1. Introduction
Medical image segmentation is a critical component of modern healthcare [
1], enabling clinicians and researchers to extract valuable insights from complex visual data [
2,
3]. Accurate and reliable segmentation is vital for tasks such as disease diagnosis, treatment planning, and the assessment of treatment outcomes [
4,
5,
6,
7]. However, the inherent challenges in this process, including noise, non-uniform illumination, and variations in tissue intensity, have spurred ongoing research into more effective techniques [
8,
9,
10].
Medical images, acquired through techniques like MRI, CT-scan, and X-ray [
11], often exhibit variations in contrast, brightness, and texture [
12]. These variations can obscure critical anatomical structures and pathologies, making accurate segmentation a formidable task [
13,
14]. Conventional segmentation methods, such as thresholding and region-growing [
15,
16], struggle to produce reliable results in the face of such complexities [
17]. Therefore, there is an urgent need for advanced techniques capable of handling these challenges with precision [
18,
19].
This article presents an innovative approach to address these challenges, offering a sophisticated solution for advancing medical image segmentation [
20]. Our method harnesses the power of Particle Swarm Optimization (PSO) [
21] in conjunction with Histogram Equalization (HE) [
22], aiming to significantly enhance the accuracy and robustness of image segmentation in the medical domain. PSO emerges as a promising optimization framework for addressing the intricacies of medical image segmentation [
23,
24]. Inspired by the collective behavior of swarming particles, PSO excels in finding optimal solutions in complex, multidimensional spaces [
25]. By adapting PSO to the task of medical image segmentation [
26,
27], we aim to exploit its ability to converge toward the most suitable image partitioning, considering both global and local characteristics. Histogram Equalization (HE) is a well-established image enhancement technique that adjusts the pixel intensities to achieve a more uniform distribution, thereby improving contrast and mitigating the effects of non-uniform illumination [
28]. Integrating HE into our approach complements the optimization capabilities of PSO by preparing the input images for segmentation, enhancing the visibility of critical details within medical images.
What sets our approach apart is the synergistic fusion of PSO and HE. PSO optimizes the segmentation process, finding an optimal partitioning of the image [
29], while HE pre-processes the image to enhance contrast and visibility [
30]. This combination addresses the dual challenges of accurately identifying regions of interest and improving image quality, resulting in a more robust and precise segmentation method. Throughout this article, we will delve into the theoretical underpinnings of PSO and HE, providing in-depth insights into their integration. Furthermore, we will present empirical evidence of the effectiveness of our approach through extensive experiments and comparisons with existing methods. Our aim is to demonstrate not only the novelty of our approach but also its practical significance, offering a potential paradigm shift in the field of medical image analysis.
This article is structured into five main sections.
Section 1 provides an overview of the significance of medical image segmentation and the challenges it presents. In
Section 2, we review previous research, identify gaps in existing methodologies, and establish the context for our study.
Section 3 presents the core method, where we combine Particle Swarm Optimization (PSO) with Histogram Equalization (HE). This section explains the technical aspects of our approach. In
Section 4 we delve into the analysis of empirical findings and their implications. Finally,
Section 5 summarizes the key points and discusses their potential impact on the healthcare field.
2. Related Works
The field of medical image segmentation has witnessed significant progress, with numerous approaches developed to address the challenges presented by diverse medical imaging scenarios [
31,
32]. In this section, we provide an overview of relevant literature that lays the foundation for our novel Particle Swarm Optimization (PSO)-based Histogram Equalization (HE) approach.
2.1. Traditional Segmentation Methods
Traditional segmentation methods have historically served as the building blocks of medical image analysis [
31,
33]. These methods include thresholding, region growing, and edge detection [
34,
35,
36]. Researchers conducted a comparative study of thresholding techniques for medical image segmentation [
33,
37], highlighting their simplicity and limitations in handling complex intensity variations [
38]. One facet of the research community has explored the challenges and prospects of region-growing-based segmentation [
39], particularly in the context of complex structures like brain tumor images [
40,
41]. This exploration has led to a recognized need for more robust techniques to address the subtleties and complexities of such images [
42].
Furthermore, foundational knowledge in digital image processing has underpinned many traditional segmentation methods [
43]. However, it is crucial to acknowledge that these traditional techniques often face difficulties when handling noisy or non-uniformly illuminated images [
44,
45]. This recognition has driven the exploration of more advanced and adaptable approaches.
2.2. Enhancement Techniques
Enhancement techniques play a pivotal role in elevating image quality and facilitating subsequent segmentation processes [
46]. These techniques aim to enhance contrast and improve overall image visibility, contributing to more effective segmentation outcomes [
47,
48].
One avenue of research explores the enhancement of medical image segmentation through adaptive histogram equalization [
49], highlighting its potential for contrast improvement [
22,
28,
50]. Additionally, a comprehensive survey has delved into the realm of image enhancement techniques within the field of medical imaging [
51]. This survey provides valuable insights into various methods applied to address challenges such as non-uniform illumination and noise in medical images [
52]. This foundational overview of image enhancement techniques is essential for understanding strategies to improve the quality of medical images prior to the segmentation process.
2.3. Optimization-Based Approaches
Optimization-based methods have gained significant attention within the domain of advanced medical image segmentation enhancement [
53,
54]. These approaches are designed to fine-tune and optimize segmentation algorithms [
21], aligning with the core objectives of our study. By leveraging optimization strategies, these methods hold the promise of enhancing the accuracy and efficiency of medical image segmentation [
55,
56], aligning with the central theme of our research, Advanced Medical Image Segmentation Enhancement (AMISE): the PSO-Based HE approach.
Through a comprehensive exploration of optimization-based approaches, as conducted by various researchers, their relevance in the context of enhancing the AMISE approach becomes evident. This review encompasses a broad spectrum of optimization strategies, including traditional optimization algorithms, metaheuristic approaches, and machine learning-based optimization [
57,
58,
59], all of which resonate with the innovative and advanced nature of our proposed method. These techniques aim to address the intricate challenges of medical image segmentation, ultimately optimizing algorithm parameters to improve the segmentation outcomes and, by extension [
23,
32], advancing the core concept of image enhancement that underpins our research.
The significance of optimization-based approaches aligns with the primary objective of our study, which is to refine and advance medical image segmentation through the AMISE approach. By optimizing segmentation algorithms, these techniques contribute to the precise delineation of regions of interest within medical images [
60]. Thus, they directly pertain to our research’s fundamental goal of advancing medical image segmentation by integrating Particle Swarm Optimization and Histogram Equalization, offering a valuable perspective into the strategies used to achieve this enhancement.
2.4. Particle Swarm Optimization (PSO) in Image Analysis
Particle Swarm Optimization (PSO) has emerged as a versatile and innovative optimization technique with applications spanning various domains, including image analysis [
61,
62]. Initially conceived as an optimization method inspired by collective swarm behavior, PSO’s adaptability, effectiveness, and problem-solving capabilities make it an increasingly influential tool in the field of image analysis [
63,
64]. Within the broader context of image analysis, PSO excels in tasks such as feature selection [
65], image registration [
66], image segmentation [
67,
68,
69], and object tracking [
70]. Its capacity to navigate complex search spaces and avoid local optima positions it as an attractive choice for researchers.
In the domain of medical image analysis, our research takes a pioneering step by extending the application of PSO to the specific realm of medical image segmentation [
58]. This strategic extension capitalizes on PSO’s inherent strengths in exploring intricate search spaces, aligning seamlessly with the complexities often encountered in medical images marked by noise, non-uniform illumination, and intricate structures [
71]. Understanding the pivotal role of PSO in image analysis forms the cornerstone of our research, where we harness its unique optimization capabilities to refine and enhance the accuracy of medical image segmentation, as outlined in AMISE: PSO-Based HE approach.
2.5. Fusion of Optimization and Image Enhancement
The fusion of optimization techniques with image enhancement methods signifies a rapidly evolving research area with significant implications for medical image analysis [
16,
72,
73]. This emerging field explores the seamless integration of optimization strategies and image enhancement techniques to collectively address the challenges encountered in medical image segmentation [
74]. Notably, it highlights the potential of optimization-enhanced approaches in substantially improving segmentation accuracy, particularly in the context of extracting critical structures such as brain tumors from MRI images [
75]. This avenue of research has yielded methods that enhance the visibility of structures within medical images while concurrently optimizing the precise delineation of regions of interest [
76].
At the same time, the growing interest in machine learning techniques and their applications within the domain of medical imaging has provided valuable insights into their role in computer-aided diagnosis and deep learning-based image segmentation [
77]. These advances have catalyzed innovation in image analysis techniques, offering valuable perspectives for addressing the complexities presented by intricate medical images [
78]. Our research builds upon this evolving body of knowledge, introducing a pioneering approach that synergizes PSO and HE to enhance medical image segmentation. In the following sections, we delve into the technical intricacies of our proposed method and present empirical findings, presenting a fresh and innovative perspective on addressing the challenges associated with noisy and non-uniform medical images.
