1. Introduction
Currently, brucellosis remains a major public health threat, as well as a disease of deep economic impact worldwide. Currently, brucellosis is considered as the most common re-emerging zoonotic disease in the world, causing significant human cases in endemic areas.
Currently, brucellosis remains a prominent public health menace, as well as a disease of substantial economic significance. Its regulation and eradication persist as an objective for Veterinary and health Authorities in numerous countries [
1]. Brucellosis is also a growing global One-health issue due to the increase in international commodities exchange, which allows the infection to spread from endemic areas to other parts of the world.
It is considered one of the most prevalent zoonoses in the Middle East and North African (MENA) countries with public health significance and substantial economic loses in the livestock industry. Therefore, the control and eradication of the disease is an objective for Veterinary and health Authorities in several countries of the World. This is also the case of countries as Iraq, where Competent Authorities need to standardize public and animal health intervention programs.
Brucellosis is transmitted through direct contact with infected sheep or goats and through consumption of unpasteurized milk and dairy products contaminated with the bacteria [
2]. The highest incidence of brucellosis was documented within a sizable flock, attributing this to the demanding nature of maintaining hygienic conditions within these flocks. Routine cleaning and disinfection of the paddock, involving the removal of manure and other unsanitary waste, necessitated diligent effort [
3]. In humans, the disease can affect many organ systems, and the most frequently reported symptoms include pyrexia, arthritis, and fatigue [
4]. Unspecific clinical presentation, and thus the failing in a correct primary diagnosis and treatment, often leads to disease progression with genitourinary, neurological, pulmonary, or cardiovascular system involvement, with endocarditis being the most serious disease complication [
5].
During the past two decades, studies carried out in Iraq reported an increase in prevalence of human brucellosis [
6,
7]. Previous serological surveillance studies revealed a prevalence of the disease in small ruminants, cattle, buffalo and camels as 6.51 %, 1 %, 1.84 % and 0.02%, respectively. The incidence rate of brucellosis in humans after the 2003 war was 5,347-recorded cases, representing 22.7 cases/100,000 inhabitants [
8]. The eradication of brucellosis in humans depends upon the eradication of the brucellosis in animal susceptible species [
9] and, in this framework, the isolation of Brucella strains circulating in a territory and their characterization from the genomic point of view is of utmost importance for the assessment of the epidemiological situation. Many molecular epidemiological studies have been (and currently are) conducted with different techniques, such as variable number of tandem repeat (VNTR) analysis (MLVA) and multi locus sequence typing (MLST), to uncover more genetic features and facilitating comparisons between strains. Whole genome sequence (WGS) -based phylogeny stands as the most powerful tool to discriminate Brucella strains and investigating epidemiological spreading risk factors, through the analysis of their phylogenetic relationships. This approach has gained significant traction in Brucella research due to increasing availability of Brucella genomes in publicly accessible database [
10].
This approach would improve the understanding of risk factors for the persistence and spread of the infection, thus allowing decision makers to take the appropriate corrective measures on the control strategies in place, hereby reducing the disease burden in animals and subsequently in humans.
Efficient surveillance programs are crucial for detecting and managing outbreaks, and they rely on the collection and accessibility of sound epidemiological data. Epidemiological investigations may take advantage from the presence of standardized and efficient molecular typing techniques and analytical tools that enable public health laboratories to identify the origin of an outbreak [
11].
As far as genomic characterization of strains is concerned, the discriminatory power of a single locus-specific sequence analysis is generally minor since it involves only a limited region of the genome. In Brucella genus, the phylogenetic markers such as 16S rRNA genes are highly conserved with an almost 100% identity. Therefore, molecular methods such as Multilocus Sequence Typing (MLST), Multiple Loci VNTR (Variable Number of Tandem Repeats) Analysis (MLVA), and, more recently, whole genome sequencing (WGS) typing methods are additionally used to discriminate between Brucella strains, providing a higher resolution genetic clustering, which allows to better identify the source of outbreak cases [
5].
The aim of this study was to sequence Brucella spp. strains isolated in Iraq from different domestic ruminant species, to verify whether there was a spatial or temporal clustering and, above all, to investigate how these Iraqi isolates are positioned in the phylogenetic context of Brucella spp.
