Submitted:
29 December 2023
Posted:
04 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. p38 molecular signaling
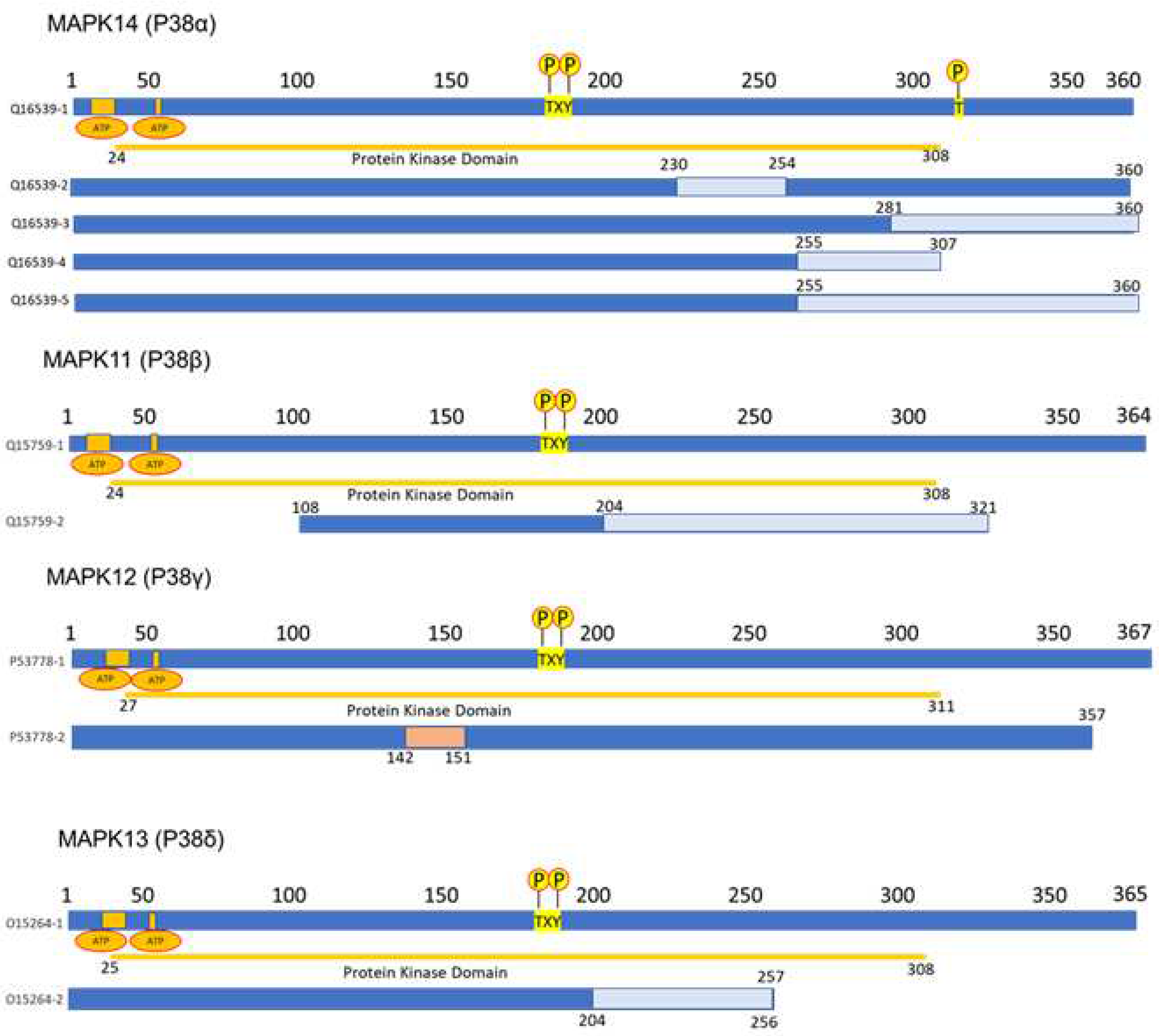
2.1. MAPK p38 structure
2.2. P38 regulation
2.2.1. Transcription factors in the regulation of p38
2.2.2. Non-coding RNAs in the regulation of p38
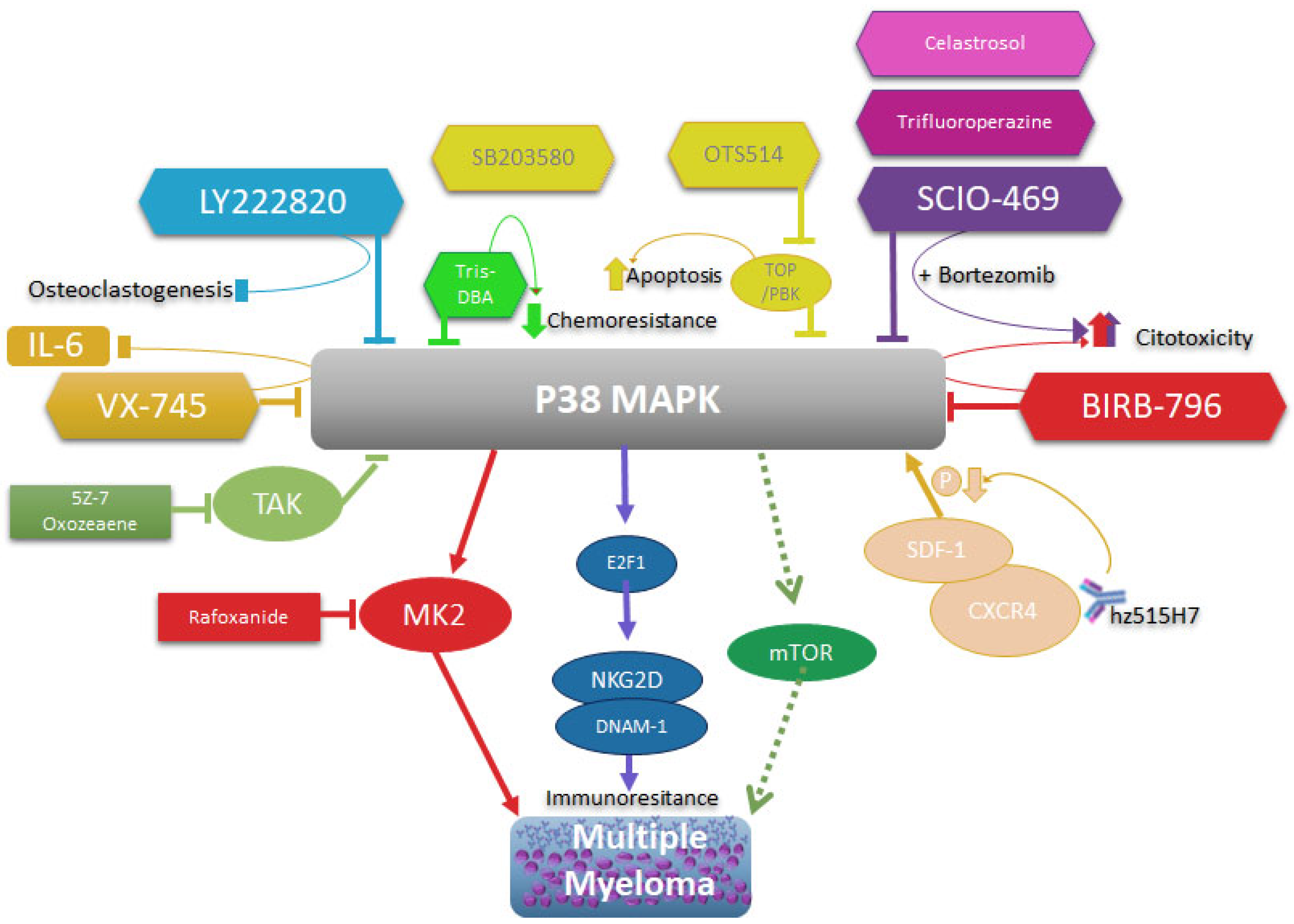
2.3. p38 chemicals inhibitors on cancer
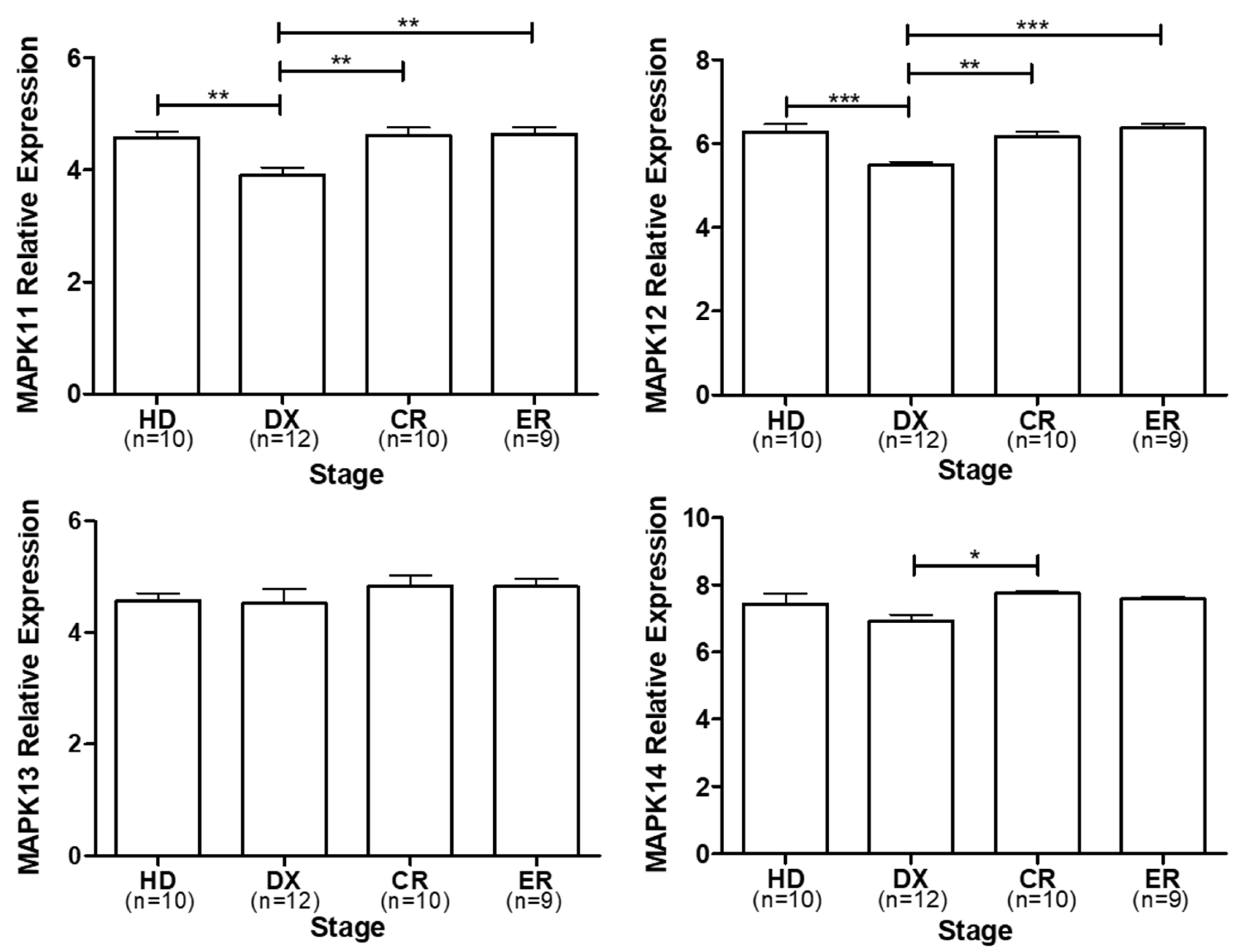
3. p38 expression in multiple myeloma
4. MAPK p38 as a molecular target in multiple myeloma therapy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ATP | Adenosine triphosphate |
| ATRA | All-trans retinoic acid |
| ATO | Arsenic trioxide |
| ATF2 | Activating Transcription Factor 2 |
| ATF1 | Activating Transcription Factor 1 |
| AP-1 | Jun Proto-Oncogene, AP-1 Transcription Factor Subunit |
| ASCL1 | Achaete-Scute Family BHLH Transcription Factor 1 |
| ATF4 | Activating Transcription Factor 4 |
| ATF7 | Activating Transcription Factor 7 |
| ARID3A | AT-Rich Interaction Domain 3A |
| ARID1A | AT-Rich Interaction Domain 1A |
| ASCL2 | Achaete-Scute Family BHLH Transcription Factor 2 |
| ARNT | Aryl Hydrocarbon Receptor Nuclear Translocator |
| ATRA | All-trans retinoic acid |
| ATO | Arsenic trioxide |
| AKT1 | AKT Serine/Threonine Kinase 1 |
| Ape/Ref-1 | apurinic endonuclease/redox factor-1 |
| BMSCs | Bone-marrow-derived mesenchymal stem cells |
| BCL6 | B-Cell Lymphoma 6 Protein |
| BACH2 | BTB Domain and CNC Homolog 2 |
| BM | Bone marrow |
| BMMNC | BM mononuclear cells. |
| BRAF | B-Raf Proto-Oncogene, Serine/Threonine Kinase |
| CML | Chronic Myeloid Leukemia |
| CRC | Colorectal cancer. |
| CDR1-AS | CDR1 Antisense RNA |
| CEBPA | CCAAT Enhancer Binding Protein Alpha |
| CTCF | CCCTC-Binding Factor |
| CTSC | cathepsin C |
| CHOP10 | DNA Damage Inducible Transcript 3 |
| CDC25 | Cell Division Cycle 25 |
| CXCL14 | C-X-C Motif Chemokine Ligand 14 |
| CXCR4 | C-X-C Motif Chemokine Receptor 4 |
| C-JUN | Jun Proto-Oncogene, AP-1 Transcription Factor Subunit |
| c-MYC | Myc Proto-Oncogene Protein |
| CREB1 | Cyclic AMP-Responsive Element-Binding Protein 1 |
| CDK | Cyclin-dependent kinase. |
| CDK7 | Cyclin Dependent Kinase 7 |
| DUSP1 | Dual Specificity Protein Phosphatase 1 |
| DUSP10 | Dual Specificity Protein Phosphatase 10 |
| DUSP16 | Dual Specificity Protein Phosphatase 16 |
| DBA | Dibenzylideneacetone |
| DCs | Dendritic cells |
| DKK-1 | Dickkopf WNT Signaling Pathway Inhibitor 1 |
| DNAM1 | CD226 Antigen |
| EPD | Eukaryotic Promotor Database |
