1. Introduction
In a recent report, annually liver disease contributes around 4% of all the deaths worldwide and accounts for over two million deaths. The global burden of liver disease is placing a significant strain on public healthcare systems. Cirrhosis and Hepatocellular Carcinoma (HCC) are major contributors including Nonalcoholic fatty liver disease (NAFLD) and Drug-induced liver injury (DILI) which significantly contributes as common cause of liver related deaths[
1].
The liver is the largest organ in the human body responsible for vital functions including metabolism of nutrients and xenobiotics. The liver is structurally made-up of small anatomical units called liver lobules. Every lobule is histologically organized in a unique hexagonal architectural arrangement of hepatocytes (parenchymal cells PCs) into three zones that exhibit a distinct functionality known as liver zonation. Venous blood from the gut mixes at portal vein with oxygenated atrial blood and flows towards central vein through sinusoids. The liver tissue area around portal triad i.e. peri-portal recognized as Zone1 (oxygen enriched) is involved in metabolic functions and specialized in gluconeogenesis whereas tissue area around central vein recognized as zone3 (less oxygen) is involved in bile acid production drug metabolism and glycolysis. The area in between i.e. mid-lobular known as Zone 2 is a transitional region where concentration of oxygenated blood, nutrients, metabolites, and gut-derived toxins varies along portal-central vein axis. Spatially heterogenous hepatocytes are distinguished by the gene expression profiles where 50% of the genes are expressed along lobular zonation axis[
2]. Zone 2 is believed to plays an important role in homeostatic renewal of hepatocytes, liver mass regeneration and proliferation upon liver injury [
3,
4]. Spatial heterogeneity of parenchymal and non- parenchymal cells (NPCs) is zone dependent which modulate the Differentially Expressed Genes (GEGs) expression and initiate several liver diseases including Nonalcoholic steatohepatitis (NASH), DILI, HCC and liver regeneration[
5].
Other than hepatocytes, Liver Sinusoidal Endothelial Cells (LSECs) contribute around 15–20% of total number of liver cells are highly specialized non parenchymal endothelial cells and act as a physical barrier between blood substrates and hepatocytes. Liver endothelial cells (ECs) includes LSEC, vascular ECs, and lymphatic ECs (LyECs)[
6,
7]. As chronic liver disease advances, hepatocyte functioning impaired by crosstalk between other liver cells which initiates an important role in regulating fibrosis. LSECs have unique fenestrae (pores) that allow for efficient clearance of pathogens, debris, and toxins from the blood, keeping the liver clean and functional. LSECs act as determinants of hepatic fibrosis where the process of capillarization precedes fibrosis in which LSECs lack fenestration and develop an organized basement membrane[
8]. LSECs are the major drivers in fibrosis[
9] and differential gene expression is observed between the different zones of the liver lobule during fibrosis[
10]. For example, the periportal zone 1 expresses genes involved in the uptake of nutrients, mid-lobular zone2 expresses genes involved in metabolism of nutrients, pericentral zone 3 expresses genes involved in the secretion of bile. Gradient of differentially expressed genes (DEG) is observed in LSECs and hepatocytes following disrupted zonation architecture and liver functionalities in most liver diseases including NAFLD, NASH and DILI. DEG manifests around pericentral zone 3 that later progresses towards advanced stages of fibrosis, bridging fibrosis and cirrhosis [
11,
12]. Liver bridging fibrosis, a specific advanced NASH feature, is a type of scarring caused by the accumulation of excess collagen around hexagonal portal triad-central vein region in the later stages of liver fibrosis. These collagen bands connect different areas of the liver and block the flow of blood and bile, which leads to liver failure. Early liver fibrosis, a condition where scar tissue builds up, can indeed be reversed if detected early in zonation at molecular level and addressed promptly. However, it's crucial to remember that this reversal is contingent on preventing the progression to more advanced stages like bridging fibrosis, where the scar tissue becomes more organized, extensive and the damage is irreversible.
It’s challenging to measure zonation-specific structural variations during early stages of fibrosis and later at bridging fibrosis level. Our knowledge is limited and poorly understood on the role of 1) zone 2 LSECs in early liver fibrosis, 2) LSECs-specific marker genes in hepatotoxic DILI conditions, 3) DEG profiles within unclear boundaries of liver zones under normal, NASH and DILI disease fibrotic patterns. In this study, efforts are made to computationally characterize and quantify spatial heterogeneity of hepatocytes at molecular level using single cell RNA seq and spatial molecular imaging techniques to develop our understanding of zonal restructuring, role of LSECs and marker genes in normal and diseased fibrotic conditions.
2. Results
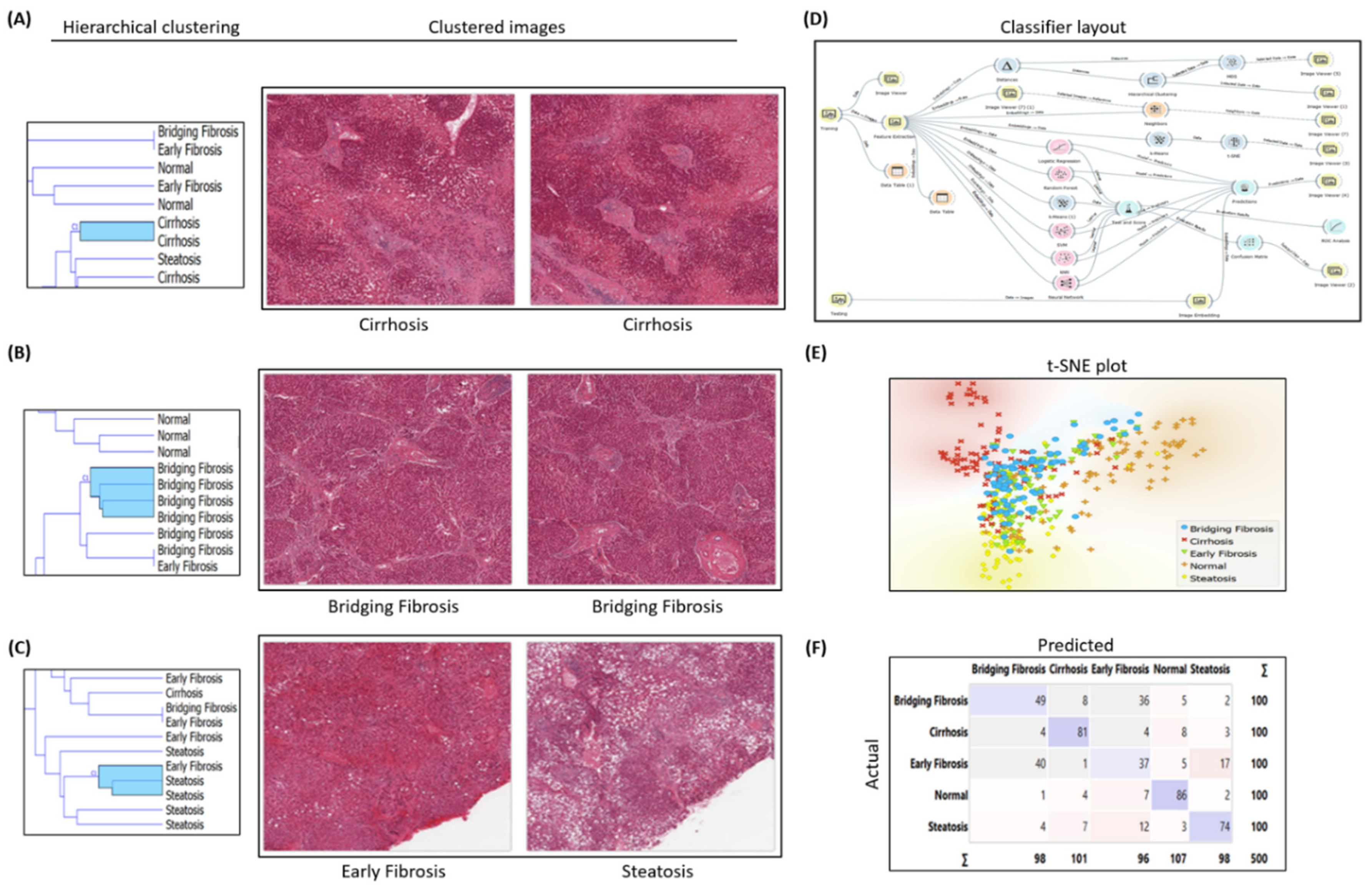
2.1. Histopathology H&E Image Classification Experimental Workflow for Early and Bridging Fibrosis
Image analytics and experimental workflow, illustrated in
Figure 1 D, is designed to classify Hematoxylin and Eosin (H&E) histopathology images under five categories i.e. normal, steatosis, early fibrosis, bridging fibrosis and cirrhosis. Machine learning widgets, shown in workflow, are executed to load, process, classify, cluster, and visualize the imaging data. After predictive analysis, images are clustered based on the common histologic disease features learned by the machine learning models, i.e. all the bridging fibrosis images grouped together under one group as shown in t-SNE plot
Figure 1E and hierarchical clustering
Figure 1B. Other category images are grouped similarly as per their disease morphology features. The Confusion matrix shown in
Figure 1 F illustrates correctly predicted images (diagonally highlighted blocks) under respective categories. Closely clustered images share common morphological disease features in early fibrosis and steatosis (NASH specific feature) where white fat droplets are present in both images, shown as an example in
Figure 1C. Classification models performance summarized in
Table 1.