2. Materials and Methods
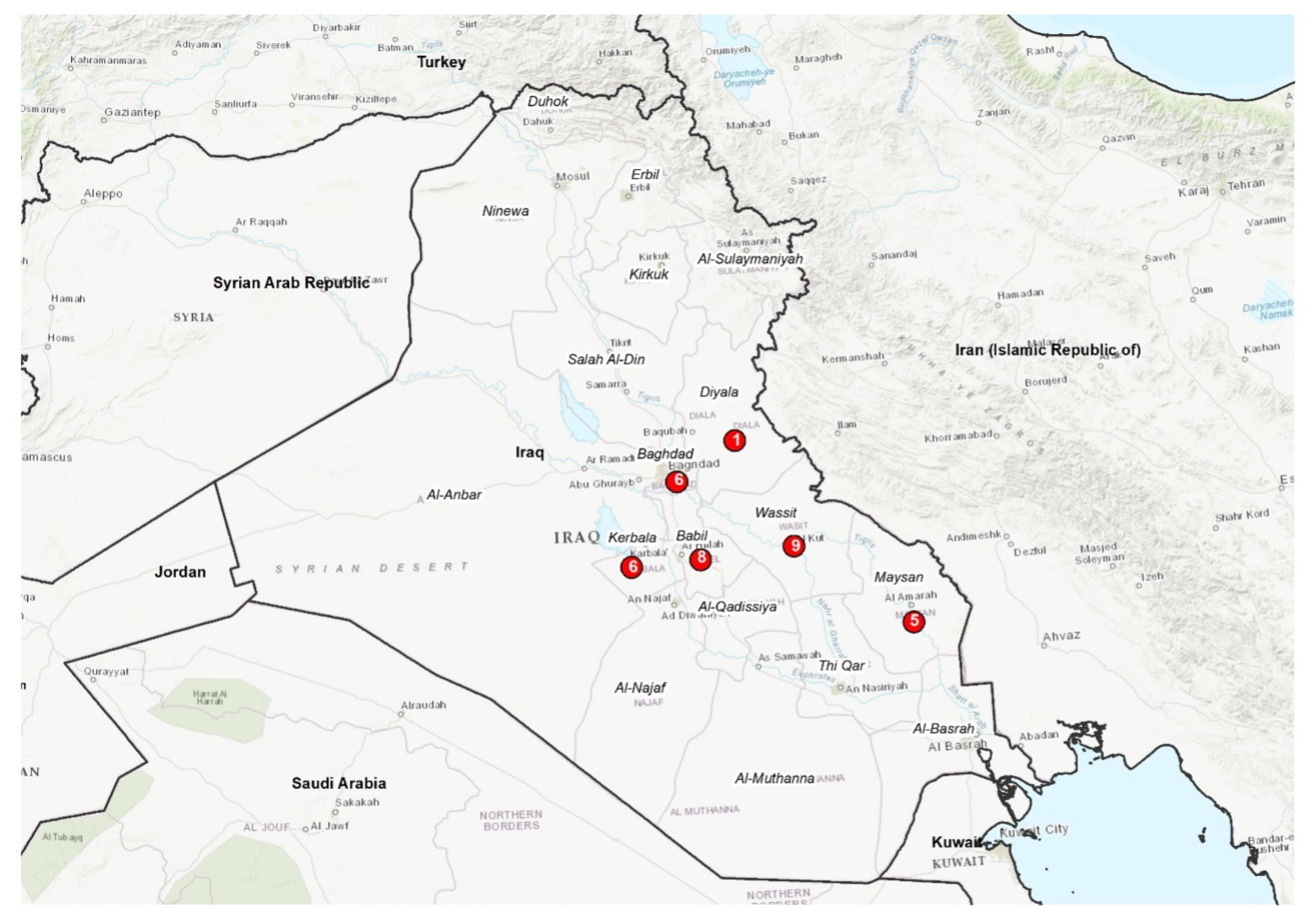
The strains under study (N=35) were isolated from abortion, milk, placenta and foetal membrane of sheep, cattle and buffalo in different governorates of Iraq (
Figure 1). They had been cultured in the centre for brucellosis and tuberculosis control in the Iraq CVL and were isolated during the 2015 - 2017 period. The DNA was extracted by means of a QUIAamp Mini Kit (Qiagen), according to manufacturer's instructions, and has been identified as B. melitensis by real-time and conventional PCR methods in the WOAH Reference Laboratory for Brucellosis, Istituto Zooprofilattico Sperimentale dell’Abruzzo e del Molise “G. Caporale”, Teramo, Italy. Overall, data on isolates collected are summarized in (
Table 1).
MLVA-16 Samples were genotyped using the MLVA-16 panel described by Le Flèche et al. [
12].
DNA extracted from each B. melitensis isolate was amplified by multiplex PCR using primers specific for 16 MLVA loci to assign specific MLVA-16 alleles, as previously described by Garofolo et al., Le Flèche et al., [
12,
13]. The amplified fragments were separated by capillary electrophoresis using an ABI 3500 Genetic Analyzer with POP-7TM Polymer, and the allele types were assigned using Genemapper 4.1 (Applied Biosystems, Carlsbad, CA).
MLVA-16 profiles were analysed using the goeBURST algorithm implemented in PHYLOViZ, version 2.0. Minimum spanning trees (MST) were created using default software settings [
14].
Whole Genome Sequencing
Total genomic DNA was quantified with the Qubit fluorometer (QubitTM DNA HS assay; Life Technologies, Thermo Fisher Scientific Inc., Waltham, MA, USA). Nextera XT library preparation kit (Illumina Inc., San Diego, CA, USA) was used to prepare the sequencing libraries according to the manufacturer's protocol and the libraries were sequenced using the Illumina NextSeq 500 instrument. Sequencing produced 150-bp paired-end sequencing reads that were demultiplexed and subsequently the adapters were removed. The reads were trimmed from 5′ and 3′ ends using Trimmomatic version 0.36 to discard the nucleotides with quality scores of less than 25 and the reads shorter than 36 bp were removed. Trimmed reads were de novo assembled using SPAdes version 3.11.1 with the careful option selected. Contigs shorter than 200 bp were removed from the draft genomes.
MLST and cgMLST
Ridom SeqSphere+, v6.0.2 (Ridom GmbH, Münster, Germany) was used to genotype the strains using MLST and cgMLST schemes available through the software. MLST profiles were assigned using Brucella 9 locus Multilocus Sequence Typing (MLST-9) scheme (
https://pubmlst.org/brucella/) and cgMLST profiles were calculated using B. melitensis task template with 2,704 target core genes as described by Janowicz et al.,2018 [
15]. Sequences containing less than 98 % of cgMLST good targets were removed and a Multiple spanning tree (MST) was generated by pairwise comparison of the target genes using default parameters. Missing values were ignored in the calculation of distance between pairs of sample profiles.