| EMT | epithelial–mesenchymal transition |
| E2F1 | E2F Transcription Factor 1 |
| EGR1 | Early Growth Response 1 |
| EP300 | E1A Binding Protein P300 |
| ELK1 | ETS Transcription Factor ELK1 |
| ESCC | Esophageal squamous cell carcinoma |
| EPD | Eukaryotic Promotor Database |
| EEF2K | Eukaryotic Elongation Factor 2 Kinase |
| ESCC | Esophageal squamous cell carcinoma |
| ERK1/2 | Extracellular Signal-Regulated Kinase 2 (MAPK1) |
| FOXP3 | Forkhead Box P3 |
| FGFR1 | Fibroblast Growth Factor Receptor 1 |
| FOXM1 | Forkhead Box M1 |
| GA | Gambogic acid |
| GDF4 | Growth Differentiation Factor 4 |
| GADD45A | Growth Arrest and DNA Damage Inducible Alpha |
| G-CSF | granulocyte colony-stimulating factor |
| HEK293 | Human Embryonic Kidney cell line |
| HIF1A | Hypoxia Inducible Factor 1 Subunit Alpha |
| HDAC3 | Histone Deacetylase 3 |
| HDAC9 | Histone Deacetylase 9 |
| HCC | Hepatocellular carcinoma |
| HO-1 | heme oxygenase-1 |
| Imp 7/3 | Importin-7 |
| Imp 9/3 | Importin-9 |
| LncRNAs | Long noncoding RNA |
| IL-6 | Interleukin 6 |
| IL-8 | Interleukin 8 |
| IL-12B | Interleukin 12β |
| IAV | Influenza A virus |
| IKK | Inhibitor of Nuclear Factor Kappa-B Kinase |
| IRAK4 | Interleukin 1 Receptor Associated Kinase 4 |
| IGF-1 | insulin-like growth factor 1 |
| JUN | Jun Proto-Oncogene, AP-1 Transcription Factor Subunit |
| JAK | Janus Kinase |
| KLF4 | Kruppel-Like Factor 4 |
| KAT2B | Lysine Acetyltransferase 2B |
| KRAS | KRAS Proto-Oncogene, GTPase |
| LINCRNA | Long intergenic non-coding RNA |
| LPS | Lipopolysaccharide |
| MM | Multiple myeloma |
| MDR | Multidrug Resistance Protein |
| MAPK | Mitogen-activated protein kinase |
| MGUS | Monoclonal gammopathy of unknown significance |
| MKKs | MAP kinase kinases |
| MW | Molecular weight |
| MEF2 | Myocyte Enhancer Factor 2 |
| MSK1 | Nuclear Mitogen- And Stress-Activated Protein Kinase 1 |
| MSK2 | Nuclear Mitogen- And Stress-Activated Protein Kinase 2 |
| MAPKAPK2 | MAPK Activated Protein Kinase 2 |
| MART | Melanoma Antigen Recognized By T-Cells 1 |
| MAP3K71P1 | TGF-Beta Activated Kinase 1 (MAP3K7) Binding Protein 1 |
| MAP2K3 | Mitogen-Activated Protein Kinase Kinase 3 |
| MKK3 | MAP kinase kinases 3 |
| MAP2K4 | Mitogen-Activated Protein Kinase Kinase 4 |
| MKK4 | MAP kinase kinases 4 |
| MAP2K6 | Mitogen-Activated Protein Kinase Kinase 6 |
| MKK6 | MAP kinase kinases 6 |
| MYB | MYB Proto-Oncogene, Transcription Factor |
| MK2 | MAPK Activated Protein Kinase 2 |
| MK3 | MAPK Activated Protein Kinase 3 |
| mRNA | Messenger RNA |