3. Materials and Methods
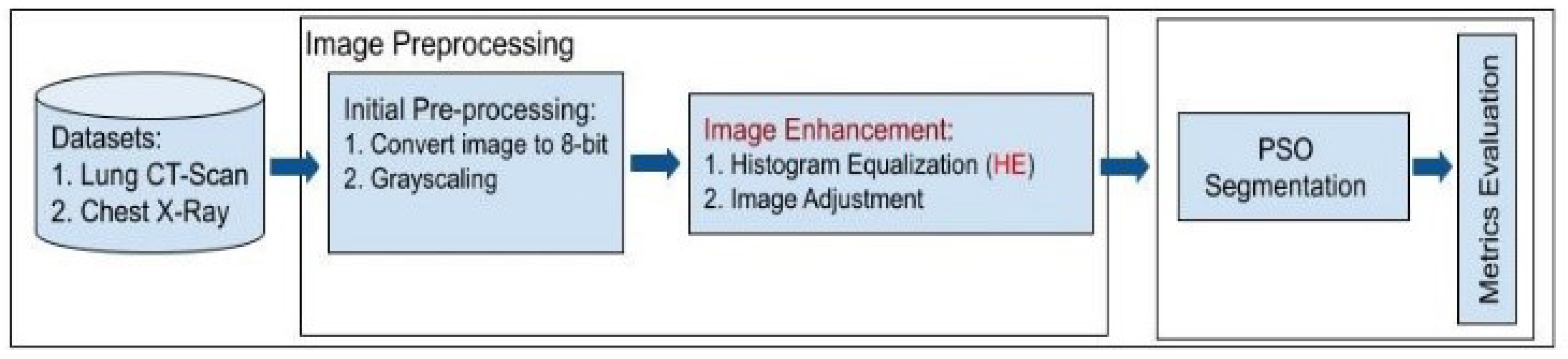
In this section, we provide a detailed description of the method employed in our novel approach, which combines Particle Swarm Optimization (PSO) with Histogram Equalization (HE) to enhance medical image segmentation. We outline the key steps involved in the proposed method, including data preprocessing, PSO-based optimization, and integration with HE. The detailed process can be seen in
Figure 1.
3.1. Dataset Selection
Our study leveraged two distinct datasets (Lung CT-scan Dataset and Chest X-ray (COVID-19) Dataset), each carefully chosen to represent different aspects of medical imaging challenges and diagnostic scenarios.
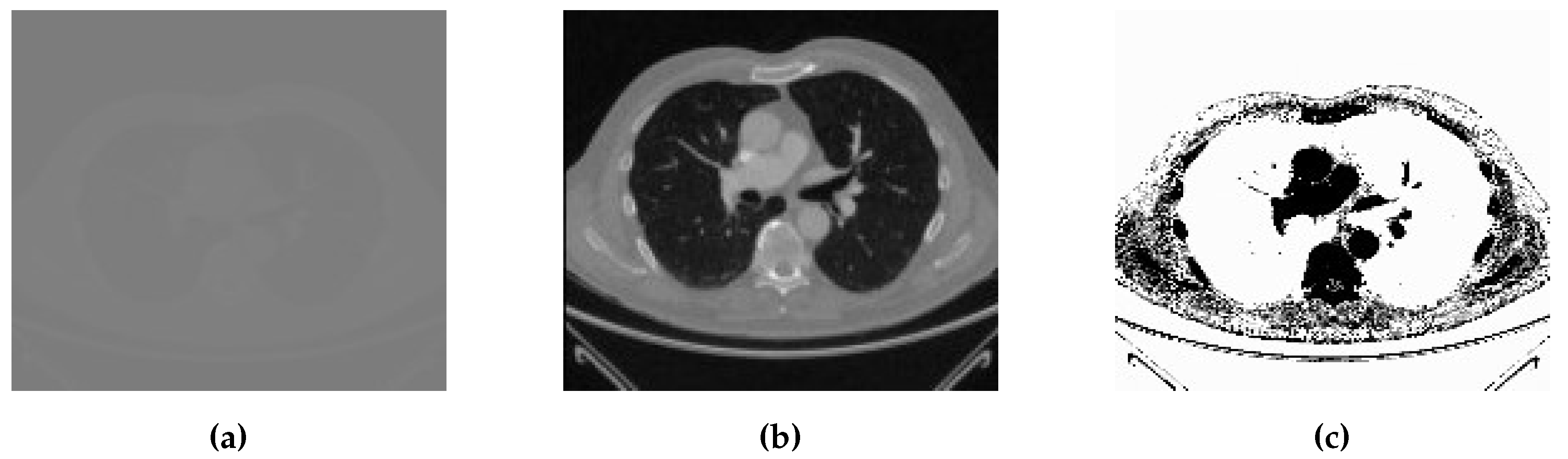
3.1.1. Lung CT-scan Dataset [79]
The Lung CT-Scan dataset consists of a total of 267 medical images. These images are representative of lung CT-Scans, a common modality used in pulmonary imaging. Lung CT-Scans provide high-resolution cross-sectional views of the chest, making them essential for diagnosing various respiratory conditions and diseases (
Figure 2).
To ensure a comprehensive evaluation of our method’s performance, we purposefully chose datasets with unique characteristics and challenges, such as the Lung CT-Scan dataset. This dataset primarily comprises high-resolution CT-Scan images of the lung, which are known for their intricate anatomical structures, including lung parenchyma, blood vessels, and airways. Challenges encountered within this dataset encompass the identification of subtle abnormalities, the management of variations in lung density, and the requirement for precise segmentation of lung regions.
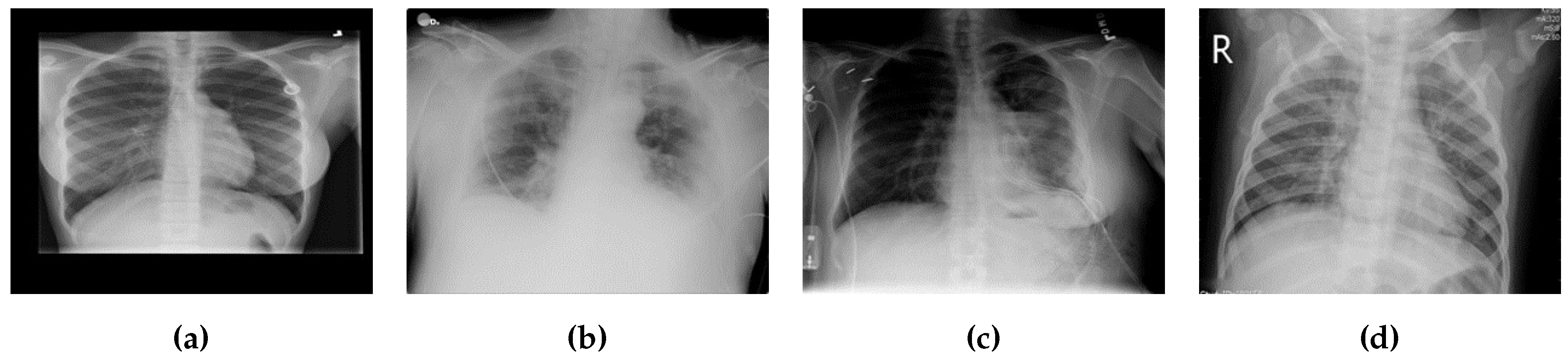
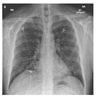
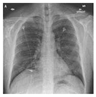
3.1.2. Chest X-ray (COVID-19) Dataset [80]
The Chest X-ray dataset encompasses a larger collection of 3616 images. These images are Chest X-rays (
Figure 3), a fundamental diagnostic tool widely used in clinical practice. This dataset reflects a diverse range of medical conditions, including those related to COVID-19, and includes a variety of anatomical regions within the chest.
As it was said above, we chose datasets with different characteristics and challenges to evaluate our method’s performance. The Chest X-ray dataset, for instance, consists of 2D X-ray images of the chest. These images introduce a unique set of challenges, characterized by lower spatial resolution and the presence of overlapping structures, such as ribs and organs. Moreover, these images, relevant to COVID-19 diagnosis, might exhibit specific patterns associated with viral pneumonia, adding another layer of complexity to the segmentation process.
3.1.3. Ground Truth Annotations
In order to rigorously assess the performance of our segmentation method, we relied on meticulously generated ground truth annotations. These annotations provide a critical reference point for evaluating the accuracy and effectiveness of our approach in delineating regions of interest within the medical images.
Our ground truth data was collected from Kaggle [
79,
80], a reputable platform for data sharing and collaboration. The annotations were crafted and meticulously reviewed by medical experts who possess specialized knowledge in the relevant domains. These experts conducted the labor-intensive task of manually outlining regions of clinical significance in the medical images, ensuring that structures and areas crucial for clinical diagnoses were precisely identified. The GT Images in
Figure 4 exemplify samples of these annotations, showcasing the quality of the reference data from both Lung CT-Scan and Chest X-ray datasets. These ground truth figures serve as invaluable benchmarks, enabling us to quantitatively measure the method’s accuracy in segmenting regions of interest within the medical images.
The utilization of this rigorously sourced and expert-validated ground truth data is of paramount importance. It offers a trustworthy foundation for evaluating the quality of our segmentation results, ensuring that our approach aligns with clinical standards and expert assessments. This alignment is essential in determining the effectiveness of our proposed PSO-based method in the context of real-world medical image analysis.
3.2. Dataset Preprocessing
Data preprocessing plays a crucial role in ensuring that the input medical images are in an optimal state for subsequent segmentation and enhancement processes [
81]. The preprocessing steps are designed to address image artifacts, enhance image quality, and simplify image representation [
82]. In this section, we provide a comprehensive description of the data preprocessing pipeline, which includes the following key steps: image conversion to 8-bit, grayscaling, image enhancement with Histogram Equalization (HE), and image adjustment.
3.2.1. Image Conversion to 8-bit
The initial step in data preprocessing involves converting the raw medical images into an 8-bit format [
83]. This conversion simplifies the pixel intensity values to a range between 0 and 255, which is more manageable for subsequent processing. The 8-bit representation ensures uniformity across all images, regardless of their original bit depths [
84]. This simplification is particularly important for consistency in the analysis and facilitates efficient memory usage.
3.2.2. Grayscaling
Following the 8-bit conversion, the images are grayscaled, ensuring that they are represented in grayscale intensity levels [
85]. Grayscaling simplifies the images by reducing them to a single channel, eliminating color information. This simplification reduces computational complexity and memory requirements while preserving essential intensity information [
86]. Grayscaling is a fundamental preprocessing step that ensures uniformity in the image data and simplifies subsequent processing [
87].
3.2.3. Image Enhancement with Histogram Equalization (HE)
One of the central aspects of data preprocessing is the application of Histogram Equalization (HE). This enhancement technique redistributes pixel intensities to achieve a more uniform histogram [
88]. Histogram Equalization (HE) is employed to improve the contrast and visibility of structures within the images [
89]. By enhancing fine details and mitigating non-uniform illumination, HE prepares the images for more accurate segmentation. This step is crucial for ensuring that subtle features and structures are well-visualized and can be accurately delineated during subsequent processing.