In the cgMLST analysis, we additionally included 105 genomes available at GenBank (with known geographical origin) and assigned to East Mediterranean clade by Janowicz et al. [
16] and 28 strains with known origin grouped in East Mediterranean clade published in Sacchini et al. [
5].
3. Results
MLVA Analysis
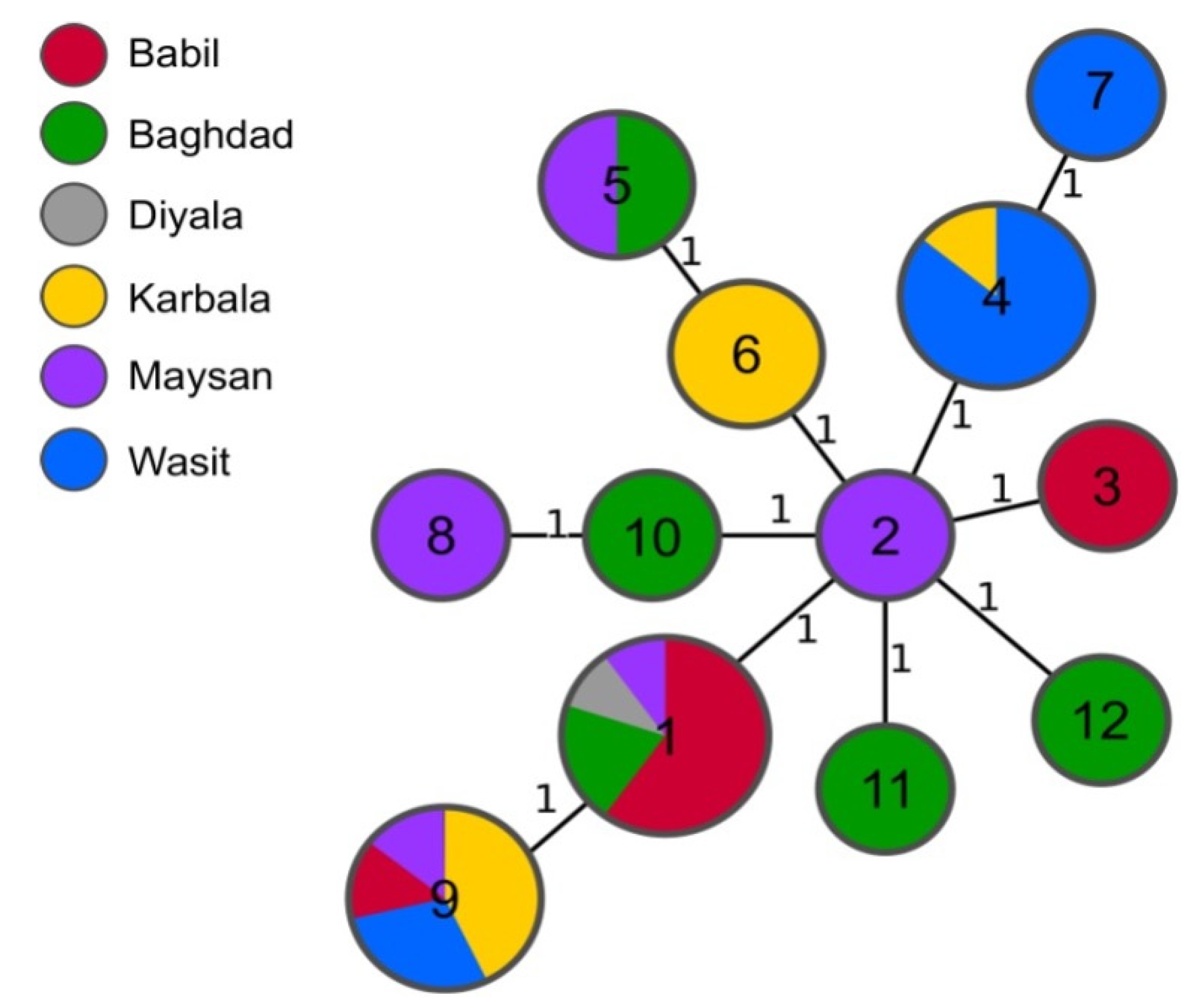
Isolates collected in the six neighbouring Iraqi governorates identified as B. melitensis were isolated from sheep (n=22), cattle (n=12) and buffalo (1). The strains were genotyped using MLVA-16 and twelve different MLVA profiles were assigned with a maximum distance between individual genotypes of two alleles. Fourteen loci were identical between all the isolates and the variation occurred only in Bruce04 and Bruce16 (
Table 2).
The most common profile (1-5-3-13-3-2-3-2-4-40-8-4-4-3-15-4) was shared by ten strains isolated from sheep and cattle in four different governorates: Babil, Baghdad, Diyala and Maysan (
Table 1,
Table 2). The second most frequently assigned profiles were 1-5-3-13-3-2-3-2-4-40-8-8-4-3-15-4 found in strains from Babil, Karbala, Maysan and Wasit and 1-5-3-13-3-2-3-2-4-40-8-4-4-3-19-4 from Karbala and Wasit. Both profiles were found in strains isolated from cattle and sheep. The minimum spanning tree of 35 Iraqi B. melitensis isolates, based on their MLVA-16 results (
Figure 2).