| MMP-2 | Matrix Metallopeptidase 2 |
| MMP-9 | Matrix Metallopeptidase 9 |
| MYC | MYC Proto-Oncogene, BHLH Transcription Factor |
| MSCs | mesenchymal stem cells |
| MKK6 | Mitogen-Activated Protein Kinase Kinase 6 |
| MDS | Myelodysplastic syndromes. |
| MCP1 | C-C Motif Chemokine Ligand 2 |
| mTOR | Mechanistic Target Of Rapamycin Kinase |
| NHL | Non-Hodgkin lymphoma |
| NSPCs | Neural stem/progenitor cells |
| NFKB1 | Nuclear Factor Kappa B Subunit 1 |
| ND8/34 | Cell Line from mouse |
| NSCLC | Non-small cell lung cancer. |
| NOTCH3 | Notch Receptor 3 |
| NKG2D | NK Cell Receptor D |
| OSCs | Osteosarcoma cells |
| PINT | p53-induced noncoding transcript. |
| PCAT1 | Prostate Cancer Associated Transcript 1 |
| POU1F1 | Pituitary Transcript Factor 1 |
| PPMID | Protein Phosphatase Magnesium-Dependent 1 Delta |
| P53 | Tumor Protein P53 |
| P38 | Mitogen-activated protein kinase p38 |
| PRKD1 | Protein Kinase D1 |
| PRAK | MAPK Activated Protein Kinase 5 |
| rRNA | Ribosomal RNA |
| RAC3 | Rac Family Small GTPase 3 |
| RAF | Raf-1 Proto-Oncogene, Serine/Threonine Kinase |
| SRF | Serum Response Factor |
| STMN1 | Stathmin 1 |
| STAT1 | Signal Transducer and Activator of Transcription 1 |
| STAT3 | Signal Transducer and Activator of Transcription 3 |
| STAT4 | Signal Transducer and Activator of Transcription 4 |
| SMAD2 | SMAD Family Member 2 |
| SMAD3 | SMAD Family Member 3 |
| SMAD4 | SMAD Family Member 4 |
| SP1 | Specificity Protein 1 |
| SYK | Spleen Associated Tyrosine Kinase |
| SNGH5 | Small Nucleolar RNA Host Gene 5 |
| SDF1 | C-X-C Motif Chemokine Ligand 12 |
| TCR | T-cell receptor |
| TAB1 | TAK1-Binding Protein 1 |
| TNF-α | Tumor necrosis Factor alfa |
| TP53 | Tumor Protein P53 |
| TGF-β | Transforming Growth Factor Beta 1 |
| TNBC | Triple-negative breast cancer. |
| Tris DBA | Tris(dibenzylideneacetone)dipalladium. |
| TAK1 | Mitogen-Activated Protein Kinase Kinase Kinase 7 |
| TOPK/PBK | cell-derived protein kinase T-LAK/PDZ-binding kinase |
| TLR5 | Toll Like Receptor 5 |
| TLR7 | Toll Like Receptor 7 |
| TLR9 | Toll Like Receptor 9 |
| VEGF | Vascular endothelial growth factor |
| VDR | Vitamin D Receptor |
| XBP1 | X-Box-Binding Protein 1 |
| YY1 | YIN-YANG-1 |
| ZAP70 | 70 KDa Zeta-Chain Associated Protein |
| ZCCHC14 | Zinc Finger CCHC Domain-Containing Protein 14 |
References
- P. G. Richardson et al., “Bortezomib or High-Dose Dexamethasone for Relapsed Multiple Myeloma,” n engl j med, vol. 352, 2005, Accessed: Sep. 27, 2023. [Online]. Available: www.nejm.org.
