remarkable is that these two genetically different subgroups are not geographically separated. Each subgroup has colonized Yunan Province and Guangdong Province. Thus, distinct lineages of
V. mandarinia appeared to have evolved sympatrically. Some of these lineages have been given subspecies status [
3,
4].
3.2. Phylogeographic patterns of V. mandarinia and the two outgroup species
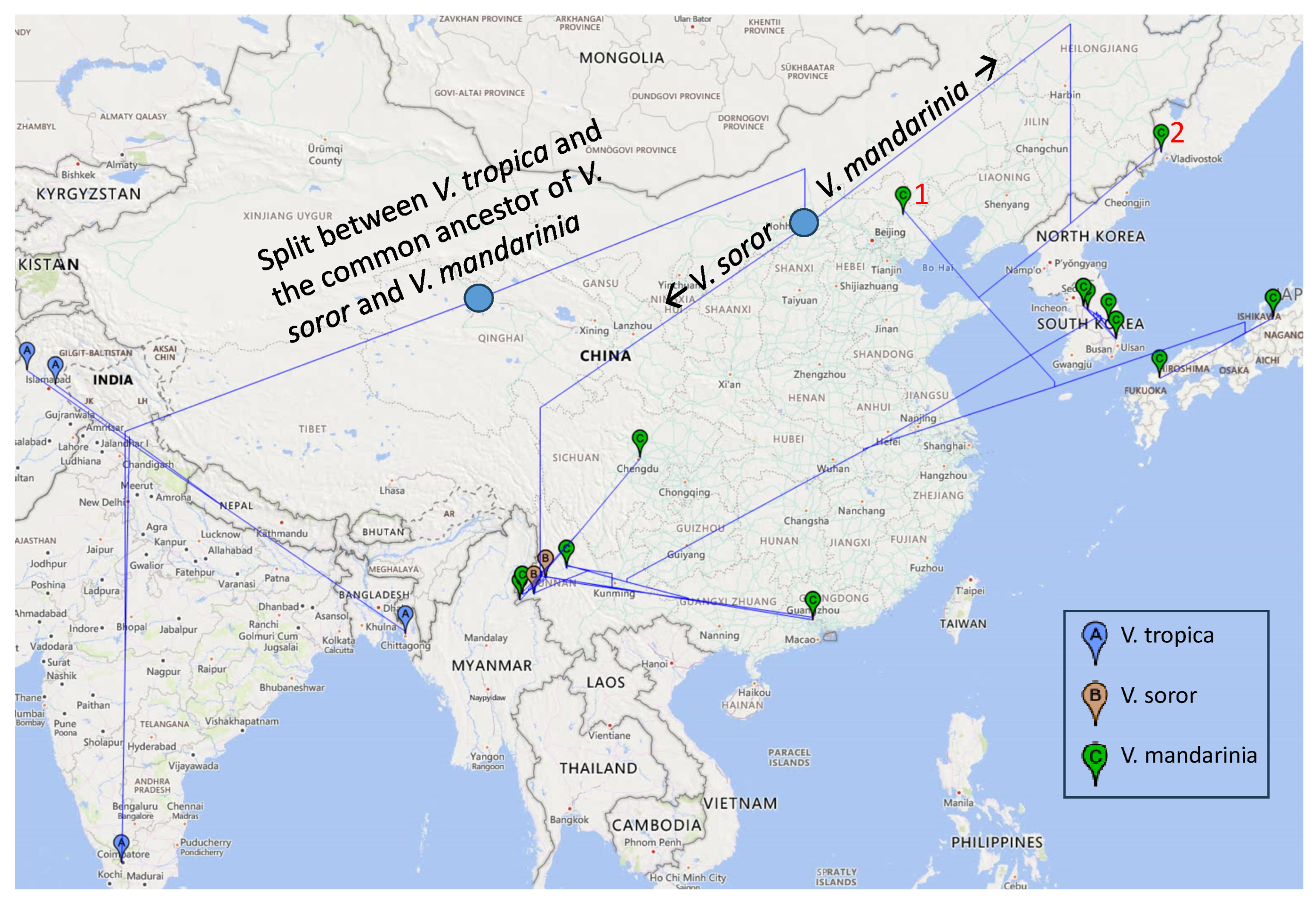
The distribution of genetic variation over both time and space is better visualized through a geophylogeny (
Figure 2). Of the two outgroup species,
V. tropica is widely distributed in tropical Asia [
4,
8,
34]. Although
V. tropica may be found occasionally in temperate regions, its nest size is much reduced as a consequence of reduced availability of food (i.e., eggs and larvae of polistine wasps) [
4].