2.2. DEG Analysis for NASH and Zonation Expression
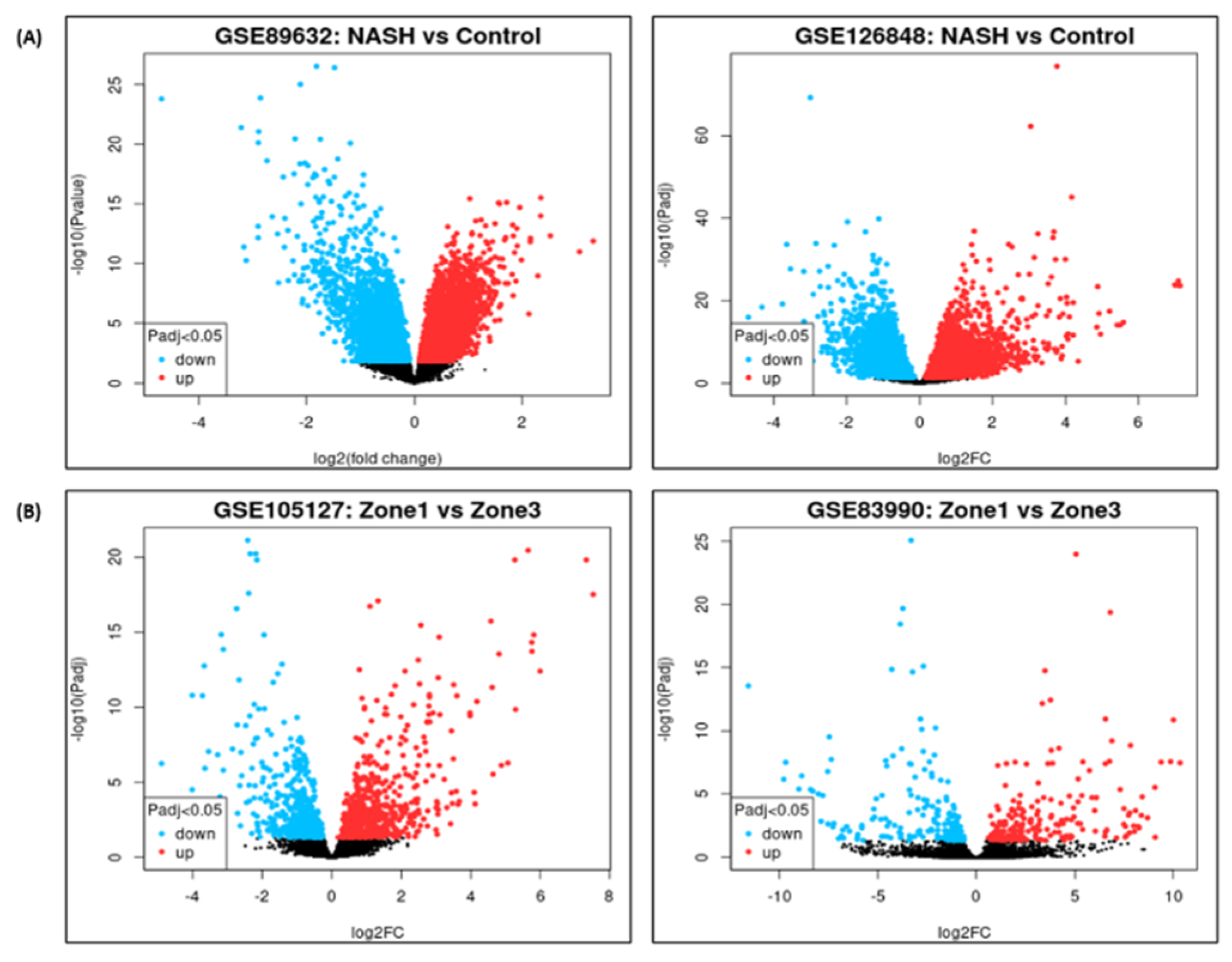
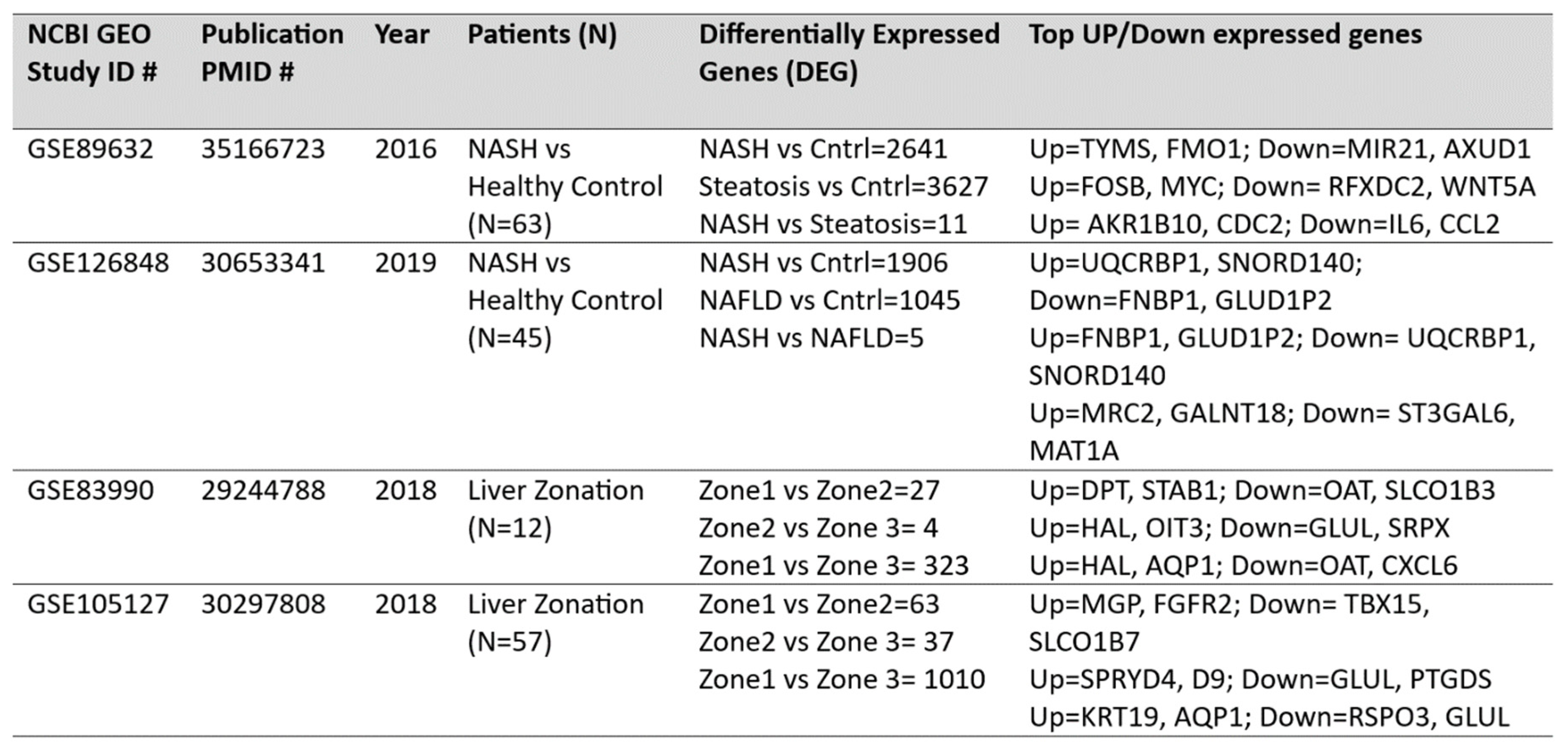
DEG analysis for NASH and zonation expression is performed over four independent studies
GSE89632, GSE126848, GSE83990, GSE105127 using NCBI GEO2R analysis tool. Volcano plots for NASH vs Control are illustrated in
Figure 2A, zone1 vs zone 3 expression are illustrated in
Figure 2B. Volcano plot displays statistical significance (-log10 P value) versus magnitude of change (log2 fold change) of differentially expressed genes in NASH vs control and zonation-specific expression studies. The most up regulated genes identified after analysis in NASH are
TYMS, FMO1, UQCRB; and in zone1 vs zone 3 analysis are HAL, AQP1 KRT19. Details of DEGs and top up/down expressed genes are summarized in
Table 2.