- R. A. Kyle et al., “Review of 1027 patients with newly diagnosed multiple myeloma,” Mayo Clin Proc, vol. 78, no. 1, pp. 21–33, Jan. 2003. [CrossRef]
- P. de la Puente and A. K. Azab, “Contemporary drug therapies for multiple myeloma,” Drugs Today (Barc), vol. 49, no. 9, p. 563, 2013. [CrossRef]
- T. Gui, Y. Sun, A. Shimokado, and Y. Muragaki, “The Roles of Mitogen-Activated Protein Kinase Pathways in TGF-β-Induced Epithelial-Mesenchymal Transition,” J Signal Transduct, vol. 2012, pp. 1–10, Jan. 2012. [CrossRef]
- A. Martínez-Limón, M. Joaquin, M. Caballero, F. Posas, and E. de Nadal, “The p38 Pathway: From Biology to Cancer Therapy,” Int J Mol Sci, vol. 21, no. 6, Mar. 2020. [CrossRef]
- V. Sahu, A. Mohan, and S. Dey, “p38 MAP kinases: plausible diagnostic and prognostic serum protein marker of non small cell lung cancer,” Exp Mol Pathol, vol. 107, pp. 118–123, Apr. 2019. [CrossRef]
- P. P. Roux and J. Blenis, “ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions,” Microbiol Mol Biol Rev, vol. 68, no. 2, pp. 320–344, Jun. 2004. [CrossRef]
- J. Robidoux et al., “Selective activation of mitogen-activated protein (MAP) kinase kinase 3 and p38alpha MAP kinase is essential for cyclic AMP-dependent UCP1 expression in adipocytes,” Mol Cell Biol, vol. 25, no. 13, pp. 5466–5479, Jul. 2005. [CrossRef]
- S. Kudaravalli, P. den Hollander, and S. A. Mani, “Role of p38 MAP kinase in cancer stem cells and metastasis,” Oncogene 2022 41:23, vol. 41, no. 23, pp. 3177–3185, Apr. 2022. [CrossRef]
- T. Zarubin and J. Han, “Activation and signaling of the p38 MAP kinase pathway,” Cell Res, vol. 15, no. 1, pp. 11–18, 2005. [CrossRef]
- J. He et al., “p38 MAPK in myeloma cells regulates osteoclast and osteoblast activity and induces bone destruction,” Cancer Res, vol. 72, no. 24, pp. 6393–6402, Dec. 2012. [CrossRef]
- T. Hideshima et al., “p38 MAPK inhibition enhances PS-341 (bortezomib)-induced cytotoxicity against multiple myeloma cells,” Oncogene, vol. 23, no. 54, pp. 8766–8776, Nov. 2004. [CrossRef]
- Z. Jing, W. Yu, A. Li, X. Chen, Y. Chen, and J. Chen, “Trifluoperazine Synergistically Potentiates Bortezomib-Induced Anti-Cancer Effect in Multiple Myeloma via Inhibiting P38 MAPK/NUPR1,” Tohoku J Exp Med, vol. 257, no. 4, pp. 315–326, 2022. [CrossRef]
- X. Wu et al., “Dihydroartemisinin Modulates Apoptosis and Autophagy in Multiple Myeloma through the P38/MAPK and Wnt/ β-Catenin Signaling Pathways,” Oxid Med Cell Longev, vol. 2020, 2020. [CrossRef]
- J. Han, J. Wu, and J. Silke, “An overview of mammalian p38 mitogen-activated protein kinases, central regulators of cell stress and receptor signaling,” F1000Res, vol. 9, 2020. [CrossRef]
- B. Canovas and A. R. Nebreda, “Diversity and versatility of p38 kinase signalling in health and disease,” Nat Rev Mol Cell Biol, vol. 22, no. 5, pp. 346–366, May 2021. [CrossRef]
- S. Uddin, J. Ah-Kang, J. Ulaszek, D. Mahmud, and A. Wickrema, “Differentiation stage-specific activation of p38 mitogen-activated protein kinase isoforms in primary human erythroid cells,” Proc Natl Acad Sci U S A, vol. 101, no. 1, pp. 147–152, Jan. 2004. [CrossRef]
- M. Deak, A. D. Clifton, J. M. Lucocq, and D. R. Alessi, “Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB,” EMBO J, vol. 17, no. 15, pp. 4426–4441, Aug. 1998. [CrossRef]
- B. Pierrat, J. Da Silva Correia, J. L. Mary, M. Tomás-Zuber, and W. Lesslauer, “RSK-B, a novel ribosomal S6 kinase family member, is a CREB kinase under dominant control of p38alpha mitogen-activated protein kinase (p38alphaMAPK),” J Biol Chem, vol. 273, no. 45, pp. 29661–29671, Nov. 1998. [CrossRef]
- J. Han, J. Wu, J. Silke, J. D. Ashwell, and G. Sabio, “An overview of mammalian p38 mitogen-activated protein kinases, central regulators of cell stress and receptor signaling,” F1000Research 2020 9:653, vol. 9, p. 653, Jun. 2020. [CrossRef]
- S. H. Lee, J. Park, Y. Che, P.-L. Han, and J.-K. Lee, “Constitutive Activity and Differential Localization of p38 and p38 MAPKs in Adult Mouse Brain,” J. Neurosci. Res, vol. 60, pp. 623–631, 2000. [CrossRef]
- H. Enslen, J. Raingeaud, and R. J. Davis, “Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6,” J Biol Chem, vol. 273, no. 3, pp. 1741–1748, Jan. 1998. [CrossRef]
- X. Qi et al., “p38alpha antagonizes p38gamma activity through c-Jun-dependent ubiquitin-proteasome pathways in regulating Ras transformation and stress response,” J Biol Chem, vol. 282, no. 43, pp. 31398–31408, Oct. 2007. [CrossRef]
- I. Corre, F. Paris, and J. Huot, “The p38 pathway, a major pleiotropic cascade that transduces stress and metastatic signals in endothelial cells,” Oncotarget, vol. 8, no. 33, pp. 55684–55714, 2017. [CrossRef]
- N. Matesanz et al., “p38α blocks brown adipose tissue thermogenesis through p38δ inhibition,” PLoS Biol, vol. 16, no. 7, Jul. 2018. [CrossRef]
- X. Wang et al., “Involvement of the MKK6-p38gamma cascade in gamma-radiation-induced cell cycle arrest,” Mol Cell Biol, vol. 20, no. 13, pp. 4543–4552, Jul. 2000. [CrossRef]
- Y. Shi and M. Gaestel, “In the cellular garden of forking paths: how p38 MAPKs signal for downstream assistance,” Biol Chem, vol. 383, no. 10, pp. 1519–1536, Oct. 2002. [CrossRef]
- A. Cuadrado and A. R. Nebreda, “Mechanisms and functions of p38 MAPK signalling,” Biochem J, vol. 429, no. 3, pp. 403–417, Aug. 2010. [CrossRef]
- K. S. Robinson et al., “ZAKα-driven ribotoxic stress response activates the human NLRP1 inflammasome,” Science, vol. 377, no. 6603, pp. 328–335, Jul. 2022. [CrossRef]
- G. G. Vega et al., “P38 MAPK expression and activation predicts failure of response to CHOP in patients with Diffuse Large B-Cell Lymphoma,” BMC Cancer, vol. 15, no. 1, Oct. 2015. [CrossRef]
- Y. Feng, J. Wen, and C. C. Chang, “p38 Mitogen-activated protein kinase and hematologic malignancies,” Arch Pathol Lab Med, vol. 133, no. 11, pp. 1850–1856, Nov. 2009. [CrossRef]
- N. A. Bhowmick et al., “Transforming growth factor-β1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism,” Mol Biol Cell, vol. 12, no. 1, pp. 27–36, Oct. 2001. [CrossRef]
- L. Mao, L. Yuan, L. M. Slakey, F. E. Jones, M. E. Burow, and S. M. Hill, “Inhibition of breast cancer cell invasion by melatonin is mediated through regulation of the p38 mitogen-activated protein kinase signaling pathway,” Breast Cancer Research, vol. 12, no. 6, pp. 1–14, Dec. 2010. [CrossRef]