3.2.4. Image Adjustment
In addition to the above steps, image adjustment techniques are applied to fine-tune specific image properties. These adjustments may include brightness and contrast adjustments to optimize the visual representation of the images. Image adjustment ensures that the preprocessed images are well-suited for subsequent segmentation and optimization processes by fine-tuning the image properties to achieve optimal visualization.
3.3. Particle Swarm Optimization (PSO)
Particle Swarm Optimization (PSO) is a powerful optimization technique inspired by the collective behavior of swarming particles [
90]. It has gained prominence in various fields, including image analysis and medical image segmentation, due to its ability to efficiently explore complex search spaces and find optimal solutions. In this section, we provide an in-depth description of the application of PSO in our method for medical image segmentation.
At the core of PSO lies the concept of simulating the social behavior of particles within a swarm. Each particle in the swarm represents a potential solution to an optimization problem [
91]. The behavior of particles is guided by their position and velocity in a multidimensional search space. PSO has been widely applied in image analysis tasks, including image registration, feature selection, and image segmentation [
92]. In the context of medical imaging, PSO is particularly valuable due to its ability to address the challenges posed by noisy and complex images [
93]. Its capacity to explore complex, multidimensional spaces efficiently makes it a valuable tool for optimizing image processing and segmentation algorithms.
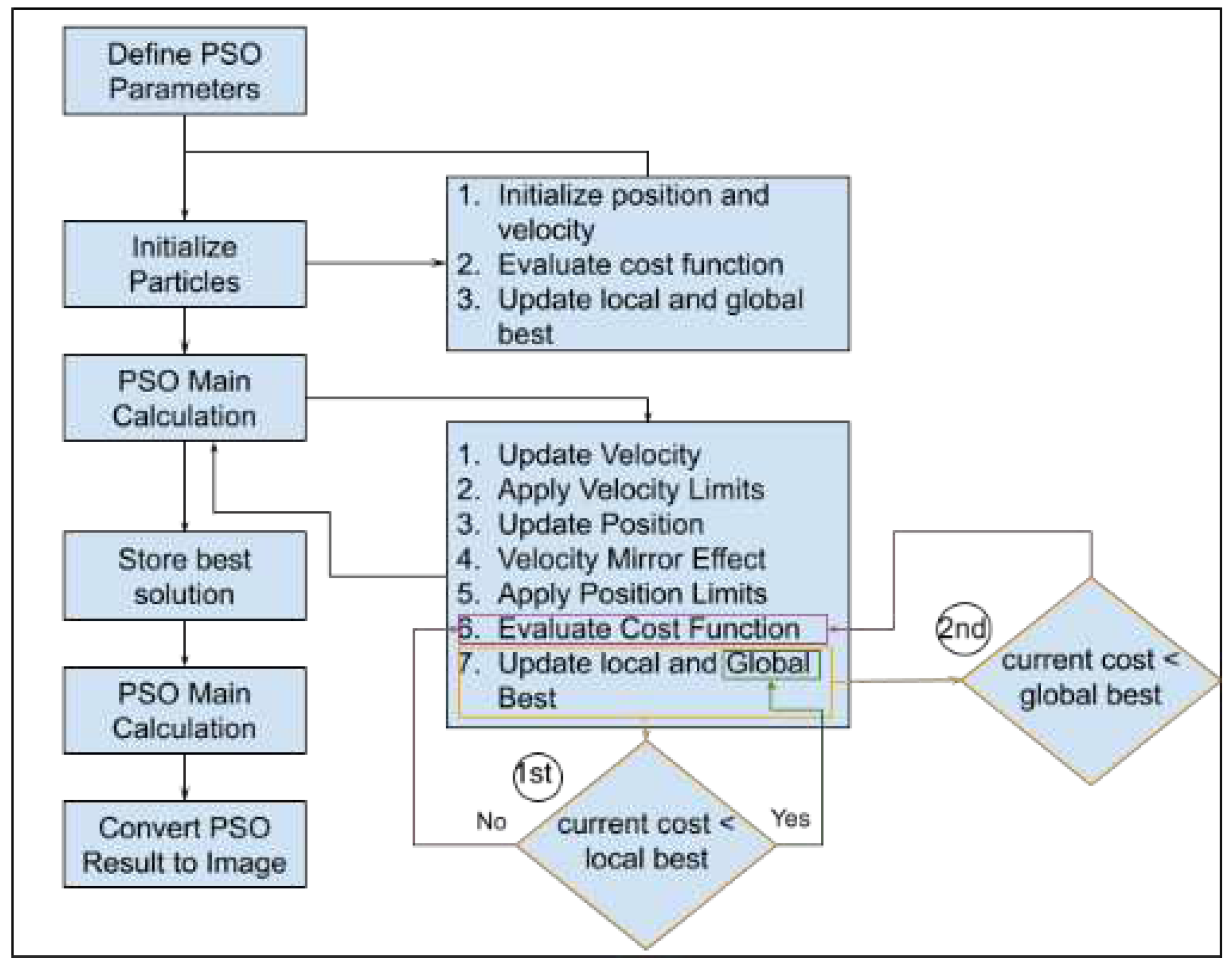
The core of our method lies in the utilization of Particle Swarm Optimization (PSO) to optimize the image segmentation process. The PSO algorithm is applied to partition the preprocessed image into distinct regions. The steps involved in PSO-based segmentation (
Figure 5) are as follows:
- a)
Particle Initialization: We initialize a swarm of particles, where each particle represents a potential segmentation solution. The particles are assigned positions in the search space, corresponding to potential image partitions. In the context of medical image segmentation, each partition represents a potential delineation of regions of interest within the image.
- b)
Objective Function: We define an objective function that quantifies the quality of a given image partition. The objective function considers factors such as intensity, gradient information, and region connectivity. The goal is to find the partition that minimizes this function, effectively identifying the optimal image segmentation.
- c)
Optimization Iterations: The PSO algorithm iteratively updates the positions of particles based on their previous best positions and the best positions found by neighboring particles. Particles adjust their positions in search of the optimal image partition. The optimization process continues until convergence criteria are met or a specified number of iterations is reached. Each particle keeps track of two pivotal fitness values: “local best (pbest)” and “global best (gbest).” “pbest” signifies the best value achieved by an individual particle throughout the optimization process, recalculated iteratively at each time step. In contrast, “global best” represents the overarching best value attained among all particles’ “pbest” values up to that specific time step.
- d)
Optimal Partition Extraction: The final result of the PSO optimization is the image partition that minimizes the objective function. This partition represents the segmentation of the input medical image into distinct regions of interest. The partition is chosen based on the collective behavior of particles, which adapt and explore the search space to find the best segmentation. Particle positions and velocities evolve dynamically based on the equations (
1) and (
2).
In Particle Swarm Optimization (PSO), key variables and parameters shape the optimization process [
27]. These include positions (
and
) and velocities
of particles, as well as essential factors like the inertia weight (
w), cognitive component (
), social component (
), and random values (
and
) within the range
. Additionally, PSO tracks two significant values:
, the best position found by a particle at time step
t, and
, the best position found across the entire swarm at time step
t. These elements collectively steer particle dynamics and the quest for optimal solutions.
The application of PSO in medical image segmentation offers several advantages, including its ability to handle non-convex and non-linear objective functions [
94], its simplicity of implementation, and its capacity to explore a diverse range of solutions. However, like other optimization techniques, PSO is sensitive to its parameter settings and may converge to local optima if not carefully tuned [
21,
56].
In our method, PSO plays a central role in optimizing the image segmentation process. It is seamlessly integrated with Histogram Equalization (HE), which enhances the quality of the segmented regions. This integration addresses the limitations associated with traditional segmentation methods and offers a novel and effective solution for the challenges associated with medical image segmentation. By incorporating PSO as a core component, our methods optimize the segmentation process, seeking the best partitioning of the input medical image. The synergy between PSO and HE enhances the accuracy and robustness of the segmentation results, ultimately improving the quality of the segmented regions and making them more suitable for subsequent analysis and clinical applications.
4. Results and Discussion
In this section, we present the results of our proposed method, which combines Particle Swarm Optimization (PSO) with Histogram Equalization (HE) to enhance medical image segmentation. We discuss the empirical findings, compare our approach with existing methods, and provide a comprehensive analysis of the outcomes.
4.1. Experimental Setup
In this section, we provide a comprehensive overview of the experimental setup used to evaluate the performance of the proposed method. A well-designed experimental setup is crucial for assessing the effectiveness and robustness of our approach.