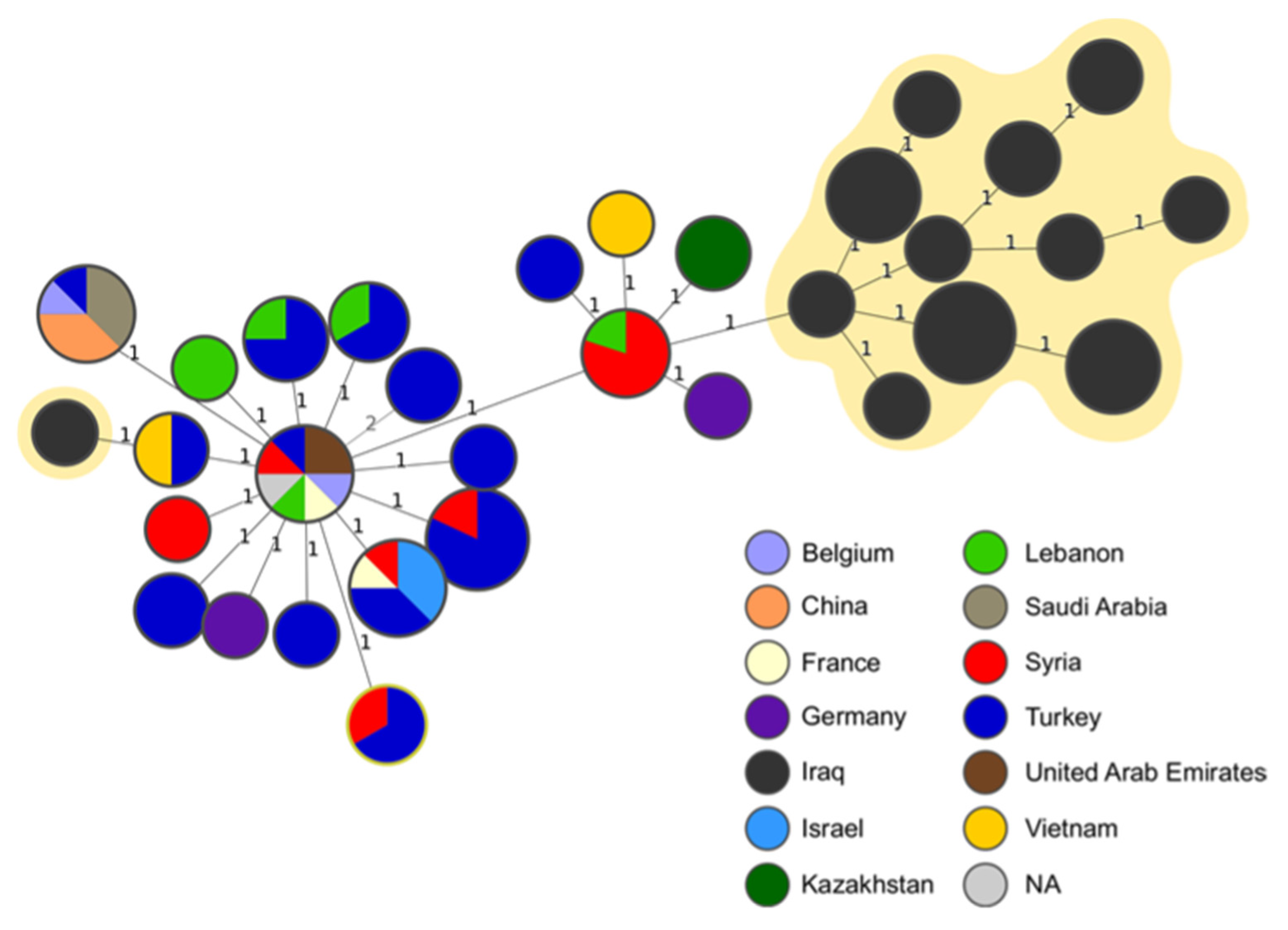
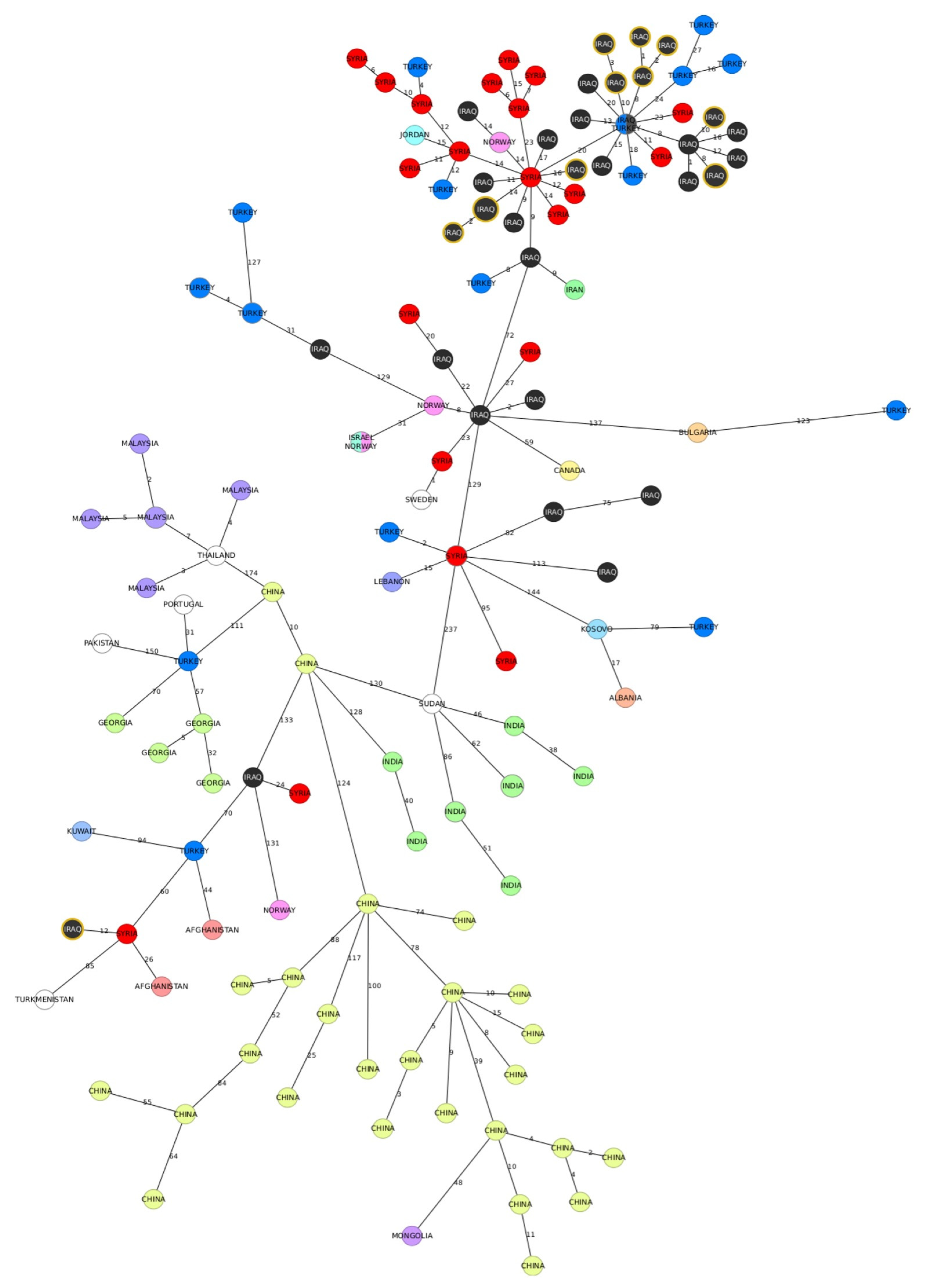
The comparison of MLVA-16 profiles of the strains analysed in this study with the profiles obtained from the public database did not reveal any shared genotypes. All Iraqi isolates from our study clustered within East Mediterranean clade and, except from one strain, all clustered together in same branch of MST tree.
The MST analysis showed the minimum distance of one allele between our isolates and the other strains and the most frequent connection was found with the MLVA profiles assigned to the isolates originating in Syria (
Figure 3). One strain, obtained from buffalo, was placed farther away from the rest of the isolates from our study and was linked to strains from Turkey and Vietnam (
Figure 3).
WGS
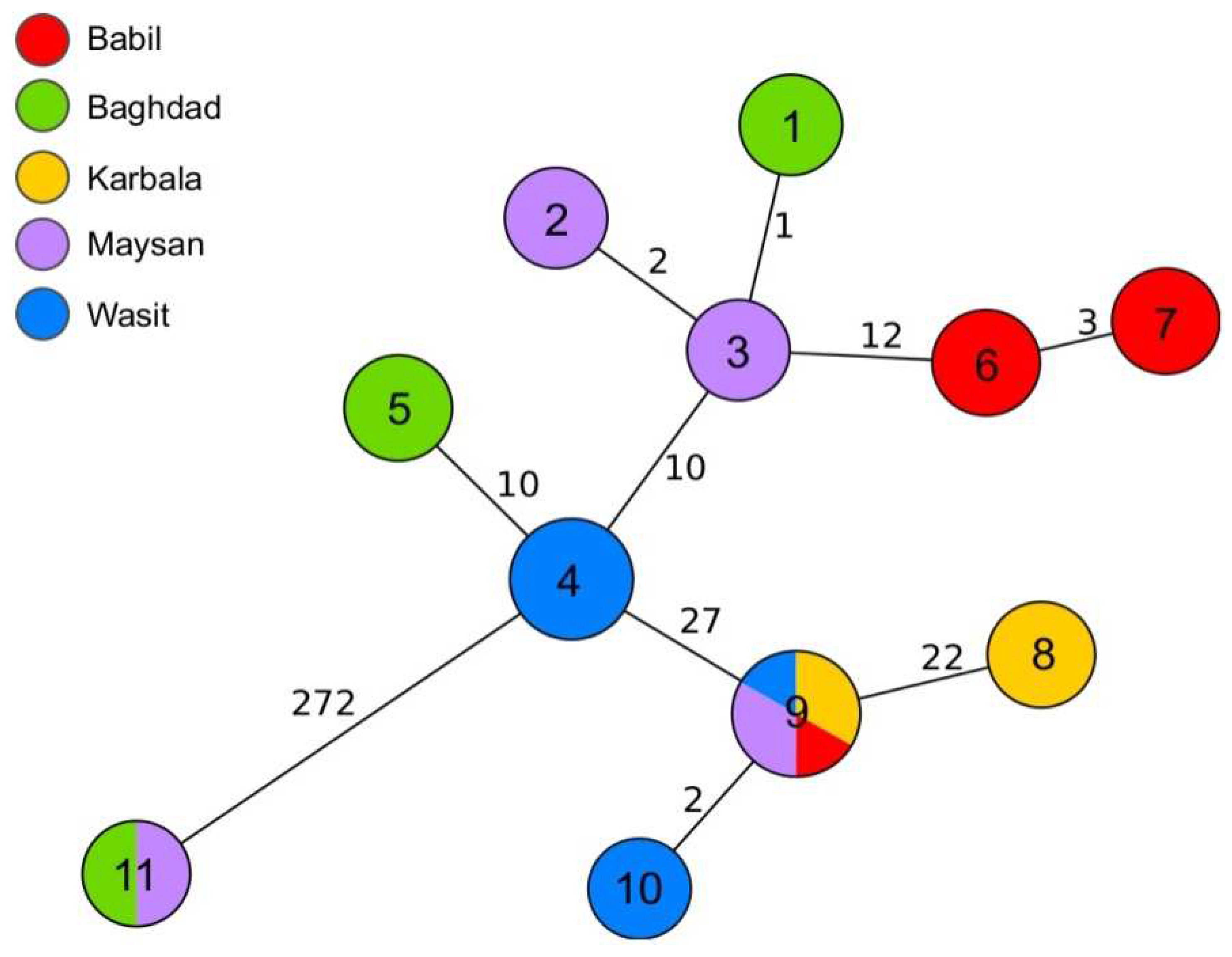
Out of 35 samples analysed by WGS, 10 passed the cgMLST quality threshold and were assigned cgMLST profiles (
Table 2). The majority of sequences grouped together within 35 allele distance (
Figure 4). Several more closely related clusters were also observed. Out of these, the biggest, which contained cgMLST profiles 9 and 10, was assigned to isolates from four governorates, Babil, Karbala, Maysan and Wasit. Six strains from these clusters shared the same MLVA-16 profiles. The second biggest cluster grouped strains with cgMLST profile 4, all collected in Wasit. Additionally, no more than three alleles of difference were observed between profiles 1, 2, and 3 from Baghdad and Maysan, and profiles 6 and 7 from Babil. Interestingly, two strains collected from sheep in Baghdad and Maysan were distant by 272 alleles from all the other genotypes. Similar relation was not identified by MLVA-16 typing (
Figure 4).
Then, we compared the strains from our study to publicly avaiable sequences from East mediterranean clade. The majority of strains was grouped within a large cluster predominantly comprising genotypes from the Middle East. (
Figure 5). The minimum distance between our sequences and other available genotypes was eight alleles, and the closest related genomes included sequences from Iraq, Syria and Turkey. The two Iraqi strains that were previously shown to be distant by 272 alleles from the other strains from the dataset, were positioned in a separate branch of the MST, containing few genotypes. The closest neighbouring strain originating in Syria differed by 12 cgMLST alleles from the two strains from our dataset.