- T. T. T. Phan, N. V. Truong, W. G. Wu, Y. C. Su, T. S. Hsu, and L. Y. Lin, “Tumor suppressor p53 mediates interleukin-6 expression to enable cancer cell evasion of genotoxic stress,” Cell Death Discov, vol. 9, no. 1, Dec. 2023. [CrossRef]
- H. An, X. Lu, D. Liu, and W. G. Yarbrough, “LZAP inhibits p38 MAPK (p38) phosphorylation and activity by facilitating p38 association with the wild-type p53 induced phosphatase 1 (WIP1),” PLoS One, vol. 6, no. 1, 2011. [CrossRef]
- V. B. Pillai, N. R. Sundaresan, S. A. Samant, D. Wolfgeher, C. M. Trivedi, and M. P. Gupta, “Acetylation of a conserved lysine residue in the ATP binding pocket of p38 augments its kinase activity during hypertrophy of cardiomyocytes,” Mol Cell Biol, vol. 31, no. 11, pp. 2349–2363, Jun. 2011. [CrossRef]
- J. C. Lee et al., “A protein kinase involved in the regulation of inflammatory cytokine biosynthesis,” Nature, vol. 372, no. 6508, pp. 739–746, 1994. [CrossRef]
- A. S. Zervos, L. Faccio, J. P. Gatto, J. M. Kyriakis, and R. Brent, “Mxi2, a mitogen-activated protein kinase that recognizes and phosphorylates Max protein,” Proc Natl Acad Sci U S A, vol. 92, no. 23, pp. 10531–10534, Nov. 1995. [CrossRef]
- T. Sudo, Y. Yagasaki, H. Hama, N. Watanabe, and H. Osada, “Exip, a new alternative splicing variant of p38α, can induce an earlier onset of apoptosis in HeLa cells,” Biochem Biophys Res Commun, vol. 291, no. 4, pp. 838–843, 2002. [CrossRef]
- P. Wang, P. Yu, P. Gao, T. Shi, and D. Ma, “Discovery of novel human transcript variants by analysis of intronic single-block EST with polyadenylation site,” BMC Genomics, vol. 10, Nov. 2009. [CrossRef]
- G. Lominadze, M. J. Rane, M. Merchant, J. Cai, R. A. Ward, and K. R. McLeish, “Myeloid-related protein-14 is a p38 MAPK substrate in human neutrophils,” J Immunol, vol. 174, no. 11, pp. 7257–7267, Jun. 2005. [CrossRef]
- J. Raingeaud et al., “Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine,” J Biol Chem, vol. 270, no. 13, pp. 7420–7426, 1995. [CrossRef]
- U. Mahlknecht, J. Will, A. Varin, D. Hoelzer, and G. Herbein, “Histone deacetylase 3, a class I histone deacetylase, suppresses MAPK11-mediated activating transcription factor-2 activation and represses TNF gene expression,” J Immunol, vol. 173, no. 6, pp. 3979–3990, Sep. 2004. [CrossRef]
- T. Ota et al., “Complete sequencing and characterization of 21,243 full-length human cDNAs,” Nat Genet, vol. 36, no. 1, pp. 40–45, Jan. 2004. [CrossRef]
- J. E. Collins et al., “A genome annotation-driven approach to cloning the human ORFeome,” Genome Biol, vol. 5, no. 10, p. R84, 2004. [CrossRef]
- N. W. Court, C. G. Dos Remedios, J. Cordell, and M. A. Bogoyevitch, “Cardiac expression and subcellular localization of the p38 mitogen-activated protein kinase member, stress-activated protein kinase-3 (SAPK3),” J Mol Cell Cardiol, vol. 34, no. 4, pp. 413–426, Apr. 2002. [CrossRef]
- C. Lechner, M. A. Zahalka, J. F. Giot, N. P. H. Møller, and A. Ullrich, “ERK6, a mitogen-activated protein kinase involved in C2C12 myoblast differentiation,” Proc Natl Acad Sci U S A, vol. 93, no. 9, pp. 4355–4359, Apr. 1996. [CrossRef]
- D. S. Gerhard et al., “The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC),” Genome Res, vol. 14, no. 10B, pp. 2121–2127, 2004. [CrossRef]
- L. New et al., “PRAK, a novel protein kinase regulated by the p38 MAP kinase,” EMBO J, vol. 17, no. 12, pp. 3372–3384, Jun. 1998. [CrossRef]
- G. Pantoja-Escobar, M. Morales-Martínez, G. G. Vega, G. Castro-Escarpulli, and M. I. Vega, “Cytotoxic effect caspase activation dependent of a genetically engineered fusion protein with a CD154 peptide mimetic (OmpC-CD154p) on B-NHL cell lines is mediated by the inhibition of bcl-6 and YY1 through MAPK p38 activation,” Leuk Lymphoma, vol. 60, no. 4, pp. 1062–1070, Mar. 2019. [CrossRef]
- M. Zhao et al., “Regulation of the MEF2 family of transcription factors by p38,” Mol Cell Biol, vol. 19, no. 1, pp. 21–30, Jan. 1999. [CrossRef]
- S.-H. Yang, A. Galanis, and A. D. Sharrocks, “Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors,” Mol Cell Biol, vol. 19, no. 6, pp. 4028–4038, Jun. 1999. [CrossRef]
- N. Umasuthan, S. D. N. K. Bathige, J. K. Noh, and J. Lee, “Gene structure, molecular characterization and transcriptional expression of two p38 isoforms (MAPK11 and MAPK14) from rock bream (Oplegnathus fasciatus),” Fish Shellfish Immunol, vol. 47, no. 1, pp. 331–343, Nov. 2015. [CrossRef]
- M. Zhou et al., “Prioritizing candidate disease-related long non-coding RNAs by walking on the heterogeneous lncRNA and disease network †,” 760 | Mol. BioSyst, vol. 11, p. 760, 2015. [CrossRef]
- Q. Yang, X. Shen, Z. Su, and S. Ju, “Emerging roles of noncoding RNAs in multiple myeloma: A review,” J Cell Physiol, vol. 234, no. 6, pp. 7957–7969, Jun. 2019. [CrossRef]
- L. Chen, X. Gong, and M. Huang, “YY1-Activated Long Noncoding RNA SNHG5 Promotes Glioblastoma Cell Proliferation Through p38/MAPK Signaling Pathway,” Cancer Biother Radiopharm, vol. 34, no. 9, pp. 589–596, Nov. 2019. [CrossRef]
- O. Marín-Béjar et al., “Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2,” Genome Biol, vol. 14, no. 9, Sep. 2013. [CrossRef]
- A. Rocci, C. C. Hofmeister, and F. Pichiorri, “The potential of miRNAs as biomarkers for multiple myeloma,” Expert Rev Mol Diagn, vol. 14, no. 8, pp. 947–959, Nov. 2014. [CrossRef]
- C. Gu et al., “Integrative analysis of signaling pathways and diseases associated with the miR-106b/25 cluster and their function study in berberine-induced multiple myeloma cells,” Funct Integr Genomics, vol. 17, no. 2–3, pp. 253–262, May 2017. [CrossRef]
- S. Li, J. Zhu, J. Li, S. Li, and B. Li, “MicroRNA-141 inhibits proliferation of gastric cardia adenocarcinoma by targeting MACC1,” Arch Med Sci, vol. 14, no. 3, pp. 588–596, 2018. [CrossRef]
- L. Hui et al., “p38α suppresses normal and cancer cell proliferation by antagonizing the JNK–c-Jun pathway,” Nature Genetics 2007 39:6, vol. 39, no. 6, pp. 741–749, Apr. 2007. [CrossRef]
- E. K. Kim and E. J. Choi, “Pathological roles of MAPK signaling pathways in human diseases,” Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, vol. 1802, no. 4, pp. 396–405, Apr. 2010. [CrossRef]
- Y. Gao et al., “The Roles of MicroRNA-141 in Human Cancers: From Diagnosis to Treatment,” Cell Physiol Biochem, vol. 38, no. 2, pp. 427–448, 2016. [CrossRef]
- R. M. Schultz, “Potential of p38 MAP kinase inhibitors in the treatment of cancer,” Prog Drug Res, vol. 60, pp. 59–92, 2003. [CrossRef]