2.3. Single Cell Clustering and DEGs Expression Profiles in DILI and NASH ECs
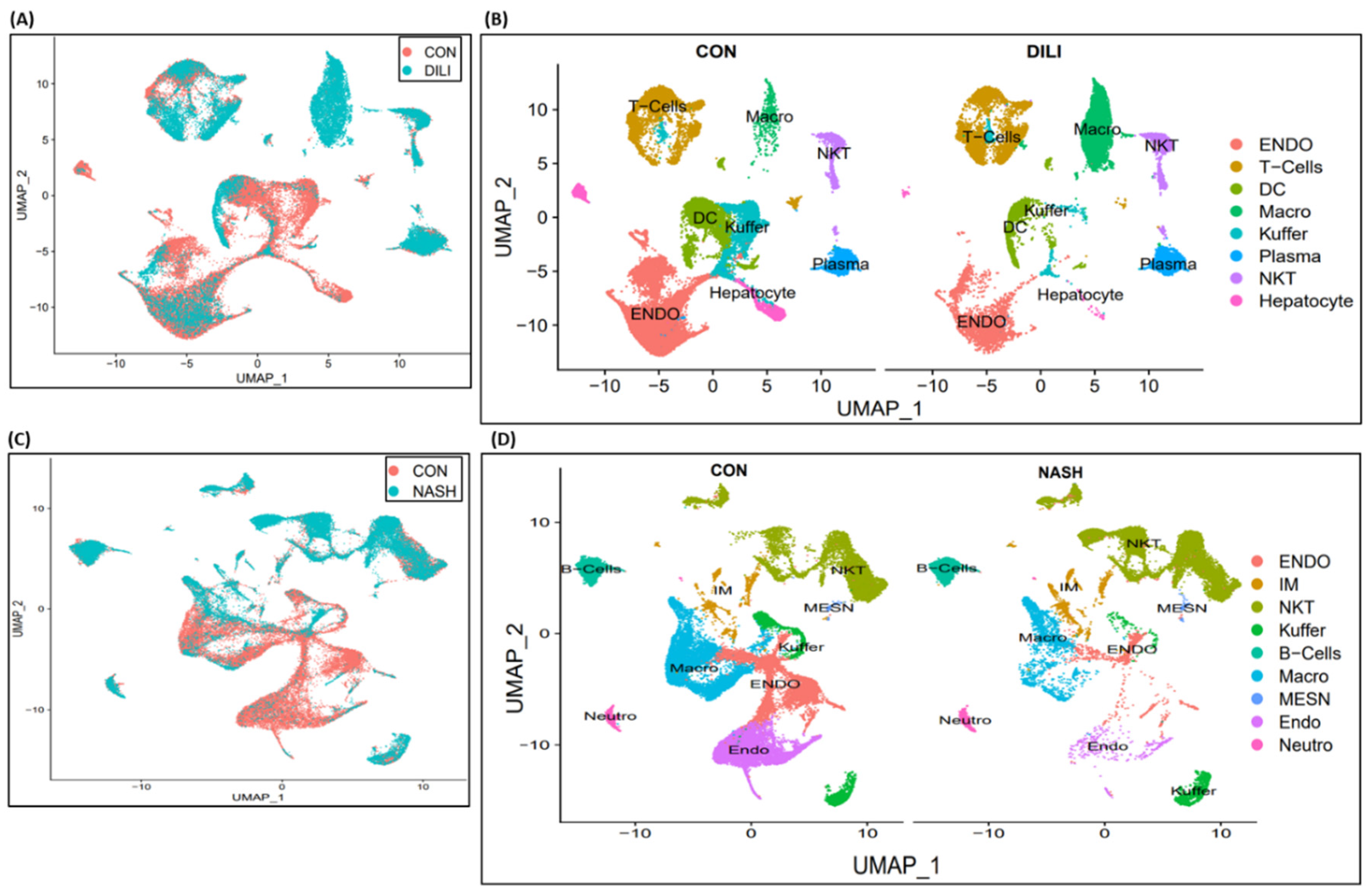
Single-cell transcriptomic analysis is performed over liver non-parenchymal cells (NPCs) in control, DILI and NASH datasets available at
NCBI GEO GSE166178[
13] and analyzed for the heterogeneity of inter- and intra-group endothelial cells in healthy and diseased mouse livers. Cell clustering results are illustrated in
Figure 3A-B for control vs DILI, and control vs NASH illustrated in
Figure 3C-D. DEGs expression is performed for eight commonly expressed genes
STAB2, OIT3, F8, AQP1, TEK, TIMP3, TIE1, CTSL in DILI and NASH ECs populations, illustrated in Supplemental
Figure S1
2.4. Spatial Transcriptomics Data Analysis for Zonation Marker Genes
2.4.1. 10x Genomics Visium Image Analysis for Spatial Distribution of Zonation Expression Markers
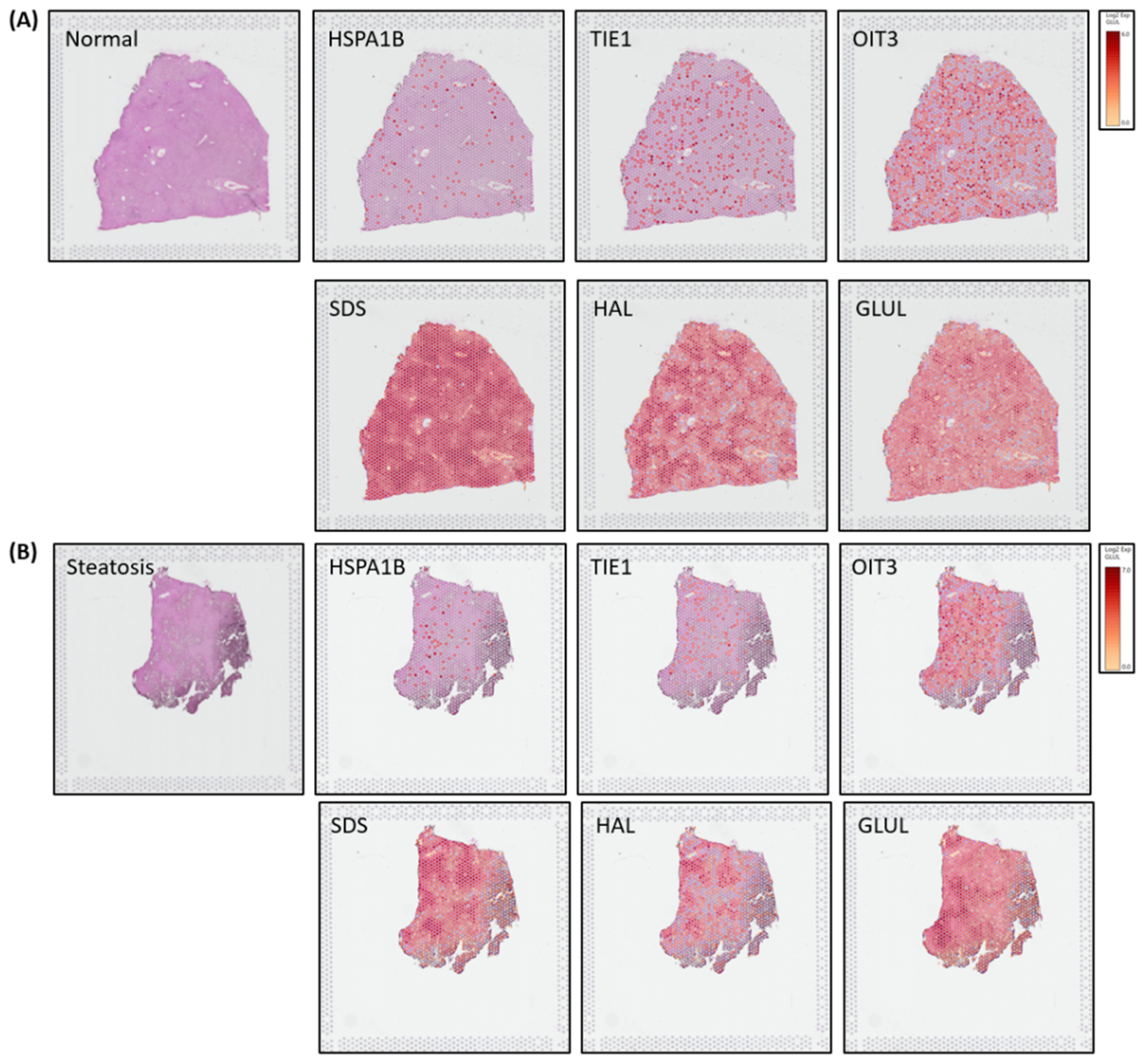