4.1.1. Preprocessing Parameters
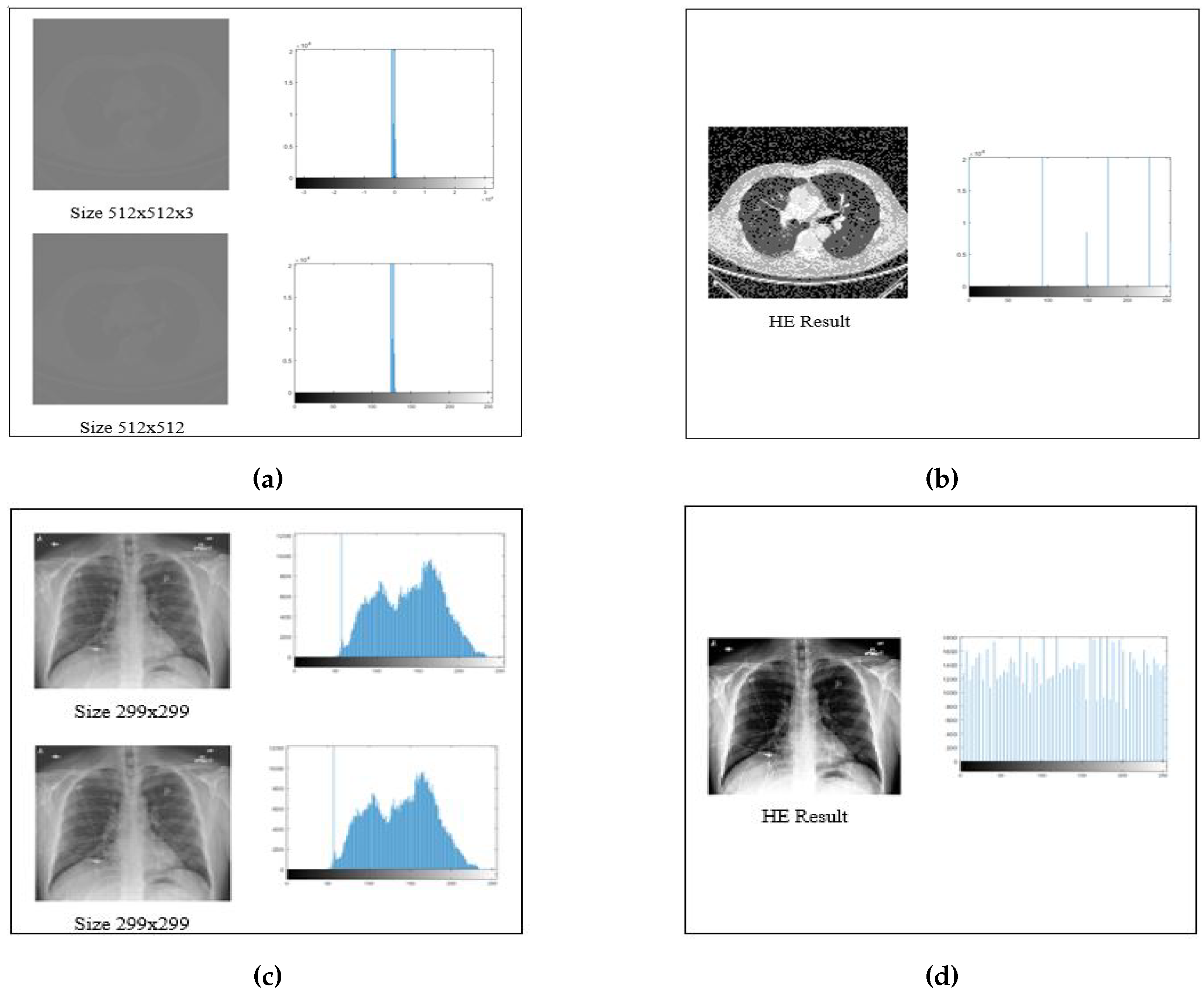
In the preparation of the datasets, a standardized set of preprocessing parameters was meticulously and consistently applied to all images. These preprocessing procedures encompassed the conversion of images into 8-bit format and grayscaling to ensure uniformity in the dataset. Additionally, Histogram Equalization (HE) and image adjustment parameters were meticulously calibrated to enhance the quality and suitability of images for subsequent segmentation and optimization processes.
The initial step of image conversion into 8-bit format was imperative due to the inherent variability within the dataset. This dataset consisted of images with both 8-bit and 16-bit depth. To maintain uniformity in image representation and facilitate consistent processing, all images were converted into 8-bit format. In the 8-bit representation, each pixel is defined within the range of 0–255, spanning across 256 levels, ensuring consistent data quality for subsequent processing.
Moreover, as some datasets featured images with three color channels (despite that their appearance was similar to grayscale), they were transformed into grayscale representations with a single color channel. This harmonization of image attributes across the dataset facilitated standardized preprocessing procedures, ensuring that all subsequent segmentation and optimization tasks were conducted on images optimized for analysis.
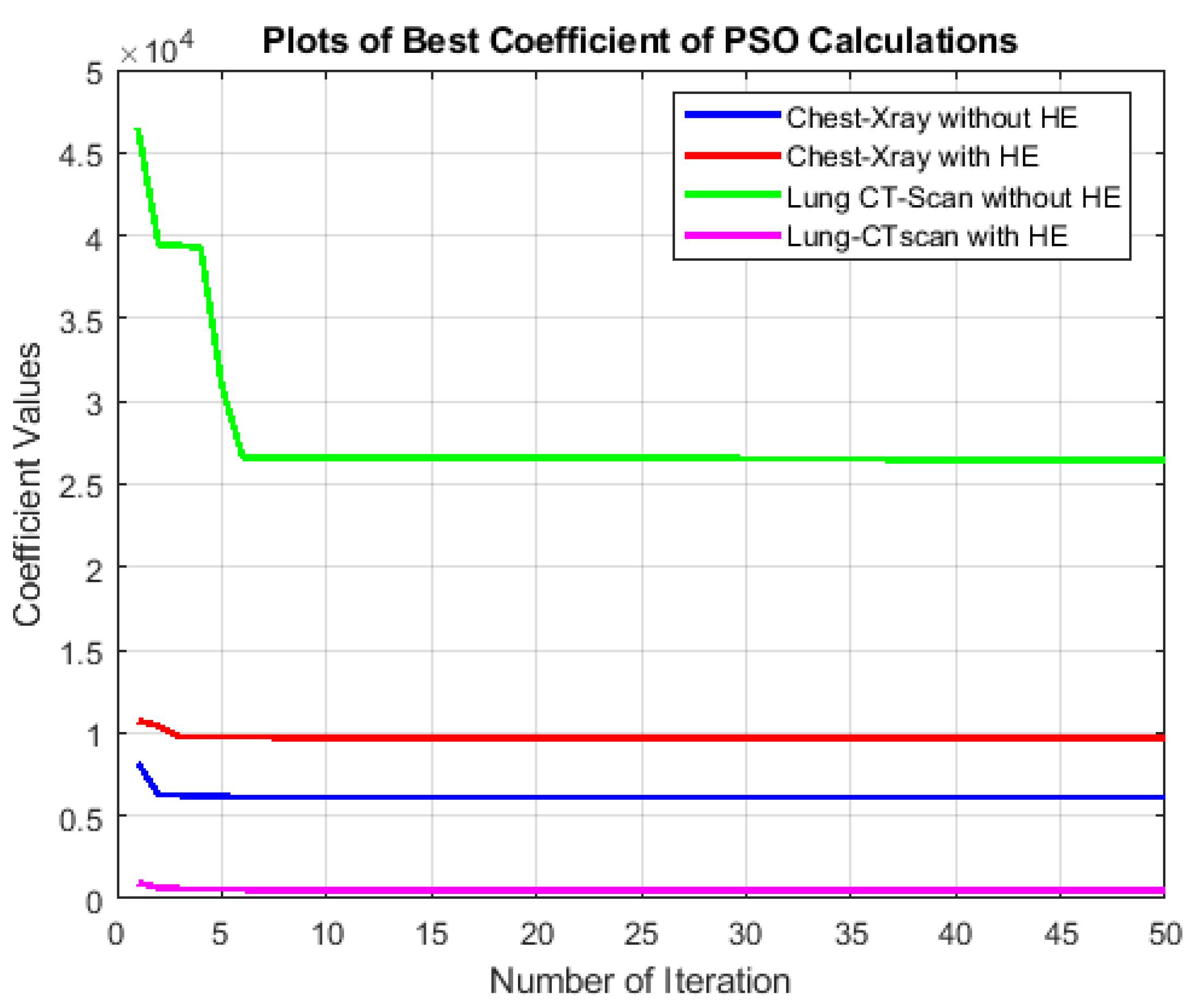
The effectiveness of these preprocessing measures can be visually appreciated in
Figure 6, which demonstrates the results of Histogram Equalization (HE) applied to Lung CT-Scan and Chest X-ray images along with their corresponding histogram distributions. This rigorous preprocessing lays the foundation for achieving consistent and high-quality results in the subsequent medical image segmentation processes, ultimately enhancing the accuracy and reliability of healthcare applications.
4.1.2. PSO-Segmentation Parameters
In our method, we tailor Particle Swarm Optimization (PSO) parameters to the specific characteristics of the datasets and the challenges associated with lung CT-Scans and Chest X-rays. The number of segments (k) is a pivotal parameter that determines how the images are partitioned, with k set to 2 segments in our experiments. The cost function (CostFunction) serves as the guiding force in the PSO optimization process, directing the partitioning of medical images based on the optimization objective.
The decision variables matrix size (VarSize) is specified as , providing insight into the size of the matrix involved in decision variables. Furthermore, the number of decision variables (nVar) is calculated by the product of the dimensions of VarSize, signifying the total number of decision variables considered in the optimization. To ensure that particles adhere to suitable ranges during optimization, we establish lower and upper bounds of variables (VarMin and VarMax) by considering the minimum and maximum values in the image data.
A series of critical PSO parameters influence the optimization process. The maximum number of iterations (MaxIt) is set to 50, thereby controlling the duration of the optimization procedure. Population size (nPop), which aligns with the swarm size in PSO, is set to 5. The inertia weight (w) is initially defined as 1, regulating the balance between a particle’s current velocity and its best-known position. An inertia weight damping ratio () of 0.99 dictates the rate at which the inertia weight diminishes over each iteration. Personal learning coefficient () and global learning coefficient () are assigned values of 1.5 and 2.5, respectively. These parameters govern the impact of a particle’s best-known position and the global best-known position on its movement within the optimization process.
Velocity limits (VelMax and VelMin) are instrumental in preserving the velocity of particles within acceptable boundaries. These limits play a pivotal role in ensuring that particle velocities do not exceed reasonable levels during optimization. By harmonizing these parameters, our method achieves a balance between exploration and exploitation, thereby seeking reliable solutions while avoiding overfitting to the training data.
4.1.3. Evaluation Metrics
In this section, we delve into the assessment of our Particle Swarm Optimization (PSO) method’s segmentation performance. To gauge the effectiveness of the PSO-based approach, we employ a range of fundamental evaluation metrics. These metrics provide us with a detailed perspective on the method’s segmentation precision, recall, and overall accuracy [
95].