- C. Schultz, A. Link, and M. Leost, “Paullones, a series of cyclin-dependent kinase inhibitors: synthesis, evaluation of CDK1/cyclin B inhibition, and in vitro antitumor activity,” Journal of medicinal …, vol. 6, pp. 2909–2919, 1999. [CrossRef]
- M. I. Vega, S. Huerta-Yepaz, H. Garban, A. Jazirehi, C. Emmanouilides, and B. Bonavida, “Rituximab inhibits p38 MAPK activity in 2F7 B NHL and decreases IL-10 transcription: Pivotal role of p38 MAPK in drug resistance,” Oncogene, vol. 23, no. 20, pp. 3530–3540, Apr. 2004. [CrossRef]
- R. J. Gum et al., “Acquisition of sensitivity of stress-activated protein kinases to the p38 inhibitor, SB 203580, by alteration of one or more amino acids within the ATP binding pocket,” Journal of Biological Chemistry, vol. 273, no. 25, pp. 15605–15610, Jun. 1998. [CrossRef]
- A. Cuenda et al., “SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1,” FEBS Lett, vol. 364, no. 2, pp. 229–233, May 1995. [CrossRef]
- M. Goedert, A. Cuenda, M. Craxton, R. Jakes, and P. Cohen, “Activation of the novel stress-activated protein kinase SAPK4 by cytokines and cellular stresses is mediated by SKK3 (MKK6); comparison of its substrate specificity with that of other SAP kinases,” EMBO J, vol. 16, no. 12, pp. 3563–3571, Jun. 1997. [CrossRef]
- I. E. Zohn, Y. Li, E. Y. Skolnik, K. V. Anderson, J. Han, and L. Niswander, “p38 and a p38-Interacting Protein Are Critical for Downregulation of E-Cadherin during Mouse Gastrulation,” Cell, vol. 125, no. 5, pp. 957–969, Jun. 2006. [CrossRef]
- J. Wen et al., “P38 MAPK inhibition enhancing ATO-induced cytotoxicity against multiple myeloma cells,” Br J Haematol, vol. 140, no. 2, pp. 169–180, Jan. 2008. [CrossRef]
- R. M. Koerber et al., “Analysis of the anti-proliferative and the pro-apoptotic efficacy of Syk inhibition in multiple myeloma,” Exp Hematol Oncol, vol. 4, no. 1, p. 21, Aug. 2015. [CrossRef]
- L. Lei et al., “Resistance of osteosarcoma cells to the proapoptotic effects of carfilzomib involves activation of mitogen activated protein kinase pathways,” Exp Physiol, vol. 106, no. 2, pp. 438–449, Feb. 2021. [CrossRef]
- P. Dey, S. Biswas, R. Das, S. Chatterjee, and U. Ghosh, “p38 MAPK inhibitor SB203580 enhances anticancer activity of PARP inhibitor olaparib in a synergistic way on non-small cell lung carcinoma A549 cells,” Biochem Biophys Res Commun, vol. 670, pp. 55–62, Aug. 2023. [CrossRef]
- S. P. Davies, H. Reddy, M. Caivano, and P. Cohen, “Specificity and mechanism of action of some commonly used protein kinase inhibitors,” Biochem J, vol. 351, no. Pt 1, pp. 95–105, Oct. 2000. [CrossRef]
- J. Saklatvala, “The p38 MAP kinase pathway as a therapeutic target in inflammatory disease,” Curr Opin Pharmacol, vol. 4, no. 4, pp. 372–377, 2004. [CrossRef]
- H. S. Sharma et al., “Pathophysiology of blood-brain barrier in brain tumor. Novel therapeutic advances using nanomedicine,” Int Rev Neurobiol, vol. 151, pp. 1–66, Jan. 2020. [CrossRef]
- Y. Wu, X. Duan, Z. Gao, N. Yang, and F. Xue, “AICAR attenuates postoperative abdominal adhesion formation by inhibiting oxidative stress and promoting mesothelial cell repair,” PLoS One, vol. 17, no. 9, Sep. 2022. [CrossRef]
- J. P. Duffy et al., “The Discovery of VX-745: A Novel and Selective p38α Kinase Inhibitor,” ACS Med Chem Lett, vol. 2, no. 10, pp. 758–763, Oct. 2011. [CrossRef]
- T. Hideshima et al., “Targeting p38 MAPK inhibits multiple myeloma cell growth in the bone marrow milieu,” Blood, vol. 101, no. 2, pp. 703–705, Jan. 2003. [CrossRef]
- C. Ding, “Drug evaluation: VX-702, a MAP kinase inhibitor for rheumatoid arthritis and acute coronary syndrome,” Current Opinion in Investigational Drugs, vol. 7, no. 11, pp. 1020–1025, Jan. 2006, Accessed: Dec. 07, 2023. [Online]. Available: /articles/journal_contribution/Drug_evaluation_VX-702_a_MAP_kinase_inhibitor_for_rheumatoid_arthritis_and_acute_coronary_syndrome/23215691/1.
- V. R. Wydra, R. B. Ditzinger, N. J. Seidler, F. W. Hacker, and S. A. Laufer, “A patent review of MAPK inhibitors (2018 - present),” Expert Opin Ther Pat, vol. 33, no. 6, pp. 421–444, 2023. [CrossRef]
- V. Gandhi et al., “Compound GW506U78 in refractory hematologic malignancies: relationship between cellular pharmacokinetics and clinical response,” J Clin Oncol, vol. 16, no. 11, pp. 3607–3615, 1998. [CrossRef]
- K. Ghias, C. Ma, V. Gandhi, L. C. Platanias, N. L. Krett, and S. T. Rosen, “8-Amino-adenosine induces loss of phosphorylation of p38 mitogen-activated protein kinase, extracelluar signal-regulated kinase 1/2, and Akt kinase: Role in induction of apoptosis in multiple myeloma,” Mol Cancer Ther, vol. 4, no. 4, pp. 569–577, Apr. 2005. [CrossRef]
- S. Medicherla et al., “p38α-Selective MAP Kinase Inhibitor Reduces Tumor Growth in Mouse Xenograft Models of Multiple Myeloma,” Anticancer Res, vol. 28, no. 6A, pp. 3827–3833, Nov. 2008, Accessed: Sep. 27, 2023. [Online]. Available: https://ar.iiarjournals.org/content/28/6A/3827.
- T. A. Navas et al., “Inhibition of p38alpha MAPK enhances proteasome inhibitor-induced apoptosis of myeloma cells by modulating Hsp27, Bcl-X(L), Mcl-1 and p53 levels in vitro and inhibits tumor growth in vivo,” Leukemia, vol. 20, no. 6, pp. 1017–1027, 2006. [CrossRef]
- K. Vanderkerken et al., “Inhibition of p38α mitogen-activated protein kinase prevents the development of osteolytic bone disease, reduces tumor burden, and increases survival in murine models of multiple myeloma,” Cancer Res, vol. 67, no. 10, pp. 4572–4577, May 2007. [CrossRef]
- A. N. Nguyen et al., “Normalizing the bone marrow microenvironment with p38 inhibitor reduces multiple myeloma cell proliferation and adhesion and suppresses osteoclast formation,” Exp Cell Res, vol. 312, no. 10, pp. 1909–1923, Jun. 2006. [CrossRef]
- D. S. Siegel et al., “Phase II Trial of SCIO-469 as Monotherapy (M) or in Combination with Bortezomib (MB) in Relapsed Refractory Multiple Myeloma (MM).,” Blood, vol. 108, no. 11, p. 3580, Nov. 2006. [CrossRef]
- H. Yasui et al., “BIRB 796 enhances cytotoxicity triggered by bortezomib, heat shock protein (Hsp) 90 inhibitor, and dexamethasone via inhibition of p38 mitogen-activated protein kinase/Hsp27 pathway in multiple myeloma cell lines and inhibits paracrine tumour growth,” Br J Haematol, vol. 136, no. 3, pp. 414–423, Feb. 2007. [CrossRef]
- M. J. HOLTZMAN, A. G. ROMERO, B. J. GEROVAC, Z. HAN, S. P. KEELER, and K. WU, “MITOGEN-ACTIVATED PROTEIN KINASE INHIBITORS, METHODS OF MAKING, AND METHODS OF USE THEREOF,” Dec. 2019.