Spatial transcriptomics (ST) image analysis is performed on human liver H&E stained healthy and steatosis images using 10x genomic`s Visium platform to identify zonation patterns based on known markers[
14]. Expressed dots(red) are the 55-micron Visium spots from where transcriptomes (RNA) are extracted for studying spatial cellular populations present and gene expressions at that specific tissue location.
Figure 4A-4B illustrates zonation-specific expressed genes such as
HSPA1B, TIE1, OIT3, GLUL, HAL, SDS overlaid Visium liver tissue image[
15]. Expression profiles unraveled zonation patterns are highly dynamic varying from mild, mid to high in normal and steatosis samples.
2.4.2. Spatial Molecular Imaging to Demonstrate Liver Zonation Architecture
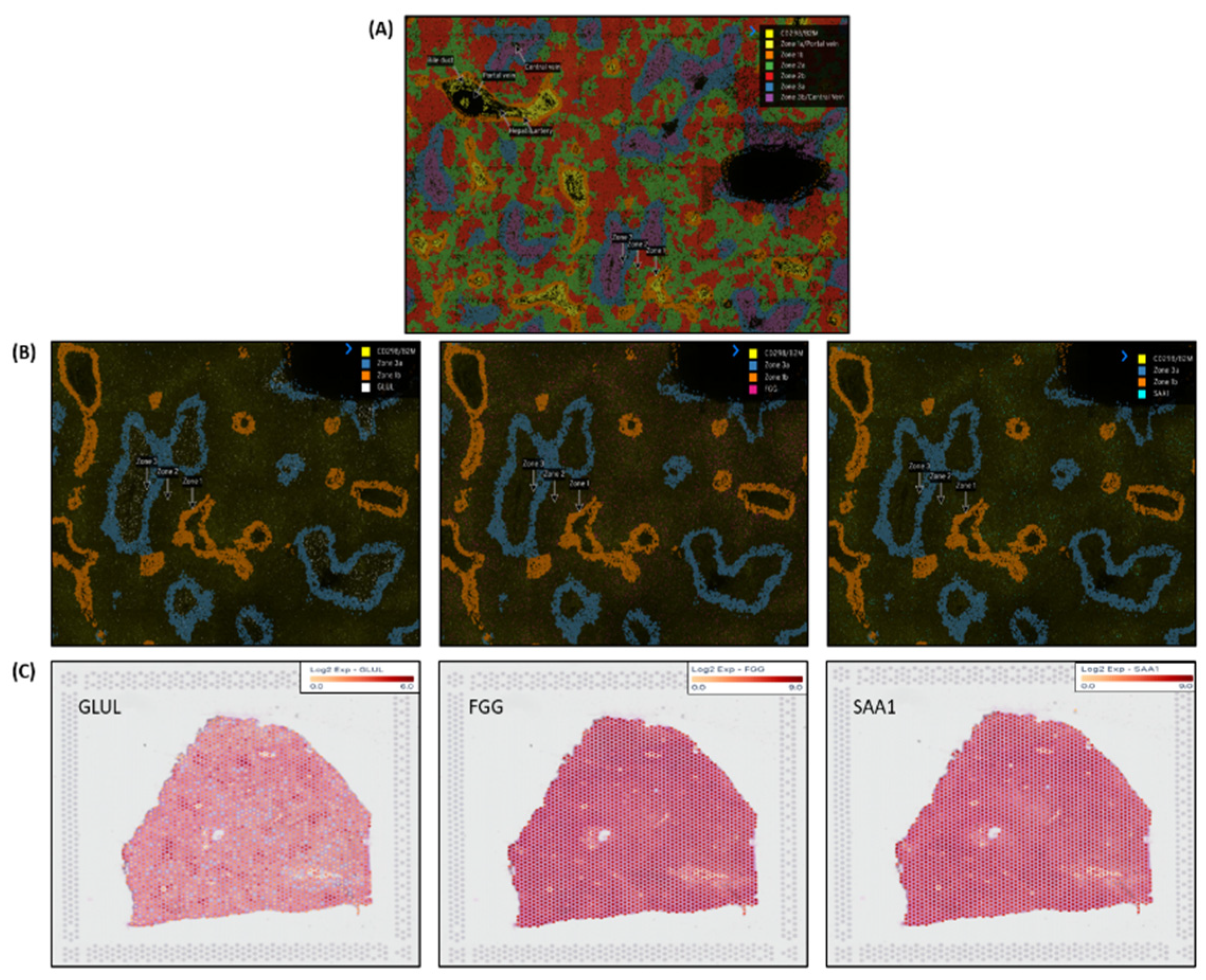
Nanostring`s CosMx
TM Spatial Molecular Imager (SMI) platform, illustrated in
Figure 5A is a high-plex spatial multiomics image generated from a 5 µm thick human formalin-fixed paraffin-embedded (FFPE) liver section stained for both protein and RNA analytes to demonstrate zone1, zone2 and zone 3 architecture.