V. soror inhabits the tropical and subtropical regions of Southeastern Asia including China [
3,
4,
8,
16]. The speciation initiated with the tropical
V. tropica splitting from the common ancestor of
V. soror and
V. mandarinia (
Figure 2). This common ancestor likely moved from the tropical to subtropical habitats, with
V. soror remaining in the subtropical region but
V. mandarinia invading the temperate region (
Figure 2). If we may interpret the geophylogeny (
Figure 2) liberally, then the ancestral lineages of
V. mandarinia first inhabited the northern temperate region, represented by the early lineages marked with red-colored 1 in Northern China) and 2 in the Russian Far East (
Figure 2). From these northern lineages were derived the Japanese lineage of
V. mandarinia (
Figure 1 and
Figure 2). The South Korean clade and the Southern Chian clade form a monophyletic clade (
Figure 1 and
Figure 2). Thus, the geographic expansion and the association appears to be from the tropical
V. tropica to the subtropical
V. soror and to the temperate
V. mandarinia. Only two vespine wasps (
V. mandarinia and
V. crabro) have expanded as far north as Hokkaido in Japan, while all other vespine wasps, such as
V. tropica, are restricted to southern Japan [
3,
4]. The adaptation of
V. mandarinia to temperate climate would increase its chance for survival and reproduction in the coastal regions of British Columbia and Washington State.
The geophylogeny in
Figure 2 highlights a strong insufficiency in sample collection in China. There is no specimen sequenced in the coastal regions of East China facing Japan and South Korea across the East China Sea and Yellow Sea, respectively. If the
V. mandarinia sequences from these coastal regions were different from those in Japan and South Korea, then we would be more confident that the Canadian specimen was from Japan and the USA specimen was from South Korea. The lack of sequence data in these coastal regions in East China does not allow us to exclude the possibility that the specimens in Canada and USA may be from these coastal
V. mandarinia populations.
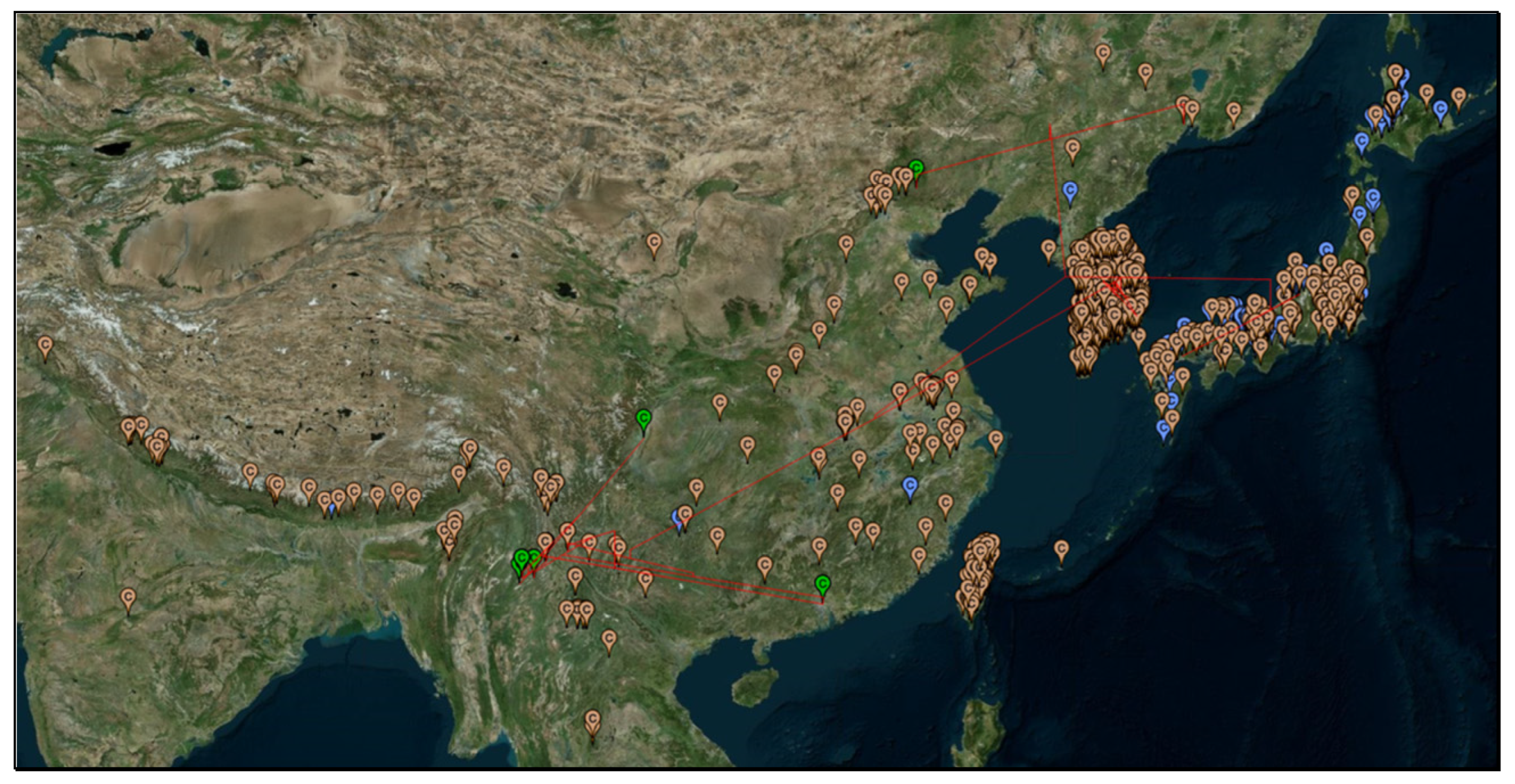
To better visualize the natural habitats of
V. mandarinia, we have generated a geophylogeny for
V. mandarinia specimens alone (
Figure 3) and supplemented the geophylogeny with recorded presence of
V. mandarinia compiled in the GBIF database [
35]. It is clear that
V. mandarinia occupies a wide temperate zone, but its natural habitats tend to be humid and forested, which resembles the environment in the coastal regions of British Columbia in Canada and Washington States in USA. If all the recorded sites of