- R. M. Campbell et al., “Characterization of LY2228820 dimesylate, a potent and selective inhibitor of p38 MAPK with antitumor activity,” Mol Cancer Ther, vol. 13, no. 2, pp. 364–374, Feb. 2014. [CrossRef]
- D. Bhattacharjee et al., “Inhibition of a lower potency target drives the anticancer activity of a clinical p38 inhibitor,” Cell Chem Biol, vol. 30, no. 10, pp. 1211-1222.e5, Oct. 2023. [CrossRef]
- M. Lepore Signorile et al., “c-MYC Protein Stability Is Sustained by MAPKs in Colorectal Cancer,” Cancers (Basel), vol. 14, no. 19, Oct. 2022. [CrossRef]
- K. Ishitsuka et al., “p38 mitogen-activated protein kinase inhibitor LY2228820 enhances bortezomib-induced cytotoxicity and inhibits osteoclastogenesis in multiple myeloma; therapeutic implications,” Br J Haematol, vol. 141, no. 5, pp. 598–606, Jun. 2008. [CrossRef]
- A. L. T. A. P. K. B. M. R. D. W. G. S. J. C. M. F. M. E. S. M. J. L. M. T. F. C. E. J. M. P. C. X. C. R. S. G. I. R. R. Patnaik A, “First-in-human Phase I Study of Copanlisib (BAY 80-6946), an Intravenous Pan-Class I Phosphatidylinositol 3-kinase Inhibitor, in Patients With Advanced Solid Tumors and non-Hodgkin’s Lymphomas,” Annals of Oncology, vol. 27, no. 10, pp. 1928–1940, 2016. [CrossRef]
- J. Yang et al., “Constitutive activation of p38 MAPK in tumor cells contributes to osteolytic bone lesions in multiple myeloma,” Leukemia, vol. 26, no. 9, p. 2114, Sep. 2012. [CrossRef]
- K. Ghias, C. Ma, V. Gandhi, L. C. Platanias, N. L. Krett, and S. T. Rosen, “8-Amino-adenosine induces loss of phosphorylation of p38 mitogen-activated protein kinase, extracellular signal-regulated kinase 1/2, and Akt kinase: role in induction of apoptosis in multiple myeloma,” Mol Cancer Ther, vol. 4, no. 4, pp. 569–577, Apr. 2005. [CrossRef]
- H. Peng et al., “Characterization of p38 MAPK isoforms for drug resistance study using systems biology approach,” Bioinformatics, vol. 30, no. 13, pp. 1899–1907, Jul. 2014. [CrossRef]
- M. Hallek, P. L. Bergsagel, and K. C. Anderson, “Multiple Myeloma: Increasing Evidence for a Multistep Transformation Process,” Blood, vol. 91, no. 1, p. 3, Jan. 1998. [CrossRef]
- G. Pratt, “Molecular aspects of multiple myeloma,” Mol Pathol, vol. 55, no. 5, pp. 273–283, Oct. 2002. [CrossRef]
- L. C. Platanias, “Map kinase signaling pathways and hematologic malignancies,” Blood, vol. 101, no. 12, pp. 4667–4679, Jun. 2003. [CrossRef]
- P. Liu et al., “Activating Mutations of N- and K-ras in Multiple Myeloma Show Different Clinical Associations: Analysis of the Eastern Cooperative Oncology Group Phase III Trial,” Blood, vol. 88, no. 7, pp. 2699–2706, Oct. 1996. [CrossRef]
- B. Zhang and R. G. Fenton, “Proliferation of IL-6-independent multiple myeloma does not require the activity of extracellular signal-regulated kinases (ERK1/2),” J Cell Physiol, vol. 193, no. 1, pp. 42–54, Oct. 2002. [CrossRef]
- D. Ma et al., “Crucial role of heme oxygenase-1 in the sensitivity of acute myeloid leukemia cell line Kasumi-1 to ursolic acid,” Anticancer Drugs, vol. 25, no. 4, pp. 406–414, 2014. [CrossRef]
- W. Wu et al., “Potential crosstalk of the interleukin-6-heme oxygenase-1-dependent mechanism involved in resistance to lenalidomide in multiple myeloma cells,” FEBS J, vol. 283, no. 5, pp. 834–849, Mar. 2016. [CrossRef]
- J. Wang et al., “Bone marrow stromal cell-derived exosomes as communicators in drug resistance in multiple myeloma cells,” Blood, vol. 124, no. 4, pp. 555–566, Jul. 2014. [CrossRef]
- E. Håland et al., “TAK1-inhibitors are cytotoxic for multiple myeloma cells alone and in combination with melphalan,” Oncotarget, vol. 12, no. 21, pp. 2158–2168, Oct. 2021. [CrossRef]
- J. Zhang et al., “Synergistic action of 5Z-7-oxozeaenol and bortezomib in inducing apoptosis of Burkitt lymphoma cell line Daudi,” Tumour Biol, vol. 37, no. 1, pp. 531–539, Jan. 2016. [CrossRef]
- M. Guo et al., “Targeting MK2 Is a Novel Approach to Interfere in Multiple Myeloma,” Front Oncol, vol. 9, no. AUG, 2019. [CrossRef]
- W. Xiao et al., “Rafoxanide, an organohalogen drug, triggers apoptosis and cell cycle arrest in multiple myeloma by enhancing DNA damage responses and suppressing the p38 MAPK pathway,” Cancer Lett, vol. 444, pp. 45–59, Mar. 2019. [CrossRef]
- A. Soriani et al., “p38 MAPK differentially controls NK activating ligands at transcriptional and post-transcriptional level on multiple myeloma cells,” Oncoimmunology, vol. 6, no. 1, Jan. 2016. [CrossRef]
- R. M. Koerber et al., “Analysis of the anti-proliferative and the pro-apoptotic efficacy of Syk inhibition in multiple myeloma,” Exp Hematol Oncol, vol. 4, no. 1, Aug. 2015. [CrossRef]
- A. Li, X. Chen, Z. Jing, and J. Chen, “Trifluoperazine induces cellular apoptosis by inhibiting autophagy and targeting NUPR1 in multiple myeloma,” FEBS Open Bio, vol. 10, no. 10, pp. 2097–2106, Oct. 2020. [CrossRef]
- R. E. Tiedemann et al., “Identification of a potent natural triterpenoid inhibitor of proteosome chymotrypsin-like activity and NF-kappaB with antimyeloma activity in vitro and in vivo,” Blood, vol. 113, no. 17, pp. 4027–4037, 2009. [CrossRef]
- X. M. Xu et al., “[Effect of Celastrol Based on IRAK4/ERK/p38 Signaling Pathway on Proliferation and Apoptosis of Multiple Myeloma Cells],” Zhongguo Shi Yan Xue Ye Xue Za Zhi, vol. 30, no. 1, pp. 175–182, Feb. 2022. [CrossRef]
- S. S. Bhandarkar et al., “Tris (dibenzylideneacetone) dipalladium, a N-myristoyltransferase-1 inhibitor, is effective against melanoma growth in vitro and in vivo,” Clin Cancer Res, vol. 14, no. 18, pp. 5743–5748, Sep. 2008. [CrossRef]
- P. de la Puente, F. Azab, B. Muz, M. Luderer, J. Arbiser, and A. K. Azab, “Tris DBA palladium overcomes hypoxia-mediated drug resistance in multiple myeloma,” Leuk Lymphoma, vol. 57, no. 7, pp. 1677–1686, Jul. 2016. [CrossRef]
- A. T. Stefka, D. Johnson, S. Rosebeck, J. H. Park, Y. Nakamura, and A. J. Jakubowiak, “Potent anti-myeloma activity of the TOPK inhibitor OTS514 in pre-clinical models,” Cancer Med, vol. 9, no. 1, pp. 324–334, Jan. 2020. [CrossRef]
- J. He et al., “Therapeutic effects of the novel subtype-selective histone deacetylase inhibitor chidamide on myeloma-associated bone disease,” Haematologica, vol. 103, no. 8, pp. 1369–1379, Jul. 2018. [CrossRef]