Accuracy is a pivotal metric that measures the general precision of our segmentation method. It provides an insight into how well the PSO-based approach performs in accurately classifying regions of interest within the medical images.
Precision, on the other hand, enables us to assess the method’s ability to pinpoint and delineate regions of interest with precision and accuracy. It highlights the extent to which the approach can identify relevant regions while minimizing false positives.
Recall, also known as sensitivity, examines the method’s effectiveness in capturing all relevant regions within the medical images. It provides an indication of the method’s capacity to identify the entirety of regions that require attention.
F-Score offers a balanced perspective, taking into account both precision and recall. It serves as a combined metric to evaluate the method’s overall performance in achieving accurate and comprehensive segmentation.
Specificity evaluates the capability of our method to exclude non-relevant regions. It highlights the extent to which the method effectively filters out areas that are not of interest.
Additionally, we employ two overlap-based metrics: Dice Similarity Coefficient (DSC) and the Jaccard Index (JI). These metrics provide insights into the degree of overlap between the segmented regions and the ground truth data. They are crucial for assessing the method’s ability to align with reference standards.
These evaluation metrics can be expressed mathematically as follows:
This comprehensive array of evaluation metrics offers a holistic perspective on the PSO method’s segmentation quality and alignment with ground truth data. It allows us to scrutinize its precision, recall, and overall accuracy, thus facilitating a robust evaluation of its performance.
4.2. Segmentation Performance
Segmentation performance evaluation is a critical aspect of any medical image processing methodology. In this section, we delve into a comprehensive analysis of the Particle Swarm Optimization (PSO) approach’s performance when applied to the segmentation of two distinct medical imaging datasets: Lung CT-Scan and Chest X-ray. We examine the impact of preprocessing, specifically Histogram Equalization (HE), on the PSO algorithm’s performance, investigating its ability to enhance image quality and segmentation accuracy. Furthermore, we compare the outcomes of the PSO-Segmentation approach with alternative methods such as Otsu, Watershed, and K-means. To achieve a comprehensive evaluation, we consider both quantitative measures and graphical representations, enabling us to gain insights into the method’s strengths and limitations when applied to different medical imaging modalities.
The Best Cost values and evaluation metrics such as Accuracy, Precision, Recall, F-Score, Specificity, Dice, and Jaccard provide a multifaceted assessment of the segmentation quality. This analysis aims to shed light on the effectiveness of the PSO-Segmentation approach and the significance of selecting appropriate preprocessing techniques tailored to the specific characteristics of each imaging dataset.
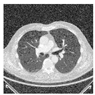
4.2.1. Analysis of Best Cost Values
The Best Cost values, as depicted in
Figure 7, offer valuable insights into the optimization process of the Particle Swarm Optimization (PSO) algorithm applied to medical image segmentation. We consider two distinct datasets, Lung CT-Scan and Chest X-ray, each with and without preprocessing using Histogram Equalization (HE). This analysis aims to understand how different combinations of the PSO algorithm and preprocessing methods influence the convergence behavior and efficiency of segmentation.
-
Chest X-ray Segmentation
Without HE, the Best Cost values exhibit fluctuations within a relatively narrow range. This indicates that the PSO algorithm converges effectively to a stable solution. The consistency in Best Cost values can be attributed to the nature of Chest X-ray images, which already possess a certain level of contrast and grayscale distribution suitable for segmentation. When HE is applied, the Best Cost values slightly increase. This is likely due to HE redistribution of pixel intensities and emphasizing the overall contrast, which can create additional complexity in the segmentation process. However, despite the slight increase, the Best Cost values remain relatively stable.
-
Lung CT-Scan Segmentation
In the case of Lung CT-Scan images, the Best Cost values demonstrate more significant variations. Without HE, the algorithm exhibits periodic dips and peaks. The complexity in these images, characterized by intricate anatomical structures like lung parenchyma, blood vessels, and airways, contributes to the periodic fluctuations. These variations might indicate the algorithm’s challenges in settling on a precise segmentation solution. With HE preprocessing, the Best Cost values exhibit a notable reduction in fluctuations. The redistribution of pixel intensities through HE improves the overall contrast, resulting in a smoother convergence pattern. It is important to note that, in this context, a lower Best Cost value reflects a more accurate segmentation.
The comparison between Chest X-ray and Lung CT-Scan datasets highlights the significance of dataset characteristics in influencing the convergence behavior of the PSO algorithm. Moreover, the impact of HE on the optimization process is more pronounced in Lung CT-Scan images, emphasizing the need for preprocessing methods tailored to the dataset’s characteristics.
4.2.2. Analysis of Evaluation Metrics
The detailed examination of the evaluation metrics in
Table 1 provides valuable insights into the performance of the PSO-Segmentation approach in comparison to alternative methods such as Otsu, Watershed, and K-means. These metrics offer a comprehensive understanding of the effectiveness and limitations of each approach in different scenarios, particularly when the Histogram Equalization (HE) technique is applied.
In CT-Lung images without HE, the PSO-Segmentation approach demonstrates notable performance, achieving an accuracy of 0.91893. This score suggests that it correctly identifies lung structures in these images, supported by a precision score of 0.90036, indicating a low rate of false positives. The recall score of 0.97545 emphasizes the ability to capture most relevant structures effectively. The F-Score, at 0.9364, strikes a balance between precision and recall. Furthermore, high Dice and Jaccard indices (0.9364 and 0.82984, respectively) reinforce the quality of segmentation.
The sensitivity score of 0.88041 highlights its capacity to identify true positives while maintaining a favorable balance with false positives. These results indicate the strength of the PSO-Segmentation approach in accurately segmenting CT-Lung images.
The integration of HE in CT-Lung images leads to even higher performance, with the accuracy of 0.9563 and the F-Score value of 0.967. This significant improvement can be attributed to the enhanced contrast and visibility in the images. The Dice and Jaccard indices also witness substantial gains, further underlining the effectiveness of HE. These results underscore the importance of employing HE as a preprocessing step to enhance segmentation accuracy, especially for CT-Lung images.
In Chest X-ray images without HE, the segmentation performance remains commendable, with an accuracy of 0.90335 and an F-Score of 0.90202. The precision score of 0.87424 suggests relatively few false positives. The recall score of 0.93162 indicates effective identification of relevant structures. However, the Dice and Jaccard indices slightly drop, reflecting some difficulty in delineating fine details. This can be attributed to the complex nature of Chest X-ray images.
The addition of HE to Chest-Xray images marginally improves segmentation performance, resulting in an accuracy of 0.90363 and the F-Score value of 0.90173. The precision score of 0.86891 implies that it maintains a low false positive rate, and the recall score of 0.93714 indicates effective identification of relevant structures. The Dice and Jaccard indices remain stable, with only slight improvements. It is worth noting that the complexity of Chest-Xray images presents inherent challenges for segmentation, which even HE cannot entirely mitigate.
When compared to Otsu’s segmentation approach, the PSO-Segmentation approach exhibits competitive performance. This highlights the strength of PSO in medical image segmentation. In contrast to the Watershed approach, the PSO-Segmentation approach consistently outperforms it. Watershed, known for over-segmentation issues, lags behind in terms of accuracy and F-Score. K-means, an alternative approach, shows comparable performance with the PSO-Segmentation approach in CT-Lung images without HE. However, HE significantly enhances K-means’ performance, making it a viable alternative. In Chest-Xray images, the PSO-Segmentation approach continues to perform more consistently.
The detailed analysis of evaluation metrics reveals the PSO-Segmentation approach’s proficiency, especially when HE is applied. The choice of using or not using HE should be tailored to the specific characteristics of the medical images. These insights offer guidance to practitioners in selecting suitable segmentation methods for various medical imaging scenarios. While these results are promising, further research may explore hybrid approaches that combine segmentation algorithms, preprocessing techniques, and machine learning for even more robust and accurate medical image segmentation.
4.3. Segmentation Results Based-on the Cropping Object
In this section, we focus on the segmentation results by specifically examining the cropping of detected objects and comparing them with their corresponding ground truth images (GT). We recognize the pivotal role played by preprocessing techniques in enhancing the visual quality of segmented images, and we aim to elucidate the profound influence of preprocessing on image clarity and precision. Furthermore, we conducted a comprehensive comparative assessment, evaluating our Particle Swarm Optimization (PSO) approach against alternative segmentation methods, including Otsu, Watershed, and K-means. This multifaceted analysis offers valuable insights into the quality of medical image segmentation achieved by our proposed approach.
4.3.1. Quality Enhancement with Preprocessing
Our exploration begins by emphasizing the critical role of preprocessing in shaping the quality of segmented images. As can be seen from
Table 2, images subjected to segmentation without any preprocessing closely resemble their original counterparts. However, the PSO segmentation process introduces scattered noise into the images, potentially obscuring vital image details and diminishing diagnostic accuracy. In contrast, images subjected to preprocessing, notably employing Histogram Equalization (HE) and our proposed adjustments, yield substantially improved segmentation outcomes. These preprocessed images exhibit greater clarity and more precise object boundaries.
The distinctive improvements in image quality, particularly when comparing images with and without HE preprocessing, underscore the significance of employing appropriate preprocessing methods to enhance the quality of medical image segmentation results. The sharper delineation of object boundaries, as demonstrated in segmented and cropped images, holds substantial value for medical practitioners, contributing to more precise diagnoses and well-informed clinical decisions.
4.3.2. Comparison with Other Segmentation Methods
Additionally, we conducted a comparative analysis between the PSO segmentation method and alternative segmentation techniques, such as Otsu, Watershed, and K-means. The findings are presented in
Table 3, and they showcase a substantial difference in the segmentation outcomes achieved by our proposed method compared to other techniques. The results suggest that the PSO segmentation approach, when coupled with preprocessing techniques, delivers images with superior clarity and accuracy.
In particular, the Dice Similarity Coefficient (DSC) and Jaccard Index values are highly indicative of the segmentation method’s performance. The values approaching 1 indicate a remarkable similarity between the segmented objects and their ground truth, demonstrating the high accuracy and precision of the proposed PSO approach. This reinforces the potential of our method for application in medical image segmentation, where precise delineation of anatomical structures is of paramount importance.