Figure 5B illustrates differential gene expression with three target markers i.e.
GLUL, FGG &
SAA1 to highlight the zonation boundaries in a healthy liver tissue at molecular resolution. Similar equivalent patterns demonstrated in 10x genomics Visium images for the same three zonation-specific genes which highlights zonation boundaries in a normal human liver. However, in the diseased and chronic liver injury conditions, this zonation disruption exhibits varying DEG expression.
2.5. Liver Cell Clustering and LSECs Markers
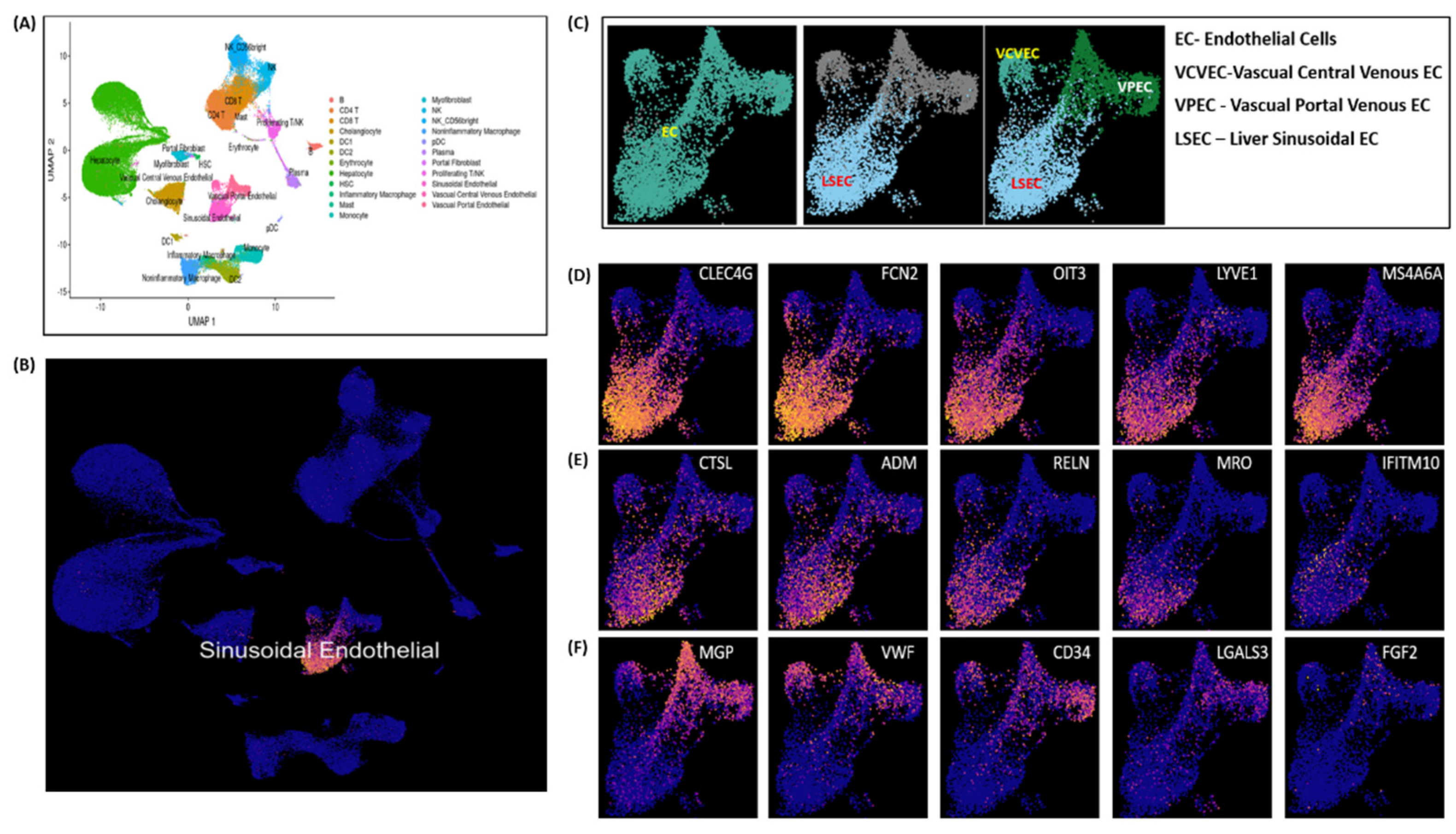
Endothelial cell (ECs) clustering data analysis is executed here to demonstrate LSECs gene expression markers on an integrated scRNA-seq data of 28 healthy human liver samples. Single-cell scRNA-seq data used here was acquired from separate five independent studies reported elsewhere in [
16].
Figure 6A illustrates UMAP clustering of all the normal liver cells. LSECs cluster shown in
Figure 6B are picked as subset of clustered ECs cells shown in
Figure 6A. Vascual Central Venous ECs (VCVEC) and Vascual Portal Venous ECs (VPEC) are clustered along-with LSECs, shown in
Figure 6C which further separated as subsets and highlighted using ECs and LSECs separately with
CLEC4G, FCN2, OIT3, LYVE1 expression marker genes illustrated in
Figure 6D-6E. VCVEC and VPEC clusters further separated and highlighted using
MGP, VWF, CD34 expression marker genes, illustrated in
Figure 6F.
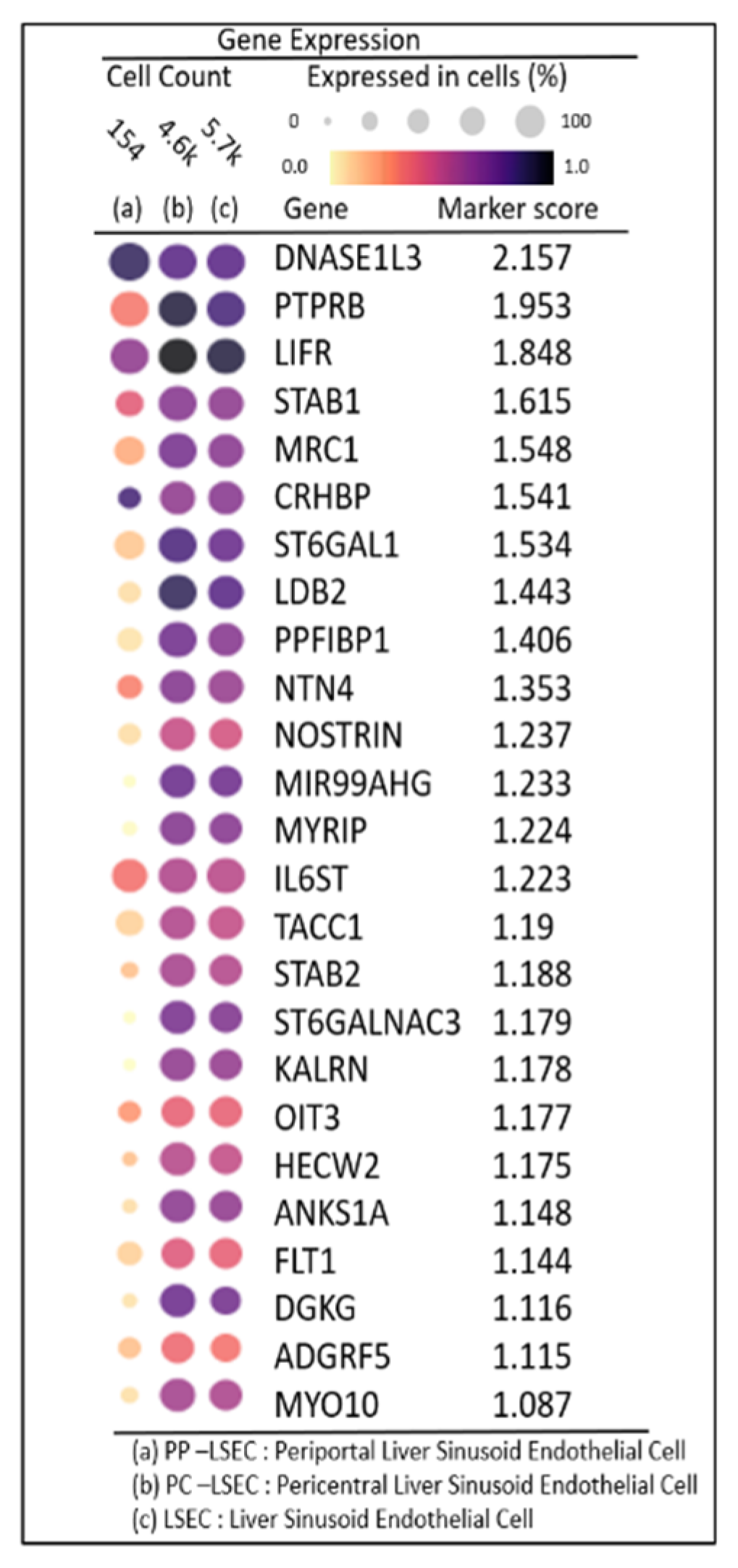
2.6. DEG Expression Profiles in Zonated LSECs
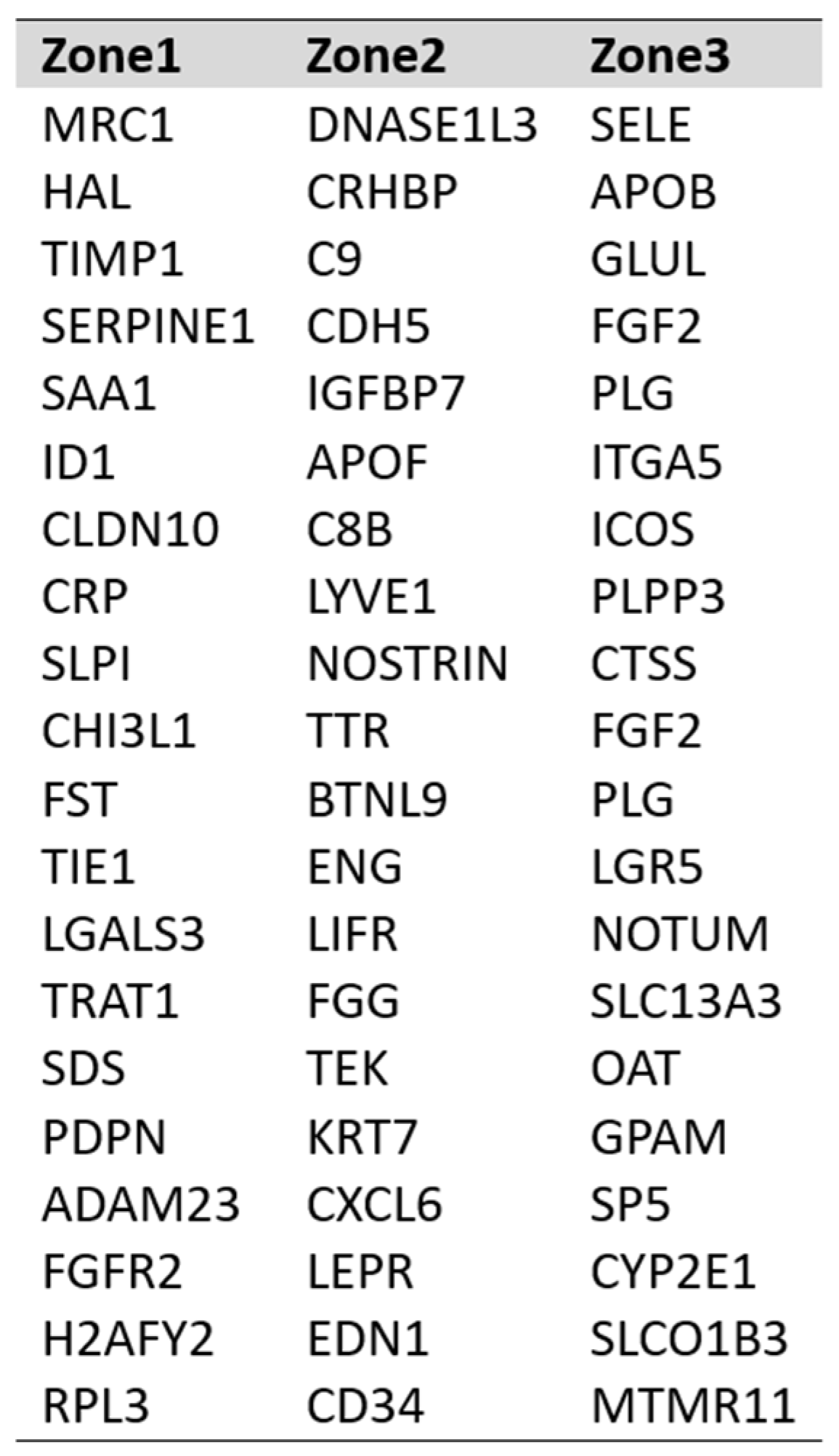
DEG expression profiles identified the top 25 highly expressed genes with their marker score in PP–LSEC: Periportal Liver Sinusoid Endothelial Cells, PC–LSEC: Pericentral Liver Sinusoid Endothelial Cells and LSECs cells illustrated in
Figure 7. A marker score is the 10th percentile of the effect sizes across all comparisons for each gene. DEG analysis is performed at cellxgene web-tool, a well curated, standardized, wide collection of interoperable single-cell transcriptomic data platform available at cellxgene.cziscience.com[
17]. Five most commonly expressed genes