4. Discussion
About 12 million Iraqis reside in rural areas and depend on agriculture for their livelihoods. Cattle, sheep and goats are farmed for the production of meat, wool, milk and leather. In Iraq, after agricultural production, livestock is the second largest sector providing income to the general population. Years of conflict in Iraq have destroyed or severely damaged crops, equipment, livestock, seeds, and food reserves, leaving 3.2 million Iraqis food insecure [
17].
Ninewa Governorate has been deeply affected by the conflict since ISIS (Islamic State of Iraq and Syria) took control of central and north-western Iraq in 2014. The conflict has displaced one million people, the majority of whom remain within the borders of the Ninewa governorate, in host communities or in reception camps. Many families have fled their homes taking their animals with them, most of which have not been vaccinated for the common infectious diseases of livestock since ISIS took control of the area. These animals could carry highly contagious epidemic diseases that can spread rapidly across national and international borders, to other animals and humans, with serious socio-economic and health consequences, brucellosis included.
Brucellosis is one of the most contagious epidemic diseases in ruminants, especially in Iraq were it disease poses a significant threat to public for both animals and humans [
18]. Records of the infection as endemic at such high levels among the different species is also indicative of the need of improving the control program. Moreover, different Brucella species may play a significant role in the transmission of brucellosis to human, mainly B. melitensis, spreading through unpasteurized milk or associated dairy products [
19].
Brucellosis’s spread in Iraq can also be linked to travel activities, as individuals can carry the disease to endemic area to different destinations, posing the risk of this dissemination [
20].