The results suggest that preprocessing techniques, especially HE and the proposed adjustments, contribute significantly to the quality of medical image segmentation. Moreover, the PSO segmentation method, when compared to conventional approaches, demonstrates its prowess in producing images with enhanced clarity and accuracy. This is particularly valuable in medical imaging scenarios, where precise segmentation is crucial for diagnosis and treatment planning.
5. Conclusions
In this study, we investigated the potential of Particle Swarm Optimization (PSO) as a medical image segmentation method, specifically focusing on Lung CT-Scan and Chest X-ray images. Our results underscore the significant role of preprocessing techniques, particularly Histogram Equalization (HE), in shaping the quality and precision of segmented images. HE, when applied, led to a notable improvement in accuracy and F-Score in Lung CT-Scan images, elevating the overall segmentation quality. Challenges encountered with Chest X-ray images, characterized by lower spatial resolution and the presence of overlapping structures, emphasize the importance of choosing the appropriate segmentation method, especially when HE is not employed. Here, certain alternative techniques demonstrate competitive performance, highlighting the need to consider image-specific characteristics in the selection process.
Comparative assessments against other segmentation methods, including Otsu, Watershed, and K-means, accentuate the ability of the PSO-Segmentation approach to generate images with superior clarity and accuracy, particularly when preprocessing is integrated. The Dice Similarity Coefficient (DSC) and Jaccard Index values approaching 1 indicate the high accuracy and precision of the proposed PSO method, signifying its potential for medical image segmentation tasks. In future work, the domain of medical image segmentation can benefit from further exploration and integration of machine learning and deep learning approaches. These methodologies, equipped with robust algorithms and vast datasets, have the potential to further enhance the accuracy and efficiency of medical image segmentation, providing invaluable support to medical practitioners and researchers in image analysis and diagnosis.
Author Contributions
Conceptualization, Shoffan Saifullah and Rafał Dreżewski; Data curation, Shoffan Saifullah; Formal analysis, Shoffan Saifullah and Rafał Dreżewski; Funding acquisition, Rafał Dreżewski; Investigation, Shoffan Saifullah; Methodology, Shoffan Saifullah; Project administration, Rafał Dreżewski; Resources, Shoffan Saifullah; Software, Shoffan Saifullah; Supervision, Rafał Dreżewski; Validation, Shoffan Saifullah and Rafał Dreżewski; Visualization, Shoffan Saifullah; Writing – original draft, Shoffan Saifullah and Rafał Dreżewski; Writing – review & editing, Shoffan Saifullah and Rafał Dreżewski. All authors have read and agreed to the published version of the manuscript.
Funding
The research presented in this paper was partially supported by the funds of Polish Ministry of Education and Science assigned to AGH University of Krakow.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Karagiannis, S.; Magkos, E.; Ntantogian, C.; Cabecinha, R.; Fotis, T. Cybersecurity and Medical Imaging: A Simulation-Based Approach to DICOM Communication. Applied Sciences 2023, 13, 10072. [Google Scholar] [CrossRef]
- Waili, A.R.A. Using Convolutional Neural Networks for Edge Detection in Medical Images to Determine Surgery Instrument Tools. Journal of Artificial Intelligence, Machine Learning and Neural Network 2023, 3, 13–25. [Google Scholar] [CrossRef]
- Hao, X.; Yin, L.; Li, X.; Zhang, L.; Yang, R. A Multi-Objective Semantic Segmentation Algorithm Based on Improved U-Net Networks. Remote Sensing 2023, 15, 1838. [Google Scholar] [CrossRef]
- Ansari, M.Y.; Abdalla, A.; Ansari, M.Y.; Ansari, M.I.; Malluhi, B.; Mohanty, S.; Mishra, S.; Singh, S.S.; Abinahed, J.; Al-Ansari, A.; Balakrishnan, S.; Dakua, S.P. Practical utility of liver segmentation methods in clinical surgeries and interventions. BMC Medical Imaging 2022, 22, 97. [Google Scholar] [CrossRef] [PubMed]
- Urban, R.; Haluzová, S.; Strunga, M.; Surovková, J.; Lifková, M.; Tomášik, J.; Thurzo, A. AI-Assisted CBCT Data Management in Modern Dental Practice: Benefits, Limitations and Innovations. Electronics 2023, 12, 1710. [Google Scholar] [CrossRef]
- Asiri, A.F.; Altuwalah, A.S. The role of neural artificial intelligence for diagnosis and treatment planning in endodontics: A qualitative review. The Saudi Dental Journal 2022, 34, 270–281. [Google Scholar] [CrossRef] [PubMed]
- Vobugari, N.; Raja, V.; Sethi, U.; Gandhi, K.; Raja, K.; Surani, S.R. Advancements in Oncology with Artificial Intelligence—A Review Article. Cancers 2022, 14, 1349. [Google Scholar] [CrossRef]
- KV, R.; Prasad, K.; Peralam Yegneswaran, P. Segmentation and Classification Approaches of Clinically Relevant Curvilinear Structures: A Review. Journal of Medical Systems 2023, 47, 40. [Google Scholar] [CrossRef]
- Iqbal, S.; Khan, T.M.; Naveed, K.; Naqvi, S.S.; Nawaz, S.J. Recent trends and advances in fundus image analysis: A review. Computers in Biology and Medicine 2022, 151, 106277. [Google Scholar] [CrossRef]
- Wu, R.; Liang, C.; Li, Y.; Shi, X.; Zhang, J.; Huang, H. Self-supervised transfer learning framework driven by visual attention for benign–malignant lung nodule classification on chest CT. Expert Systems with Applications 2023, 215, 119339. [Google Scholar] [CrossRef]
- Kaur, C.; Garg, U. Artificial intelligence techniques for cancer detection in medical image processing: A review. Materials Today: Proceedings 2023, 81, 806–809. [Google Scholar] [CrossRef]
- Liu, F.; Chen, L.; Lu, L.; Ahmad, A.; Jeon, G.; Yang, X. Medical image fusion method by using Laplacian pyramid and convolutional sparse representation. Concurrency and Computation: Practice and Experience 2020, 32. [Google Scholar] [CrossRef]
- Junn, J.C.; Soderlund, K.A.; Glastonbury, C.M. Imaging of Head and Neck Cancer With CT, MRI, and US. Seminars in Nuclear Medicine 2021, 51, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Dara, O.A.; Lopez-Guede, J.M.; Raheem, H.I.; Rahebi, J.; Zulueta, E.; Fernandez-Gamiz, U. Alzheimer’s Disease Diagnosis Using Machine Learning: A Survey. Applied Sciences 2023, 13, 8298. [Google Scholar] [CrossRef]
- Chakraborty, S.; Chatterjee, S.; Das, A.; Mali, K. Penalized Fuzzy C-Means Enabled Hybrid Region Growing in Segmenting Medical Images. Studies in Computational Intelligence 2020, 841, 41–65. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, K.; Kumar, G.; Swapna, K.; Datta, D.; Rajest, S. A Review of Medical Image Segmentation Algorithms. EAI Endorsed Transactions on Pervasive Health and Technology 2021, 7, 169184. [Google Scholar] [CrossRef]
- Oulefki, A.; Agaian, S.; Trongtirakul, T.; Kassah Laouar, A. Automatic COVID-19 lung infected region segmentation and measurement using CT-scans images. Pattern Recognition 2021, 114, 107747. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, F.; Dehmeshki, J.; Bisdas, S.; Roettger-Dupont, D.; Kubassova, O.; Aziz, M.; Awan, O. Artificial Intelligence-Based Clinical Decision Support Systems Using Advanced Medical Imaging and Radiomics. Current Problems in Diagnostic Radiology 2021, 50, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Mall, P.K.; Singh, P.K.; Srivastav, S.; Narayan, V.; Paprzycki, M.; Jaworska, T.; Ganzha, M. A comprehensive review of deep neural networks for medical image processing: Recent developments and future opportunities. Healthcare Analytics 2023, 4, 100216. [Google Scholar] [CrossRef]
- Faragallah, O.S.; El-Hoseny, H.; El-Shafai, W.; El-Rahman, W.A.; El-Sayed, H.S.; El-Rabaie, E.S.M.; El-Samie, F.E.A.; Geweid, G.G.N. A Comprehensive Survey Analysis for Present Solutions of Medical Image Fusion and Future Directions. IEEE Access 2021, 9, 11358–11371. [Google Scholar] [CrossRef]
- Shang, H.; Zhao, S.; Du, H.; Zhang, J.; Xing, W.; Shen, H. A new solution model for cardiac medical image segmentation. Journal of Thoracic Disease 2020, 12, 7298–7312. [Google Scholar] [CrossRef]
- Saifullah, S.; Dreżewski, R. Enhanced Medical Image Segmentation using CNN based on Histogram Equalization. 2023 2nd International Conference on Applied Artificial Intelligence and Computing (ICAAIC) 2023, pp. 121–126. [CrossRef]
- Allioui, H.; Sadgal, M.; Elfazziki, A. Optimized control for medical image segmentation: improved multi-agent systems agreements using Particle Swarm Optimization. Journal of Ambient Intelligence and Humanized Computing 2021, 12, 8867–8885. [Google Scholar] [CrossRef]