DNASE1L3,LIFR,STAB1,MRC1,CRHBP are detected using cellxgene analysis and confirmed in another datasets[
14]. Also, identified top 20 expressed genes in each zone are summarized in
Table 3. Commonly expressed genes in this group are
MRC1, HAL, TIMP1 (zone1),
DNASE1L3, CRHBP, C9 (zone2) and
SELE, APOB, GLUL (zone3). Their zonation-specific expression is confirmed using the human hepatocyte zonation browser tool, another open-source web platform publicly available web tool [
18], illustrated in supplementary
Figure S2.
3. Discussion
In this study, efforts are made to develop our understanding towards complex interplay of LSECs in mid-lobular zone2 and their contributions in initiation of early fibrosis and regeneration. LSECs presence in zone 2 is influential and play a critical role in determining whether injured hepatocytes will regenerate or susceptible to fibrosis. Their anti-fibrotic ability to support regeneration is crucial for a successful liver repair process. LSECs secrete extracellular matrix (ECM) components like laminins, providing a supporting microenvironment for regeneration and ensuring proper tissue restructuring [
19].
It’s important to understand the zonation LSCEs functioning at cellular and molecular level and to quantify those architectural changes spatially in normal healthy and diseased liver conditions. Scarring tissue in NASH and DILI disease models exhibit common features of liver fibrosis. In this study,
OIT3 is computationally identified as commonly expressed marker gene in mid-lobular zone2 by LSECs cells population both in NASH and DILI datasets, which is also reported elsewhere in a study as a hallmark gene expressed in ECs [
20]. The second most prominent gene identified is
DNASE1L3. Other zonation-specific marker genes identified in our study are
MRC1, HAL (zone1),
CRHBP, LYVE1(zone2) and
SELE, GLUL (zone3). Protein
F8 secreted by LSECs[
21] plays important role in blood clotting is another marker gene detected in our experiment on
GSE166178 dataset as highly expressed zone 2 marker gene and confirmed by zonation browser tool. Also found in this series
STAB2,CLEC4G, LYVE1 genes as mid-lobular zone 2 markers and confirmed as expressed healthy human liver[
6,
7].