Brucellosis control needs a series of actions to be implemented in the field, and these actions should be set at different levels, according to the epidemiological situation of the territory:
in free areas, or in areas with low prevalence, periodic screening of farms is needed to identify infected animals to be eliminated in a short time;
in endemic areas, and according to the prevalence of the disease, vaccination of healthy animals should be considered as an option, either as mass vaccination or only as a young animals vaccination;
educational campaigns aimed at the population in relation to both eating habits (consumption of pasteurized or boiled milk), and to the adoption of good hygienic practices (avoid contact and handling of organic material at risk) are of paramount importance for decreasing the impact of brucellosis on public health. In parallel, it is important also to carry out educational campaigns aimed at professionally exposed categories of workers (i.e. farmers, veterinarians, butchers, laboratory workers) so that they gain sufficient information allowing them to adopt the most appropriate hygiene rules in their professional context.
The findings of this study suggests that even relatively small investigations can provide sufficient evidence to justify changes in existing control strategies, especially by using genomic analysis. The same genotypes have been found in neighbouring governorates, and this finding can be related to breeding practices, which are similar across the region, making the epidemiological situation of brucellosis in neighbouring governorates unlikely to differ significantly. However, distantly related genotypes have also been detected, some of them related to strains isolated in foreign countries, such as Syria and Lebanon (one allele difference in MLVA-16,
Figure 3) or Turkey, Vietnam and Kazakhstan (two alleles difference in MLVA-16,
Figure 3). This finding underscores the necessity to investigate the international connections of Iraqi strains to identify potential risk factors for the disease's introduction. This would involve scrutinizing major relationships with neighboring countries like Turkey and Syria (possibly due to infections from shared pastures) or more distant nations (likely associated with the trade of live animals).