- L. H, H. J, and Y. J, “Tumor cell p38 MAPK: A trigger of cancer bone osteolysis,” Cancer Cell Microenviron, vol. 2, no. 1, Jan. 2015. [CrossRef]
- L. Trentin et al., “Multiple myeloma plasma cells show different chemokine receptor profiles at sites of disease activity,” Br J Haematol, vol. 138, no. 5, pp. 594–602, Sep. 2007. [CrossRef]
- M. Broussas et al., “A New Anti-CXCR4 Antibody That Blocks the CXCR4/SDF-1 Axis and Mobilizes Effector Cells,” Mol Cancer Ther, vol. 15, no. 8, pp. 1890–1899, Aug. 2016. [CrossRef]
- M. K. Pandey et al., “Gambogic acid inhibits multiple myeloma mediated osteoclastogenesis through suppression of chemokine receptor CXCR4 signaling pathways,” Exp Hematol, vol. 42, no. 10, pp. 883–896, Oct. 2014. [CrossRef]
- J. Li, J. Zhu, B. Cao, and X. Mao, “The mTOR signaling pathway is an emerging therapeutic target in multiple myeloma,” Curr Pharm Des, vol. 20, no. 1, pp. 125–135, Jan. 2014. [CrossRef]
- K. Han et al., “SC06, a novel small molecule compound, displays preclinical activity against multiple myeloma by disrupting the mTOR signaling pathway,” Sci Rep, vol. 5, Sep. 2015. [CrossRef]
- H. Y. Cho and S. W. Lee, “TLR5 activation by flagellin induces doxorubicin resistance via interleukin-6 (IL-6) expression in two multiple myeloma cells,” Cell Immunol, vol. 289, no. 1–2, pp. 27–35, 2014. [CrossRef]
- J. Abdi, T. Mutis, J. Garssen, and F. Redegeld, “Characterization of the Toll-like receptor expression profile in human multiple myeloma cells,” PLoS One, vol. 8, no. 4, Apr. 2013. [CrossRef]
- G. Jego, D. Chiron, K. Berthenet, and C. Pellat-Deceunynck, “Modulation of normal and malignant plasma cells function by toll-like receptors,” Front Biosci (Elite Ed), vol. 4, no. 6, pp. 2289–2301, Jan. 2012. [CrossRef]
- Z. Liu, T. Li, K. Jiang, Q. Huang, Y. Chen, and F. Qian, “Induction of chemoresistance by all-trans retinoic acid via a noncanonical signaling in multiple myeloma cells,” PLoS One, vol. 9, no. 1, Jan. 2014. [CrossRef]
- T. Otsuki, H. Sakaguchi, T. Hatayama, P. Wu, A. Takata, and F. Hyodoh, “Effects of all-trans retinoic acid (ATRA) on human myeloma cells,” Leuk Lymphoma, vol. 44, no. 10, pp. 1651–1656, Oct. 2003. [CrossRef]
- C. Simard, M. Cloutier, and S. Néron, “Feasibility study: phosphospecific flow cytometry enabling rapid functional analysis of bone marrow samples from patients with multiple myeloma,” Cytometry B Clin Cytom, vol. 86, no. 2, pp. 139–144, Mar. 2014. [CrossRef]
| Transcription factor | Putative binding site positions relative to TSS | |||
| MAPK14_1 | MAPK11_1 | MAPK12_1 | MAPK13_1 | |
| ARNT: HIF1A | -466, -455, -260 | -93, | -40, -88 | -120, 32 |
| ASCL1 | -933, -901, -880, -878, -785, -603, -602, -392, -391, -248, -59, 22, 2 | 74, 73, -459, -506, -550, -575, -609, -610, -755, -830, -937 | 44, 42, -2, -3, -190, -214, -215, -309, -310, -410, -411, -518, -523, -666, -688, -810, -812, -882, -883, -968, -982 | -993, -810, -809, -551, -278, -264, -254, -185, -184, -103, -102, -87, -40, -17, 25, 26 |
| ATF4 | -823, -777, -774 | |||
| ATF7 | -824, -823 | -92 | ||
| Ahr: Arnt | -466, -455, -334, -260 | -93 | -120, 32 | |
| Arid3a | -860 | -929, -490, -485 | ||
| Ascl2 | -901, -900, -785, -392, -391, -381, -380, -249, -248, -59, 24, 25 | -380, -829 | -2, -214, -309, -523, -524, -882, -883 | -809, -580, -278, -185, -184, -87 |
| Atf1 | -823 | -579 | -92 | -697 |
| BACH2 | -633, -629 | -392 | -508, -504 | |
| Bcl6 | -874 | -356, -497, -672, -732, -884 | -382, -927 | |
| CEBPA | -979, -823, -549 | -548 | -947 | |
| CTCF | -768, -513, -275, -209, 1 | -178, -219, -221, -253, -383, -584, -848 | 27, -52, -76, -146, -270, -336, -601, -672, -799, -891 | -870, -87, -54, -14 |
| E2F1 | -597, -596, -270, -269, -218, -217 | -183, -184, -290, -543, -544, -660 | 35, 34 | 15 |
| EGR1 | -465, -394, -28 | 34, -8, -252, -783, -903, -949 | 30, -256, -335, -602, -744, -917 | -866, -835, -604, -384, -278, -246, -147, -61, -39, -14 |
| ELF1 | -62, -28, 33 | 82, -126, -154, -489, -880 | 24, -120, -438 | -155, 73, 76 |
| JUN | -887, -822 | -609, -460 | ||
| KLF4 | -770, -587, -453, -334 | -91, -684 | -94, -290, -906 | -380, -122, -58 |
| MYC | -942, -785, -603, -602, -444, -392, -391, -266 | -230, -506 | -3, -87, -88, -518, -523, -643, -644 | -984, -606, -199, -185, -184, -103, -102, -87 |
| SMAD2:SMAD3: SMAD4 | -896, -464, -251, -204 | -147, -211, -926 | -260, -266, -563, -625, -829, -865, -910 | |
| YY1 | -977, -962, -547, -98 | -949, -814, -672 | ||
| XBP1 | -943 | -578 | -738 | |
| TP53 | -759, -758 | -578 | -754 | -959, -958, -437, -125, -124 |
| Stat4 | -419 | -672, -676 | -395 | |
| SP1 | -925, -881, -659, -633, -499, -225, -186, -141, -114, 72 | 63, -12, -33, -46, -87, -141, -165, -192, -200, -279, -294, -347, -508, -527, -543, -598, -650, -683, -759, -905 | 78, 28, 16, -19, -47, -103, -147, -164, -195, -206, -267, -289, -317, -592, -670, -740, -803, -822, -853, -905 | -868, -837, -666, -522, -329, -290, -257, -205, -59, -44, -25, -9 |
| POU1F1 | -575 | -407 | -919, -490, -395 | |
| NFKB1 | -982, -938, -552, -506, -32 | -69, -101, -102, -788, -945 | 34, 5, -62, -314, -513, -586, -899, -961 | -988, -944, -467, -466, -232, -211 |
| . | MAPK11 | MAPK12 | MAPK13 | MAPK14 |
| Predicted microRNAs (miRtarBase) | hsa-miR-122-5p, hsa-miR-124-3p, hsa-let-7a-5p | hsa-miR-18a-5p, hsa-miR-150-5p, hsa-miR-18b-5p, hsa-miR-3134, hsa-miR-3691-5p, hsa-miR-4434, hsa-miR-4516, hsa-miR-4525, hsa-miR-4531, hsa-miR-4534, hsa-miR-4690-3p, hsa-miR-4731-5p, hsa-miR-4735-3p, hsa-miR-4761-5p, hsa-miR-4773, hsa-miR-5010-5p, hsa-miR-5187-5p, hsa-miR-5589-5p, hsa-miR-5685, hsa-miR-5703, hsa-miR-6778-3p, hsa-miR-6795-5p, hsa-miR-6798-5p, hsa-miR-6814-5p, hsa-miR-6887-5p, hsa-miR-8082 | hsa-miR-124-3p, hsa-miR-24-3p, hsa-miR-199a-3p, hsa-miR-200a-3p, hsa-miR-141-3p, hsa-miR-125b-5p, hsa-miR-214-3p, hsa-miR-155-5p, hsa-miR-17-5p, hsa-miR-106a-5p | |
| Bio-Predicted(TargetScan) | hsa-miR-325-3p | hsa-miR-125a-5p, has-miR-125b-5p, has-miR-4319 | hsa-miR-3681-3p, hsa-miR-128-3p, has-miR-216-3p |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