V. mandarinia were represented by sequenced specimens, which is likely achievable in the next 10 years, then one would be able to identify highly accurately the source population of any invasive individuals of
V. mandarinia.
The relatively few sites recorded in mainland China and India, especially before 2000 (
Figure 3) could have two explanations. First, there was little research effort in sampling and documenting the distribution of
V. mandarinia. Second, the species is indeed much less abundant in mainland than in islands. The dramatically increased number of recorded presences after the year 2000 relative to those before the year 2000 (
Figure 3) suggests that the first hypothesis is more likely than the second.
We should emphasize here that more data are needed to establish the rooting position for
V. mandarinia (Figs. 1). With the general trend of the expansion of the
Vespa species from tropical to temperate regions (
Figure 2), it seems odd that the two northern specimens of
V. mandarinia should be the closest to the root (Figs. 1 and 2).
4. Discussion
The tentative identification of
V. mandarinia populations in Japan and South Korea as the source populations of the Canadian and USA specimens, respectively (
Figure 1) facilitates downstream research and monitoring. First, Canadian and USA border services on invasive insect species should focus more on incoming cargos from Japan and South Korea than from elsewhere. Second, in terms of habitat modelling of
V. mandarinia, the input habitat data from Japan and South Korea should carry more weight than those from elsewhere. Third, any invading individuals of
V. mandarinia that is genetically distinct from the Japanese and South Korean populations of
V. mandarinia would suggest a new source population. Tracing an invading individual to its source population is crucial in understanding its habitat requirement for survival and reproduction. Different
V. mandarinia lineages could have different habitat requirements, so it could be misleading to model the habitat requirement of invading individuals from a Japanese
V. mandarinia population by using habitat requirement of certain Chinese
V. mandarinia populations as input.
Biogeographic distribution of a species is determined mainly by three factors: 1) the dispersal ability of the species, 2) the ability to survive and reproduce in the new environment, and 3) the time needed for dispersal and adaptation to colonize a new habitat. Many failed experiments that introduced animals into seemingly suitable new habitats, or reintroduced zoo-raised animals into the natural habitat of their ancestors, highlight the importance of the ability to survive and reproduce in a new environment.