One of the strengths of this study lies on the analysis of a database containing data on Brucella spp. isolated from animals, and this is an added value with respect to other studies relying only on sequences of isolates coming from returning travellers. This allows for a more direct and comprehensive understanding of the local situation regarding animal brucellosis in Iraq. However, a study limitation could be the relatively small dataset at disposal for the study. This suggests the importance and the need for larger and more robust datasets for more accurate future research. However, this approach is costly, demanding important financial resources [
21].
Variable Number Tandem Multiple Repeat Analysis (MLVA) and Whole Genome Sequencing (WGS) analyse different parts of the genome and reflect different evolutionary processes; therefore, they can have different performances as tools for the surveillance and epidemiology of Brucella.
Both offer greater discrimination than other typing schemes. The discriminating power is one of the most important selection criteria for a genotyping method. Although, it is important to consider that the discriminating capacity of a method is influenced by other factors, such as enzymes and primers used, enzymatic and amplification conditions and epidemiological characteristics of the microorganisms tested.
Finally, it is also important to consider the epidemiological context under study; the surveillance of "local epidemic events" of bacterial infections, actually, can only be carried out using rapidly evolving markers, such as those studied with MLVA, while for population or long-term epidemiological studies, methods based on the investigation of stable and conserved markers as whole genome sequencing are preferred.
Due to the broad, nonspecific consequences of brucellosis, the actual disease burden often goes undiagnosed, resulting in a lack of prioritization of brucellosis control in the context of National control plans for animal diseases and zoonosis prevention [
22]. Large- size herds and flocks combined with pastoralism practices may also play an important role in region with limited resources.
By applying different control measures in specific locations, it may be possible to maximize public health benefits and minimize the spread of infection to areas with lower or no prevalence, providing that all the Authorities concerned will be involved in a “One-Health” approach.
An importance issue is also the need for vaccination against brucellosis in domestic ruminants. In given epidemiological situations, this approach would be an important starting point to improve animal health, food safety and to decrease the burden of economic implications related to the presence and spread of the disease. It is of utmost importance the identification of the areas and animal species in which the adoption of vaccination programs may have long-term economic benefits. It is widely acknowledged that controlling and eliminating livestock brucellosis, is crucial for reducing the disease in humans, and these actions are falling under veterinary responsibility [
23].
The FAO / WHO report on
Brucella melitensis in Eurasia and the Middle East [
24] proposes zoning / compartmentalization within a country as one of the generic disease control measures that could be applicable to
Brucella melitensis control. However, an effective compartmentalization should be accompanied by a series of biosecurity measures that could be difficult to implement in Iraq, given the intensity of non-regulated animal movements.
5. Conclusions
The findings of this study suggests that even relatively small investigations can provide sufficient evidence to justify improvements in existing control strategies. Breeding practices are similar across the region, therefore, the application of different control measures in specific locations with high prevalence, based on molecular epidemiological studies, would increase the chances to maximize public health benefits and minimize the spread of infection to areas free from the disease or with a low prevalence. In the areas under study, the focus should be on important public health preventive interventions, such as livestock vaccination, accompanied by the development of related infrastructures, and robust campaigns on the general population about disease awareness. In addition to that, there is a need for effective border control, as well as a regulatory framework to control this disease. Each region should collaborate and exchange information, knowledge and strategy about infectious disease prevention and control with the other regions of the country. Moreover, the control of brucellosis in Iraq requires a strong disease surveillance program managed by the veterinary and health authorities, increasing awareness among local health professionals, in a “One-Health” framework, which will improve the diagnosis and management of brucellosis cases, and the control and eradication of this disease.
Acknowledgments
This work was supported with funding provided by Civilian Research and Development Foundation (CRDF) Global with CVL in Iraq.
Conflicts of Interest
authors declare no conflicts of interest.
References
- Savini, L.; Candeloro, L.; Conte, A.; De Massis, F.; Giovannini, A.; et al. Development of a forecasting model for brucellosis spreading in the Italian cattle trade network aimed to prioritise the field interventions. PLoS One 2017, 12(6), e0177313. [CrossRef]
- Hadad, J.J.; Hammad, D.A.; Alaboudi, A.R. Isolation of Brucella strains from dairy products in Ninevah province, Iraq. Iraqi Journal of Veterinary Sciences 1997, 10, 39–44.