- El-Khatib, S.; Skobtsov, Y.; Rodzin, S. Improved Particle Swarm Medical Image Segmentation Algorithm for Decision Making. Studies in Computational Intelligence 2020, 869, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Eisham, Z.K.; Haque, M.M.; Rahman, M.S.; Nishat, M.M.; Faisal, F.; Islam, M.R. Chimp optimization algorithm in multilevel image thresholding and image clustering. Evolving Systems 2023, 14, 605–648. [Google Scholar] [CrossRef]
- Vijh, S.; Sharma, S.; Gaurav, P. Brain Tumor Segmentation Using OTSU Embedded Adaptive Particle Swarm Optimization Method and Convolutional Neural Network. Lecture Notes on Data Engineering and Communications Technologies 2020, 32, 171–194. [Google Scholar] [CrossRef]
- Shehanaz, S.; Daniel, E.; Guntur, S.R.; Satrasupalli, S. Optimum weighted multimodal medical image fusion using particle swarm optimization. Optik 2021, 231, 166413. [Google Scholar] [CrossRef]
- Saifullah, S.; Dreżewski, R. Modified Histogram Equalization for Improved CNN Medical Image Segmentation. Procedia Computer Science 2023, 225, 3021–3030, 27th International Conference on Knowledge Based and Intelligent Information and Engineering Sytems (KES 2023). [Google Scholar] [CrossRef]
- Lan, K.; Zhou, J.; Jiang, X.; Wang, J.; Huang, S.; Yang, J.; Song, Q.; Tang, R.; Gong, X.; Liu, K.; Wu, Y.; Li, T. Group theoretic particle swarm optimization for multi-level threshold lung cancer image segmentation. Quantitative Imaging in Medicine and Surgery 2023, 13, 1312–1322. [Google Scholar] [CrossRef]
- Naidu, S.; Quadros, A.; Natekar, A.; Parvatkar, P.; Chaman Kumar, K.; Aswale, S. Enhancement of X-ray images using various Image Processing Approaches. 2021 International Conference on Technological Advancements and Innovations (ICTAI). IEEE, 2021, pp. 115–120. [CrossRef]
- Elyan, E.; Vuttipittayamongkol, P.; Johnston, P.; Martin, K.; McPherson, K.; Moreno-García, C.F.; Jayne, C.; Mostafa Kamal Sarker, M. Computer vision and machine learning for medical image analysis: recent advances, challenges, and way forward. Artificial Intelligence Surgery 2022, 2, 24–25. [Google Scholar] [CrossRef]
- Abualigah, L.; Habash, M.; Hanandeh, E.S.; Hussein, A.M.; Shinwan, M.A.; Zitar, R.A.; Jia, H. Improved Reptile Search Algorithm by Salp Swarm Algorithm for Medical Image Segmentation. Journal of Bionic Engineering 2023, 20, 1766–1790. [Google Scholar] [CrossRef]
- Khaniabadi, S.M.; Ibrahim, H.; Huqqani, I.A.; Khaniabadi, F.M.; Sakim, H.A.M.; Teoh, S.S. Comparative Review on Traditional and Deep Learning Methods for Medical Image Segmentation. 2023 IEEE 14th Control and System Graduate Research Colloquium (ICSGRC) 2023, pp. 45–50. [CrossRef]
- Jardim, S.; António, J.; Mora, C. Image thresholding approaches for medical image segmentation - short literature review. Procedia Computer Science 2023, 219, 1485–1492. [Google Scholar] [CrossRef]
- Feng, Y.; Liu, Y.; Liu, Z.; Liu, W.; Yao, Q.; Zhang, X. A Novel Interval Iterative Multi-Thresholding Algorithm Based on Hybrid Spatial Filter and Region Growing for Medical Brain MR Images. Applied Sciences 2023, 13, 1087. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, Z.; Chen, S.; Qiu, C. Detect, Grow, Seg: A weakly supervision method for medical image segmentation based on bounding box. Biomedical Signal Processing and Control 2023, 86, 105158. [Google Scholar] [CrossRef]
- Jaglan, P.; Dass, R.; Duhan, M. A Comparative Analysis of Various Image Segmentation Techniques. Lecture Notes in Networks and Systems 2019, 46, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Sarhan, A.; Rokne, J.; Alhajj, R. Glaucoma detection using image processing techniques: A literature review. Computerized Medical Imaging and Graphics 2019, 78, 101657. [Google Scholar] [CrossRef] [PubMed]
- Bennai, M.T.; Guessoum, Z.; Mazouzi, S.; Cormier, S.; Mezghiche, M. A stochastic multi-agent approach for medical-image segmentation: Application to tumor segmentation in brain MR images. Artificial Intelligence in Medicine 2020, 110, 101980. [Google Scholar] [CrossRef]
- Biratu, E.S.; Schwenker, F.; Ayano, Y.M.; Debelee, T.G. A Survey of Brain Tumor Segmentation and Classification Algorithms. Journal of Imaging 2021, 7, 179. [Google Scholar] [CrossRef]
- Biratu, E.S.; Schwenker, F.; Debelee, T.G.; Kebede, S.R.; Negera, W.G.; Molla, H.T. Enhanced Region Growing for Brain Tumor MR Image Segmentation. Journal of Imaging 2021, 7, 22. [Google Scholar] [CrossRef]
- HM, N.; C, N.; VN, M.A. An approach for classification of lung nodules. Tumor Discovery 2023, 2, 317. [Google Scholar] [CrossRef]
- Azouz, Z.; Honarvar Shakibaei Asli, B.; Khan, M. Evolution of Crack Analysis in Structures Using Image Processing Technique: A Review. Electronics 2023, 12, 3862. [Google Scholar] [CrossRef]
- Kheradmandi, N.; Mehranfar, V. A critical review and comparative study on image segmentation-based techniques for pavement crack detection. Construction and Building Materials 2022, 321, 126162. [Google Scholar] [CrossRef]
- Aldoury, R.S.; Al-Saidi, N.M.; Ibrahim, R.W.; Kahtan, H. A new X-ray images enhancement method using a class of fractional differential equation. MethodsX 2023, 11, 102264. [Google Scholar] [CrossRef]
- Pradeep Kumar, B.P.; Rangaiah, P.K.B.; Augustine, R. Enhancing Medical Image Reclamation for Chest Samples using B-Coefficients, DT-CWT and EPS Algorithm. IEEE Access 2023, pp. 1–1. [CrossRef]
- Zebari, D.A.; Zeebaree, D.Q.; Abdulazeez, A.M.; Haron, H.; Hamed, H.N.A. Improved Threshold Based and Trainable Fully Automated Segmentation for Breast Cancer Boundary and Pectoral Muscle in Mammogram Images. IEEE Access 2020, 8, 203097–203116. [Google Scholar] [CrossRef]
- Zhang, W.; Zhuang, P.; Sun, H.H.; Li, G.; Kwong, S.; Li, C. Underwater Image Enhancement via Minimal Color Loss and Locally Adaptive Contrast Enhancement. IEEE Transactions on Image Processing 2022, 31, 3997–4010. [Google Scholar] [CrossRef]
- Majeed, S.H.; Isa, N.A.M. Adaptive Entropy Index Histogram Equalization for Poor Contrast Images. IEEE Access 2021, 9, 6402–6437. [Google Scholar] [CrossRef]
- Fan, X.; Sun, Z.; Tian, E.; Yin, Z.; Cao, G. Medical image contrast enhancement based on improved sparrow search algorithm. International Journal of Imaging Systems and Technology 2023, 33, 389–402. [Google Scholar] [CrossRef]
- Jawdekar, A.; Dixit, M. A Review of Image Enhancement Techniques in Medical Imaging. In In: Agrawal, S., Kumar Gupta, K., H. Chan, J., Agrawal, J., Gupta, M. (eds) Machine Intelligence and Smart Systems . Algorithms for Intelligent Systems; 2021; pp. 25–33. [CrossRef]
- Islam, S.M.; Mondal, H.S. Image Enhancement Based Medical Image Analysis. 2019 10th International Conference on Computing, Communication and Networking Technologies (ICCCNT). IEEE, 2019, pp. 1–5. [CrossRef]
- Wang, S.; Cao, G.; Wang, Y.; Liao, S.; Wang, Q.; Shi, J.; Li, C.; Shen, D. Review and Prospect: Artificial Intelligence in Advanced Medical Imaging. Frontiers in Radiology 2021, 1. [Google Scholar] [CrossRef] [PubMed]
- Farshi, T.R.; Drake, J.H.; Özcan, E. A multimodal particle swarm optimization-based approach for image segmentation. Expert Systems with Applications 2020, 149, 113233. [Google Scholar] [CrossRef]
- Shi, M.; Chen, C.; Liu, L.; Kuang, F.; Zhao, D.; Chen, X. A grade-based search adaptive random slime mould optimizer for lupus nephritis image segmentation. Computers in Biology and Medicine 2023, 160, 106950. [Google Scholar] [CrossRef]
- Khosla, T.; Verma, O.P. Optimal threshold selection for segmentation of Chest X-Ray images using opposition-based swarm-inspired algorithm for diagnosis of pneumonia. Multimedia Tools and Applications 2023. [Google Scholar] [CrossRef]
- Lakshman Narayana, V.; Lakshmi Patibandla, R.S.M.; Pavani, V.; Radhika, P. Optimized Nature-Inspired Computing Algorithms for Lung Disorder Detection. Studies in Computational Intelligence 2023, 1066, 103–118. [Google Scholar] [CrossRef]
- de Albuquerque, V.H.C.; Gupta, D.; De Falco, I.; Sannino, G.; Bouguila, N. Special issue on Bio-inspired optimization techniques for Biomedical Data Analysis: Methods and applications. Applied Soft Computing 2020, 95, 106672. [Google Scholar] [CrossRef]
- Shehab, M.; Abualigah, L.; Al Hamad, H.; Alabool, H.; Alshinwan, M.; Khasawneh, A.M. Moth–flame optimization algorithm: variants and applications. Neural Computing and Applications 2020, 32, 9859–9884. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, C.; Fu, Q.; Kou, R.; Huang, F.; Yang, B.; Yang, T.; Gao, M. Techniques and Challenges of Image Segmentation: A Review. Electronics 2023, 12, 1199. [Google Scholar] [CrossRef]