The liver displays unique spatial heterogeneity and functioning of hepatocytes by exhibiting distinct metabolic and functional profiles across different acinar zones (periportal, perivenous, and intermediate). Specific enzymes, transporters, and other proteins act as zonation markers, reflecting the specialized functions of each zone. 10x genomic`s Visium platform and Nanostring’s CosMx technologies used here to demonstrate these changes at molecular level and confirmed by zonation browser tool as the zonation-specific i.e. GLUL as zone3, FGG as zone2 and SAA1 as zone 1 expressed marker genes.
DEG analysis of liver zonation can provide valuable insights into the mechanisms and potential therapeutic targets for fibrosis in NASH and DILI. In NASH related fibrosis, DEGs in Zone 1 might be related to lipid metabolism and oxidative stress, while Zone 3 DEGs could be involved in inflammation and bile acid signaling. Similarly in a specific DILI drug, DEGs might be related to mitochondrial dysfunction, immune response, or direct cell injury in specific zones. Fibrosis is caused by long-term chronic liver injury and is considered a hallmark disease feature of both NASH and DILI progression. This disrupts liver architecture and function, leading to potential organ failure. Different genes might be driving fibrosis in each zone, suggesting targeted therapies for each zonated area. DEGs could serve as early diagnostic or prognostic biomarkers for fibrosis progression in NASH and DILI. DEG analysis in NCBI GEO
GSE126848 study identified UQCRB as the most up regulated gene in NASH and confirmed as a zone 2 marker, which is reported in a study elsewhere as molecular prognostic biomarker in human colorectal cancer[
22]. DEG expression profiles identified five common expressed genes are as
DNASE1L3, LIFR, STAB1, MRC1, CRHBP. Table summarized the top 25 highly expressed genes in PP–LSEC and PC-LSEC, such as
MRC1, HAL, TIMP1 (zone1),
DNASE1L3, CRHBP, C9 (zone2) and
SELE, APOB, GLUL (zone3).
4. Methods and Materials
4.1. H&E Histopathology Image Classifications
Open-source visual programming data mining framework orange toolbox is used for classification of H&E histopathology image tiles. Orange is a machine learning data mining platform (
http://orange.biolab.si) for image analysis and data visualization[
23,
24]. H&E stained Whole Slide (WSI) histopathology images were acquired from open-source National Cancer Institute (NCI) USA Biorepositories and Biospecimen Research Branch’s (BBRB) Genotype-Tissue Expression (GTEx)Tissue Image library of annotated WSI with clinical data publicly available at
https://brd.nci.nih.gov/brd/specimen/GTEX-117XS-0926 and downloaded under five categories as normal, steatosis, early fibrosis, bridging fibrosis & cirrhosis. Each WSI were cropped into tiles for training machine learning models and classified in orange tool classification layout. Experimental workflow is shown in
Figure 1D.
4.2. Spatial Transcriptomics Data Analysis
4.2.1. Visium Data Analysis and Visualization
The Visium dataset used here is available at the Gene Expression Omnibus NCBI GEO public database under accession number GSE192742. Visium 10x genomics loupe browser image files downloaded from liver cell atlas available at
https://www.livercellatlas.org/download.php The Visium ST images of normal and steatosis human liver samples were processed using Loupe Browser 7.0.1 (10x Genomics Inc.) downloaded from here
https://www.10xgenomics.com/support/software/loupe-browser/downloads. The gene expression data for the k-mean clusters generated by the Space Ranger software for up regulated genes. The data consists of the median-normalized average of gene expression, log2 fold changes, and statistical significance (p values) computed for genes with a p value < 0.05.
4.2.2. Spatial Molecular Imaging
Spatial molecular imaging highlights DEG in human liver zonation. Imaging data were acquired from an open-source publicly available multiplex dataset at
http://nanostring.com/CosMx-dataset CosMx
TM Spatial Molecular Imager which includes staining for a panel of morphological features.
4.3. Liver Cell Clustering and Analysis Tool
For liver cell clustering and LSECs marker analysis, National Institute of Health`s (NIH) USA, Human BioMolecular Atlas Program (HuBMAP) Azimuth app is used. Azimuth is a Seurat based web application tool that uses an annotated reference dataset to automate the processing, analysis, and interpretation of single-cell RNA-seq data. Azimuth utilizes ‘reference-based mapping' pipeline that accepts counts matrix file in multiple formats as input and performs normalization, visualization, cell annotation and DEG expression without any coding requirement on web cloud. All results can be visualized within the app, and can be downloadable for additional downstream analysis[
25,
26,
27,
28,
29]. The development of Azimuth is led by the New York Genome Center Mapping Component as part of the NIH HuBMAP consortium.
5. Conclusions
Mammalian healthy liver lobule is spatially well zonated and recognized with known markers based on distribution of metabolic functions that disrupts in diseased pathological conditions and liver injury. However, due to hepatocyte heterogeneity these zonation markers, such as enzymes, metabolites, and gene expression patterns vary in their expression and activity even within the same lobule. These molecular markers are not 1) zonation-specific; 2) disease -specific, for example, the expression and distribution of specific molecules within the liver changes during fibrogenesis in NASH and DILI. LSECs play an important and intricated role in the early stages of liver fibrosis and regeneration during mid-lobular zonation restructuring. Their strategic position allows them to act as critical conductors in influencing the delicate balance between tissue repair and scarring. Emerging trends in new technologies such as scRNA-seq, spatial transcriptomics and multiomics can be helpful to determine the role of LSECs in identifying the zonation marker DEGs at cellular and molecular level. LSCEs can provide valuable insights into how zonation marker variations and fibrosis are interconnected. This study identified zonation-specific mid-lobular markers of LSECs. Liver fibrosis in early stages can be reversible if detected in zonation at molecular level before moving to advanced stages like bridging fibrosis. Current diagnostic methods often lack accuracy at early stages of liver fibrosis. This is where computational biomarkers derived from integrated multi-omics, cellular and molecular data hold immense promise for providing a deeper understanding and improved diagnosis of liver fibrosis. The limitation of this study is that DEG analysis alone is not sufficient to understand early stages of fibrosis in complex diseases like NASH and DILI. Integrating it with other data, such as protein expression and metabolic profiling is required to analyze in normal and diseased conditions. More research is needed to validate DEGs as reliable zonation markers and translate them into effective diagnostic and prognostic biomarkers. Investigating LSCEs in the context of both zonation and fibrosis holds significant promise for developing novel therapeutic strategies against liver diseases. This may help to protect zonation patterns, prevent fibrosis development, and ultimately improve liver health.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Conflicts of Interest
No conflict of interest.