- Abdulhameed, M.F.; Sayhood, M.H.; Aldeewan, A.B.; Srayyih, T.H. Assessment of Seroprevalence and the Risk Factors of Sheep Brucellosis in Basrah (Southern Iraq): A Challenge to Prospectively Control Brucellosis. J Pure Appl Microbiol. 2020, 14 (4), 2543-2554. [CrossRef]
- Franc, K. A.; Krecek, R. C.; Häsler, B. N.; Arenas-Gamboa, A. M. Brucellosis remains a neglected disease in the developing world: a call for interdisciplinary action. BMC Public Health 2018, 18 (1), 125. [CrossRef]
- Sacchini, F.; Wahab, T.; Di Giannatale, E.; Zilli, K.; Abass, A.; Garofolo, G.; Janowicz, A. Whole Genome Sequencing for Tracing Geographical Origin of Imported Cases of Human Brucellosis in Sweden. Microorganisms 2019, 7 (10), 398. [CrossRef]
- Gwida, M.; Al Dahouk, S.; Melzer, F.; Rösler, U.; Neubauer, H.; Tomaso, H. Brucellosis – Regionally Emerging Zoonotic Disease? Croat medial Journal 2010, 51, 289-295. [CrossRef]
- Zhu, X.; Zhao, Z.; Ma, S.; Guo, Z.; Wang, M.; Li, Z.; Liu, Z. et al. Brucella melitensis, a latent "travel bacterium," continual spread and expansion from Northern to Southern China and its relationship to worldwide lineages. Emerg. Microbes Infect. 2020, 9 (1), 1618-1627. [CrossRef]
- Al-Koubaisy, H.N.; Lafi, S.A. Presentation of brucellosis in an endemic area; west of Iraq. Egypt. Acad. J. Biol. Sci. 2011, 3 (1), 13-18. [CrossRef]
- 9. De Massis F, Zilli K, Di Donato G, Nuvoloni R, Pelini S, Sacchini L, et al. Distribution of Brucella field strains isolated from livestock, wildlife populations, and humans in Italy from 2007 to 2015. PLoS ONE 2019, 14(3): e0213689. [CrossRef]
- Sun, M.; Liu, M.; Zhang, X.; Zhang, G.; Zhu, L.; Ding, J.; Zhang, Z.; Sun, S.; Sun, S.; Shao, W.; et al. First Identification of a Brucella abortus biovar 4 Strain from Yak in Tibet, China. Vet. Microbiol. 2020, 247, 108751. [CrossRef]
- Orsini, M.; Ianni, A.; Zinzula, L. Brucella ceti and Brucella pinnipedialis genome characterization unveils genetic features that highlight their zoonotic potential. Microbiology open 2022, 11 (5), e1329. [CrossRef]
- Le Flèche, P.; Jacques, I.; Grayon, M.; Al Dahouk, S.; Bouchon, P.; Denoeud, F.; Nöckler, K.; Neubauer, H.; Guilloteau, L.A.; Vergnaud, G. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 2006, 6, 1-14. [CrossRef]
- Garofolo, G.; Ancora, M.; Di Giannatale, E. MLVA-16 loci panel on Brucella spp. using multiplex PCR and multicolor capillary electrophoresis. J. Microbiol. Methods 2013, 92 (2), 103–107. [CrossRef]
- Nascimento, M.; Sousa, A.; Ramirez, M.; Francisco, A.P.; Carriço, J.A.; Vaz, C. PHYLOViZ 2.0: Providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 2017, 33, 128-129. [CrossRef]
- Janowicz, A.; De Massis, F.; Ancora, M.; Cammà, C.; Patavino, C.; Battisti, A.; Prior, K.; Harmsen, D.; Scholz, H.; Zilli, K. et al. Core genome multilocus sequence typing and single nucleotide polymorphism analysis in the epidemiology of Brucella melitensis infections. J. Clin. Microbiol. 2018, 56. [CrossRef]
- Janowicz, A.; De Massis, F.; Zilli, K.; Ancora, M.; Tittarelli, M.; Sacchini, F.; Di Giannatale, E.; Sahl, J.W.; Foster, J.T.; Garofolo, G. Evolutionary history and current distribution of the West Mediterranean lineage of Brucella melitensis in Italy. Microb Genom. 2020, 6 (11), mgen000446. [CrossRef]
- Food and Agriculture Organization of the United Nations (FAO). 2017. "Iraq: FAO launches emergency animal health campaign in former ISIL-held Mosul area. Vaccines and animal feed critical to safeguard livestock assets, production and food security." Rome. Available online: https://www.fao.org/newsroom/detail/Iraq-FAO-launches-emergency-animal-health-campaign-in-former-ISIL-held-Mosul-area/en.
- Dahl, M.O. Brucellosis in food-producing animals in Mosul, Iraq: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0235862. [CrossRef]
- Khalid, H.M. Seroprevalence and Associated Risk Factors of Brucellosis Among Human Population in Duhok City, Iraq. Infect Drug Resist. 2023, 16, 2805-2811. [CrossRef]
- Ameen, A.M.; Abdulaziz, N.S.; Ghaffar, N.M. Molecular Detection and Associated Risk Factors of Brucella melitensis in Aborted Sheep and Goats in Duhok Province, Iraq. Pathogens 2023, 12, 544. [CrossRef]
- Boschiroli, M.-L.; Foulongne, V.; O’Callaghan, D. Brucellosis: A Worldwide Zoonosis. Current Opinion in Microbiology 2001, 4 (1), 58–64. [CrossRef]
- Halliday, J.E.; Allan, K.J.; Ekwem, D.; Cleaveland, S.; Kazwala, R.R.; Crump, J.A. Endemic zoonoses in the tropics: a public health problem hiding in plain sight. Vet Rec. 2015, 176 (9), 220-225. [CrossRef]
- Zhang, N.; Huang, D.; Wu, W.; Liu, J.; Liang, F.; Zhou, B.; Guan, P. Animal brucellosis control or eradication programs worldwide: A systematic review of experiences and lessons learned. Prev. Vet. Med. 2018, 160, 105-115. [CrossRef]
- Food and Agriculture Organization of the United Nations (FAO). 2010. Brucella melitensis in Eurasia and the Middle East. FAO Animal Production and Health Proceedings. No. 10. Rome.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).