Vespine hornets appear to be highly capable of survival and reproduction in new environments and include multiple invasive species. Although
V. velutina is perhaps the most notorious of vespine wasps [
1,
2,
17,
21,
36,
37],
V. mandarinia is also known for attacking honeybee hives en masse and incurring high economic losses [
3,
4]. It is also well documented that Asian giant hornets carry multiple viruses with yet unknown implications on environmental health [
38]. Furthermore, it takes an average of only 59 stings by
V. mandarinia to cause human death [
5]. Thus, it is important to prevent
V. mandarinia to spread to Norther America. Accurate identification of the source population of invading individuals of
V. mandarinia would provide key information in stopping the spread of the invasive species.
DNA barcoding has been used for the identification of invasive species, especially when samples represent eggs, immature larvae or poorly preserved specimens [
39,
40]. The geographic information associated with DNA-barcoded specimens has contributed to the elucidation of colonization and speciation of Hawaiian wingless katydids [
41]. However, to identify the source population of an invading individual, much extensive sampling and sequencing information is needed. For example, the identification of the Canadian and USA specimens would have been much more accurate and confident if we have sequence information from specimens sampled along the coastal populations of
V. mandarinia in East China. One needs to DNA-barcode not only species, but also natural populations, ideally before human-mediated long-distance transportation obliterates the natural biogeographic patterns. Another problem with the current DNA barcoding data is that only a partial sequence of the mitochondrial COX1 gene is used as a DNA barcode. Such a short sequence increases the chance of the COX1 sequence from an invasive individual matching DNA barcodes from multiple populations, rendering it impossible to pinpoint which is the source population.
To be practically useful for identifying individuals of invasive species to their source populations, DNA barcoding data needs to be expanded in two ways. First, the DNA barcode should be the entire mitochondrial genome instead of a segment in the COX1 gene. Second, specimens in major populations should be sampled and sequenced.
5. Conclusions
The V. mandarinia individuals invading British Columbia in Canada and Washington State in USA have mitochondrial COX1 sequences identical to those of V. mandarinia specimens collected in Japan and South Korea, respectively. They are distinct from specimens sampled in various locations in mainland China. However, for DNA barcoding data to be practically useful for identifying invasive individuals to their source populations, longer DNA barcodes (e.g., the entire mitochondrial genome) and specimens from all representative populations should be included in relevant databases.
Funding
This research was funded by Discovery Grant from Natural Science and Engineering Research Council (NSERC) of Canada, grant number RGPIN/2018-03878.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We particularly wish to thank S. J. Wei for providing the sampling location for a V. mandarinia specimen sampled in Chengde, Hebei, China (accession KR059904), and J. Y. Xia who helped much with data compilation and analysis, as well as manuscript editing and revising.
Conflicts of Interest
The author declares no conflict of interest.
References
- Monceau, K.; Bonnard, O.; Thiéry, D. , Vespa velutina: a new invasive predator of honeybees in Europe. J. Pest Sci. 2014, 87, 1–16. [Google Scholar] [CrossRef]
- Barbet-Massin, M.; Salles, J.-M.; Courchamp, F. , The economic cost of control of the invasive yellow-legged Asian hornet. NeoBiota 2020, 55, 11–25. [Google Scholar] [CrossRef]
- Matsuura, M.; Sakagami, S.F. , A Bionomic Sketch of the Giant Hornet, Vespa mandarinia, a Serious Pest for Japanese Apiculture. Jour. Faa. Sci. Hokkaido Univ. Ser. VI, Zool. 1973, 19, 125–162. [Google Scholar]
- Matsuura, M.; Yamane, S. , Biology of the Vespine Wasps. Springer-Verlag: Heidelberg, 1990; p XIX, 323.