References
- Devarbhavi, H.; Asrani, S.K.; Arab, J.P.; Nartey, Y.A.; Pose, E.; Kamath, P.S. Global Burden of Liver Disease: 2023 Update. J. Hepatol. 2023, 79, 516–537. [CrossRef]
- Ben-Moshe, S.; Itzkovitz, S. Spatial Heterogeneity in the Mammalian Liver. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 395–410. [CrossRef]
- Itoh, T. The Truth Lies Somewhere in the Middle: The Cells Responsible for Liver Tissue Maintenance Finally Identified. Cell Regen. Lond. Engl. 2021, 10, 28. [CrossRef]
- Wei, Y.; Wang, Y.G.; Jia, Y.; Li, L.; Yoon, J.; Zhang, S.; Wang, Z.; Zhang, Y.; Zhu, M.; Sharma, T.; et al. Liver Homeostasis Is Maintained by Midlobular Zone 2 Hepatocytes. Science 2021, 371, eabb1625. [CrossRef]
- Panday, R.; Monckton, C.P.; Khetani, S.R. The Role of Liver Zonation in Physiology, Regeneration, and Disease. Semin. Liver Dis. 2022, 42, 001–016. [CrossRef]
- Verhulst, S.; van Os, E.A.; De Smet, V.; Eysackers, N.; Mannaerts, I.; van Grunsven, L.A. Gene Signatures Detect Damaged Liver Sinusoidal Endothelial Cells in Chronic Liver Diseases. Front. Med. 2021, 8, 750044. [CrossRef]
- Su, T.; Yang, Y.; Lai, S.; Jeong, J.; Jung, Y.; McConnell, M.; Utsumi, T.; Iwakiri, Y. Single-Cell Transcriptomics Reveals Zone-Specific Alterations of Liver Sinusoidal Endothelial Cells in Cirrhosis. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 1139–1161. [CrossRef]
- DeLeve, L.D. Liver Sinusoidal Endothelial Cells in Hepatic Fibrosis. Hepatol. Baltim. Md 2015, 61, 1740–1746. [CrossRef]
- Lafoz, E.; Ruart, M.; Anton, A.; Oncins, A.; Hernández-Gea, V. The Endothelium as a Driver of Liver Fibrosis and Regeneration. Cells 2020, 9, 929. [CrossRef]
- Ghallab, A.; Myllys, M.; H. Holland, C.; Zaza, A.; Murad, W.; Hassan, R.; A. Ahmed, Y.; Abbas, T.; A. Abdelrahim, E.; Schneider, K.M.; et al. Influence of Liver Fibrosis on Lobular Zonation. Cells 2019, 8, 1556. [CrossRef]
- Nagy, D.; Maude, H.; Birdsey, G.M.; Randi, A.M.; Cebola, I. RISING STARS: Liver Sinusoidal Endothelial Transcription Factors in Metabolic Homeostasis and Disease. J. Mol. Endocrinol. 2023, 71. [CrossRef]
- Ben-Moshe, S.; Veg, T.; Manco, R.; Dan, S.; Papinutti, D.; Lifshitz, A.; Kolodziejczyk, A.A.; Halpern, K.B.; Elinav, E.; Itzkovitz, S. The Spatiotemporal Program of Zonal Liver Regeneration Following Acute Injury. Cell Stem Cell 2022, 29, 973-989.e10. [CrossRef]
- Wang, Z.; Qian, J.; Lu, X.; Zhang, P.; Guo, R.; Lou, H.; Zhang, S.; Yang, J.; Fan, X. A Single-Cell Transcriptomic Atlas Characterizes Liver Non-Parenchymal Cells in Healthy and Diseased Mice; Genomics, 2021;
- Halpern, K.B.; Shenhav, R.; Matcovitch-Natan, O.; Tóth, B.; Lemze, D.; Golan, M.; Massasa, E.E.; Baydatch, S.; Landen, S.; Moor, A.E.; et al. Single-Cell Spatial Reconstruction Reveals Global Division of Labour in the Mammalian Liver. Nature 2017, 542, 352–356. [CrossRef]
- Hildebrandt, F.; Andersson, A.; Saarenpää, S.; Larsson, L.; Van Hul, N.; Kanatani, S.; Masek, J.; Ellis, E.; Barragan, A.; Mollbrink, A.; et al. Spatial Transcriptomics to Define Transcriptional Patterns of Zonation and Structural Components in the Mouse Liver. Nat. Commun. 2021, 12, 7046. [CrossRef]
- Brancale, J.; Vilarinho, S. A Single Cell Gene Expression Atlas of 28 Human Livers. J. Hepatol. 2021, 75, 219–220. [CrossRef]
- Program, C.S.-C.B.; Abdulla, S.; Aevermann, B.; Assis, P.; Badajoz, S.; Bell, S.M.; Bezzi, E.; Cakir, B.; Chaffer, J.; Chambers, S.; et al. CZ CELL×GENE Discover: A Single-Cell Data Platform for Scalable Exploration, Analysis and Modeling of Aggregated Data 2023, 2023.10.30.563174.
- Massalha, H.; Bahar Halpern, K.; Abu-Gazala, S.; Jana, T.; Massasa, E.E.; Moor, A.E.; Buchauer, L.; Rozenberg, M.; Pikarsky, E.; Amit, I.; et al. A Single Cell Atlas of the Human Liver Tumor Microenvironment. Mol. Syst. Biol. 2020, 16, e9682. [CrossRef]
- Natarajan, V.; Harris, E.N.; Kidambi, S. SECs (Sinusoidal Endothelial Cells), Liver Microenvironment, and Fibrosis. BioMed Res. Int. 2017, 2017, 4097205. [CrossRef]
- Li, Z.-W.; Ruan, B.; Yang, P.-J.; Liu, J.-J.; Song, P.; Duan, J.-L.; Wang, L. Oit3, a Promising Hallmark Gene for Targeting Liver Sinusoidal Endothelial Cells. Signal Transduct. Target. Ther. 2023, 8, 1–10. [CrossRef]
- Jamil, M.A.; Singer, H.; Al-Rifai, R.; Nüsgen, N.; Rath, M.; Strauss, S.; Andreou, I.; Oldenburg, J.; El-Maarri, O. Molecular Analysis of Fetal and Adult Primary Human Liver Sinusoidal Endothelial Cells: A Comparison to Other Endothelial Cells. Int. J. Mol. Sci. 2020, 21, 7776. [CrossRef]
- Kim, H.-C.; Chang, J.; Lee, H.S.; Kwon, H.J. Mitochondrial UQCRB as a New Molecular Prognostic Biomarker of Human Colorectal Cancer. Exp. Mol. Med. 2017, 49, e391. [CrossRef]
- Demšar, J.; Curk, T.; Erjavec, A.; Gorup, Č.; Hočevar, T.; Milutinovič, M.; Možina, M.; Polajnar, M.; Toplak, M.; Starič, A.; et al. Orange: Data Mining Toolbox in Python. J. Mach. Learn. Res. 2013, 14, 2349–2353.
- Godec, P.; Pančur, M.; Ilenič, N.; Čopar, A.; Stražar, M.; Erjavec, A.; Pretnar, A.; Demšar, J.; Starič, A.; Toplak, M.; et al. Democratized Image Analytics by Visual Programming through Integration of Deep Models and Small-Scale Machine Learning. Nat. Commun. 2019, 10, 4551. [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated Analysis of Multimodal Single-Cell Data. Cell 2021, 184, 3573-3587.e29. [CrossRef]
- Aizarani, N.; Saviano, A.; Sagar; Mailly, L.; Durand, S.; Herman, J.S.; Pessaux, P.; Baumert, T.F.; Grün, D. A Human Liver Cell Atlas Reveals Heterogeneity and Epithelial Progenitors. Nature 2019, 572, 199–204. [CrossRef]
- Ramachandran, P.; Dobie, R.; Wilson-Kanamori, J.R.; Dora, E.F.; Henderson, B.E.P.; Luu, N.T.; Portman, J.R.; Matchett, K.P.; Brice, M.; Marwick, J.A.; et al. Resolving the Fibrotic Niche of Human Liver Cirrhosis at Single-Cell Level. Nature 2019, 575, 512–518. [CrossRef]
- Zhang, M.; Yang, H.; Wan, L.; Wang, Z.; Wang, H.; Ge, C.; Liu, Y.; Hao, Y.; Zhang, D.; Shi, G.; et al. Single-Cell Transcriptomic Architecture and Intercellular Crosstalk of Human Intrahepatic Cholangiocarcinoma. J. Hepatol. 2020, 73, 1118–1130. [CrossRef]
- MacParland, S.A.; Liu, J.C.; Ma, X.-Z.; Innes, B.T.; Bartczak, A.M.; Gage, B.K.; Manuel, J.; Khuu, N.; Echeverri, J.; Linares, I.; et al. Single Cell RNA Sequencing of Human Liver Reveals Distinct Intrahepatic Macrophage Populations. Nat. Commun. 2018, 9, 4383. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).