- Zhang, L.; Lim, C.P. Intelligent optic disc segmentation using improved particle swarm optimization and evolving ensemble models. Applied Soft Computing 2020, 92, 106328. [Google Scholar] [CrossRef]
- Mandave, D.D.; Patil, L.V. Bio-inspired computing algorithms in dementia diagnosis – a application-oriented review. Results in Control and Optimization 2023, 12, 100276. [Google Scholar] [CrossRef]
- Nayak, J.; Swapnarekha, H.; Naik, B.; Dhiman, G.; Vimal, S. 25 Years of Particle Swarm Optimization: Flourishing Voyage of Two Decades. Archives of Computational Methods in Engineering 2023, 30, 1663–1725. [Google Scholar] [CrossRef]
- Dhal, K.G.; Ray, S.; Das, A.; Das, S. A Survey on Nature-Inspired Optimization Algorithms and Their Application in Image Enhancement Domain. Archives of Computational Methods in Engineering 2019, 26, 1607–1638. [Google Scholar] [CrossRef]
- Kavitha, A.; Chellamuthu, C. Brain tumour detection using self-adaptive learning PSO-based feature selection algorithm in MRI images. International Journal of Business Intelligence and Data Mining 2019, 15, 71. [Google Scholar] [CrossRef]
- Sarvamangala, D.R.; Kulkarni, R.V. A Comparative Study of Bio-inspired Algorithms for Medical Image Registration. Studies in Computational Intelligence 2019, 687, 27–44. [Google Scholar] [CrossRef]
- Kate, V.; Shukla, P. Image Segmentation of Breast Cancer Histopathology Images Using PSO-Based Clustering Technique. Lecture Notes in Networks and Systems 2020, 100, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yu, X.; Wu, H.; Zhou, Y.; Sun, X.; Yu, S.; Yu, S.; Liu, H. A Fast 2-D Otsu lung tissue image segmentation algorithm based on improved PSO. Microprocessors and Microsystems 2021, 80, 103527. [Google Scholar] [CrossRef]
- Chakraborty, R.; Sushil, R.; Garg, M.L. An Improved PSO-Based Multilevel Image Segmentation Technique Using Minimum Cross-Entropy Thresholding. Arabian Journal for Science and Engineering 2019, 44, 3005–3020. [Google Scholar] [CrossRef]
- Öztürk, c.; Ahmad, R.; Akhtar, N. Variants of Artificial Bee Colony algorithm and its applications in medical image processing. Applied Soft Computing 2020, 97, 106799. [Google Scholar] [CrossRef]
- Guo, J.; Ma, J.; García-Fernández, Á.F.; Zhang, Y.; Liang, H. A survey on image enhancement for Low-light images. Heliyon 2023, 9, e14558. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhao, D.; Yu, F.; Heidari, A.A.; Ru, J.; Chen, H.; Mafarja, M.; Turabieh, H.; Pan, Z. Performance optimization of differential evolution with slime mould algorithm for multilevel breast cancer image segmentation. Computers in Biology and Medicine 2021, 138, 104910. [Google Scholar] [CrossRef]
- Alnazer, I.; Bourdon, P.; Urruty, T.; Falou, O.; Khalil, M.; Shahin, A.; Fernandez-Maloigne, C. Recent advances in medical image processing for the evaluation of chronic kidney disease. Medical Image Analysis 2021, 69, 101960. [Google Scholar] [CrossRef] [PubMed]
- Saini, M.; Susan, S. Tackling class imbalance in computer vision: a contemporary review. Artificial Intelligence Review 2023. [Google Scholar] [CrossRef]
- Huang, Q.; Ding, H.; Razmjooy, N. Oral cancer detection using convolutional neural network optimized by combined seagull optimization algorithm. Biomedical Signal Processing and Control 2024, 87, 105546. [Google Scholar] [CrossRef]
- Karsa, A.; Punwani, S.; Shmueli, K. An optimized and highly repeatable MRI acquisition and processing pipeline for quantitative susceptibility mapping in the head-and-neck region. Magnetic Resonance in Medicine 2020, 84, 3206–3222. [Google Scholar] [CrossRef]
- Hadjiiski, L.; Cha, K.; Chan, H.; Drukker, K.; Morra, L.; Näppi, J.J.; Sahiner, B.; Yoshida, H.; Chen, Q.; Deserno, T.M.; Greenspan, H.; Huisman, H.; Huo, Z.; Mazurchuk, R.; Petrick, N.; Regge, D.; Samala, R.; Summers, R.M.; Suzuki, K.; Tourassi, G.; Vergara, D.; Armato, S.G. AAPM task group report 273: Recommendations on best practices for AI and machine learning for computer-aided diagnosis in medical imaging. Medical Physics 2023, 50. [Google Scholar] [CrossRef] [PubMed]
- Maier-Hein, L.; Eisenmann, M.; Sarikaya, D.; März, K.; Collins, T.; Malpani, A.; Fallert, J.; Feussner, H.; Giannarou, S.; Mascagni, P.; Nakawala, H.; Park, A.; Pugh, C.; Stoyanov, D.; Vedula, S.S.; Cleary, K.; Fichtinger, G.; Forestier, G.; Gibaud, B.; Grantcharov, T.; Hashizume, M.; Heckmann-Nötzel, D.; Kenngott, H.G.; Kikinis, R.; Mündermann, L.; Navab, N.; Onogur, S.; Roß, T.; Sznitman, R.; Taylor, R.H.; Tizabi, M.D.; Wagner, M.; Hager, G.D.; Neumuth, T.; Padoy, N.; Collins, J.; Gockel, I.; Goedeke, J.; Hashimoto, D.A.; Joyeux, L.; Lam, K.; Leff, D.R.; Madani, A.; Marcus, H.J.; Meireles, O.; Seitel, A.; Teber, D.; Ückert, F.; Müller-Stich, B.P.; Jannin, P.; Speidel, S. Surgical data science – from concepts toward clinical translation. Medical Image Analysis 2022, 76, 102306. [Google Scholar] [CrossRef]
- K Scott Mader. Finding and Measuring Lungs in CT Data, 2017.
- Rahman, T.; Chowdhury, M.; Khandakar, A. COVID-19 Radiography Database, 2022.
- Song, Y.; Ren, S.; Lu, Y.; Fu, X.; Wong, K.K. Deep learning-based automatic segmentation of images in cardiac radiography: A promising challenge. Computer Methods and Programs in Biomedicine 2022, 220, 106821. [Google Scholar] [CrossRef] [PubMed]
- Saifullah, S.; Drezewski, R.; Khaliduzzaman, A.; Tolentino, L.K.; Ilyos, R. K-Means Segmentation Based-on Lab Color Space for Embryo Detection in Incubated Egg. Jurnal Ilmiah Teknik Elektro Komputer dan Informatika 2022, 8, 175–185. [Google Scholar] [CrossRef]
- Masoudi, S.; Harmon, S.A.; Mehralivand, S.; Walker, S.M.; Raviprakash, H.; Bagci, U.; Choyke, P.L.; Turkbey, B. Quick guide on radiology image pre-processing for deep learning applications in prostate cancer research. Journal of Medical Imaging 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Aumann, S.; Donner, S.; Fischer, J.; Müller, F. Optical Coherence Tomography (OCT): Principle and Technical Realization. High Resolution Imaging in Microscopy and Ophthalmology 2019, pp. 59–85. [CrossRef]
- El-Shafai, W.; Almomani, I.; Ara, A.; Alkhayer, A. An optical-based encryption and authentication algorithm for color and grayscale medical images. Multimedia Tools and Applications 2023, 82, 23735–23770. [Google Scholar] [CrossRef]
- Hoque, M.Z.; Keskinarkaus, A.; Nyberg, P.; Seppänen, T. Stain normalization methods for histopathology image analysis: A comprehensive review and experimental comparison. Information Fusion 2024, 102, 101997. [Google Scholar] [CrossRef]
- Nazir, N.; Sarwar, A.; Saini, B.S.; Shams, R. A Robust Deep Learning Approach for Accurate Segmentation of Cytoplasm and Nucleus in Noisy Pap Smear Images. Computation 2023, 11, 195. [Google Scholar] [CrossRef]
- Saifullah, S.; Drezewski, R.; Yudhana, A.; Pranolo, A.; Kaswijanti, W.; Suryotomo, A.P.; Putra, S.A.; Khaliduzzaman, A.; Prabuwono, A.S.; Japkowicz, N. Nondestructive chicken egg fertility detection using CNN-transfer learning algorithms. Jurnal Ilmiah Teknik Elektro Komputer dan Informatika (JITEKI) 2023, 9, 854–871. [Google Scholar] [CrossRef]
- Saifullah, S.; Dreżewski, R. Non-Destructive Egg Fertility Detection in Incubation Using SVM Classifier Based on GLCM Parameters. Procedia Computer Science 2022, 207, 3254–3263, Knowledge-Based and Intelligent Information & Engineering Systems: Proceedings of the 26th International Conference KES2022. [Google Scholar] [CrossRef]
- Okwu, M.O.; Tartibu, L.K. Particle Swarm Optimisation. In: Metaheuristic Optimization: Nature-Inspired Algorithms Swarm and Computational Intelligence, Theory and Applications. Studies in Computational Intelligence 2021, 927, 5–13. [CrossRef]
- Gad, A.G. Particle Swarm Optimization Algorithm and Its Applications: A Systematic Review. Archives of Computational Methods in Engineering 2022, 29, 2531–2561. [Google Scholar] [CrossRef]
- Djemame, S.; Batouche, M.; Oulhadj, H.; Siarry, P. Solving reverse emergence with quantum PSO application to image processing. Soft Computing 2019, 23, 6921–6935. [Google Scholar] [CrossRef]
- Narayan, V.; Faiz, M.; Mall, P.K.; Srivastava, S. A Comprehensive Review of Various Approach for Medical Image Segmentation and Disease Prediction. Wireless Personal Communications 2023, 132, 1819–1848. [Google Scholar] [CrossRef]
- Papazoglou, G.; Biskas, P. Review and Comparison of Genetic Algorithm and Particle Swarm Optimization in the Optimal Power Flow Problem. Energies 2023, 16, 1152. [Google Scholar] [CrossRef]
- Taha, A.A.; Hanbury, A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Medical Imaging 2015, 15, 29. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).