- Yanagawa, Y.; Morita, K.; Sugiura, T.; Okada, Y. , Cutaneous hemorrhage or necrosis findings after Vespa mandarinia (wasp) stings may predict the occurrence of multiple organ injury: A case report and review of literature. Clin. Toxicol. 2007, 45, 803–807. [Google Scholar] [CrossRef] [PubMed]
- Girish Kumar, P.; Srinivasan, G. , Taxonomic Studies of Hornet Wasps (Hymenoptera : Vespidae) Vespa Linnaeus of India. Records of the Zoological Survey of India 2010, 110, 57–80. [Google Scholar] [CrossRef]
- Smith-Pardo, A.H.; Carpenter, J.M.; Kimsey, L. , The Diversity of Hornets in the Genus Vespa (Hymenoptera: Vespidae; Vespinae), Their Importance and Interceptions in the United States. Insect Systematics and Diversity 2020, 4. [Google Scholar] [CrossRef]
- Carpenter, J.M.; Kojima, J. , Checklist of the Species in the Subfamily Vespinae (Insecta: Hymenoptera: Vespidae). Nat. Hist. Bull Ibaraki Univ. 1997, 1, 51–92. [Google Scholar]
- Wilson, T.M.; Takahashi, J.; Spichiger, S.-E.; Kim, I.; van Westendorp, P. , First Reports of Vespa mandarinia (Hymenoptera: Vespidae) in North America Represent Two Separate Maternal Lineages in Washington State, United States, and British Columbia, Canada. Ann. Entomol. Soc. Am. 2020, 113, 468–472. [Google Scholar] [CrossRef]
- Alaniz, A.J.; Carvajal, M.A.; Vergara, P.M. , Giants are coming? Predicting the potential spread and impacts of the giant Asian hornet (Vespa mandarinia, Hymenoptera:Vespidae) in the USA. Pest Management Science 2021, 77, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Nuñez-Penichet, C.; Osorio-Olvera, L.; Gonzalez, V.H.; Cobos, M.E.; Jiménez, L.; DeRaad, D.A.; Alkishe, A.; Contreras-Díaz, R.G.; Nava-Bolaños, A.; Utsumi, K. , et al., Geographic potential of the world's largest hornet, Vespa mandarinia Smith (Hymenoptera: Vespidae), worldwide and particularly in North America. PeerJ 2021, 9, e10690. [Google Scholar] [CrossRef]
- 12. British Columbia Government 2019, Asian giant hornet nest eradicated in Nanaimo. https://news.gov.bc.ca/releases/2019AGRI0106-001818 (Dec. 27, 2023).
- Bérubé, C. , Giant alien insect invasion averted. Am. Bee J. 2020, 160, 209–214. [Google Scholar]
- Zhu, G.; Gutierrez Illan, J.; Looney, C.; Crowder, D.W. , Assessing the ecological niche and invasion potential of the Asian giant hornet. Proc. Natl. Acad. Sci. U S A 2020, 117, 24646–24648. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.S.; Palumbi, S.R. , Which whales are hunted? A molecular genetic approach to monitoring whaling. Science 1994, 265, 1538–1539. [Google Scholar] [CrossRef] [PubMed]
- Mattila, H.R.; Nguyen, L.T.P.; Perrard, A.; Bain, M.; Otis, G.W. , Biology of the southern giant hornet, Vespa soror: nest architecture, morphological differences among castes, and the genetic structure of colonies. Frontiers in Insect Science 2023, 3. [Google Scholar] [CrossRef]
- Mohamadzade Namin, S.; Jung, C. , Genetic diversity of genus Vespa including an invaded species of V. velutina (Hymenoptera: Vespidae) in Korea inferred from DNA barcoding data. Journal of Asia-Pacific Entomology 2020, 23, 540–545. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D. , BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Molecular ecology notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Xia, X. , DAMBE6: New Tools for Microbial Genomics, Phylogenetics, and Molecular Evolution. J Hered 2017, 108, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Perrard, A.; Pickett, K.; Villemant, C.; Kojima, J.-i.; Carpenter, J.M. , Phylogeny of hornets: a total evidence approach (Hymenoptera, Vespidae, Vespinae, Vespa). Journal of Hymenoptera Research 2013, 32, 1–15. [Google Scholar] [CrossRef]
- Urtgam, S.; Jongjitvimol, T. , Genetic Evolution of Asian Predatory Wasp, Vespa velutina, in Northernof Thailand Based on Cytochrome Oxidase Subunit I DNA Barcoding. NU. International Journal of Science 2020, 17, 101–113. [Google Scholar]
- Chen, P.Y.; Wei, S.J.; Liu, J.X. , The mitochondrial genome of the Vespa mandarinia Smith (Hymenoptera: Vespidae: Vespinae) and a phylogenetic analysis of the Vespoidea. Mitochondrial DNA A DNA Mapp Seq Anal 2016, 27, 4414–4415. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Asimenos, G.; Toh, H. , Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol 2009, 537, 39–64. [Google Scholar] [PubMed]
- Yang, Z. , Statistical properties of the maximum likelihood method of phylogenetic estimation and comparison with distance matrix method. Syst Biol 1994, 43, 329–342. [Google Scholar] [CrossRef]
- Xia, X. , Drug efficacy and toxicity prediction: an innovative application of transcriptomic data. Cell Biol Toxicol 2020, 36, 591–602. [Google Scholar] [CrossRef]
- Xia, X. , Information-theoretic indices and an approximate significance test for testing the molecular clock hypothesis with genetic distances. Mol. Phylogenet. Evol. 2009, 52, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Kishino, H.; Yano, T. , Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. , Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 1993, 10, 512–526. [Google Scholar]
- Tavaré, S. , Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences. American Mathematical Society: Providence, RI, 1986; Vol. 17, p 57-86.
- Lanave, C.; Preparata, G.; Saccone, C.; Serio, G. , A new method for calculating evolutionary substitution rates. J. Mol. Evol. 1984, 20, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. , A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Xia, X. , PGT: Visualizing temporal and spatial biogeographic patterns. Global Ecology & Biogeography 2019, 28, 1195–1199. [Google Scholar]
- Rambaut, A. FigTree., 1.4.4. http: //tree.bio.ed.ac.uk/software/figtree/, 2020. [Google Scholar]
- Barss, P. , Renal failure and death after multiple stings in Papua New Guinea. Ecology, prevention and management of attacks by vespid wasps. Med J Aust 1989, 151, 659–663. [Google Scholar] [CrossRef] [PubMed]
- GBIF.org 2024, GBIF Occurrence Download. [CrossRef]
- Bessa, A.S.; Carvalho, J.; Gomes, A.; Santarém, F. , Climate and land-use drivers of invasion: predicting the expansion of Vespa velutina nigrithorax into the Iberian Peninsula. Insect Conservation and Diversity 2016, 9, 27–37. [Google Scholar] [CrossRef]
- Takahashi, R.; Okuyama, H.; Minoshima, Y.N.; Takahashi, J.I. , Complete mitochondrial DNA sequence of the alien hornet Vespa velutina (Insecta: Hymenoptera) invading Kyushu Island, Japan. Mitochondrial DNA. Part B, Resources 2018, 3, 179–181. [Google Scholar] [CrossRef]
- Power, K.; Martano, M.; Ragusa, E.; Altamura, G.; Maiolino, P. , Detection of honey bee viruses in larvae of Vespa orientalis. Frontiers in cellular and infection microbiology 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Ball, S.L.; Armstrong, K.F. , DNA barcodes for insect pest identification: a test case with tussock moths (Lepidoptera: Lymantriidae). Canadian Journal of Forest Research 2006, 36, 337–350. [Google Scholar] [CrossRef]
- Madden, M.J.L.; Young, R.G.; Brown, J.W.; Miller, S.E.; Frewin, A.J.; Hanner, R.H. , Using DNA barcoding to improve invasive pest identification at U.S. ports-of-entry. PLOS ONE 2019, 14, e0222291. [Google Scholar] [CrossRef] [PubMed]
- Rakesh, M.; Aris-Brosou, S.; Xia, X. , Testing alternative hypotheses on the origin and speciation of Hawaiian katydids. BMC ecology and evolution 2022, 22, 83. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).