Submitted:
26 December 2023
Posted:
26 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
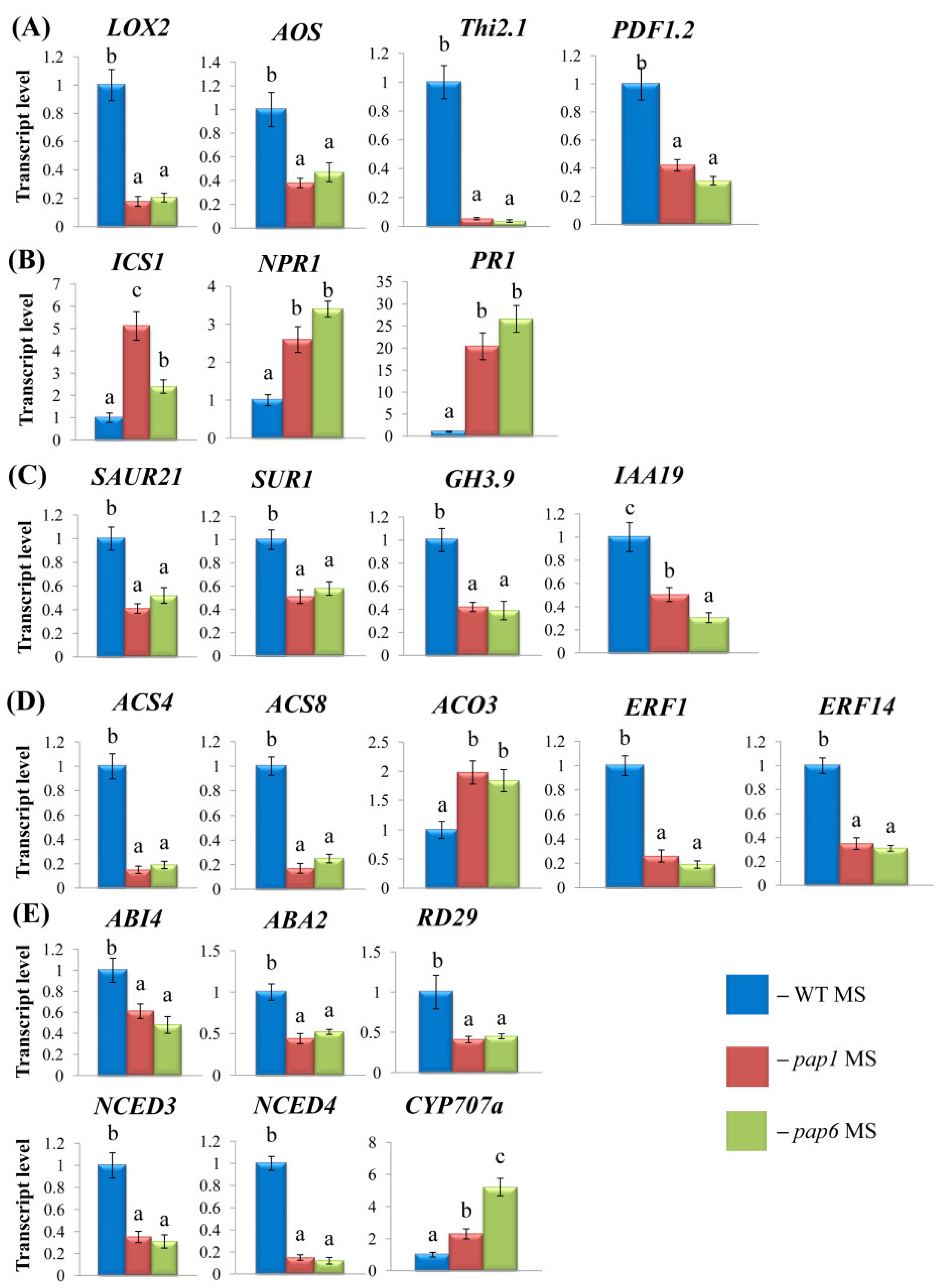
2.1. Disruption of pap genes contributes to altered hormone levels
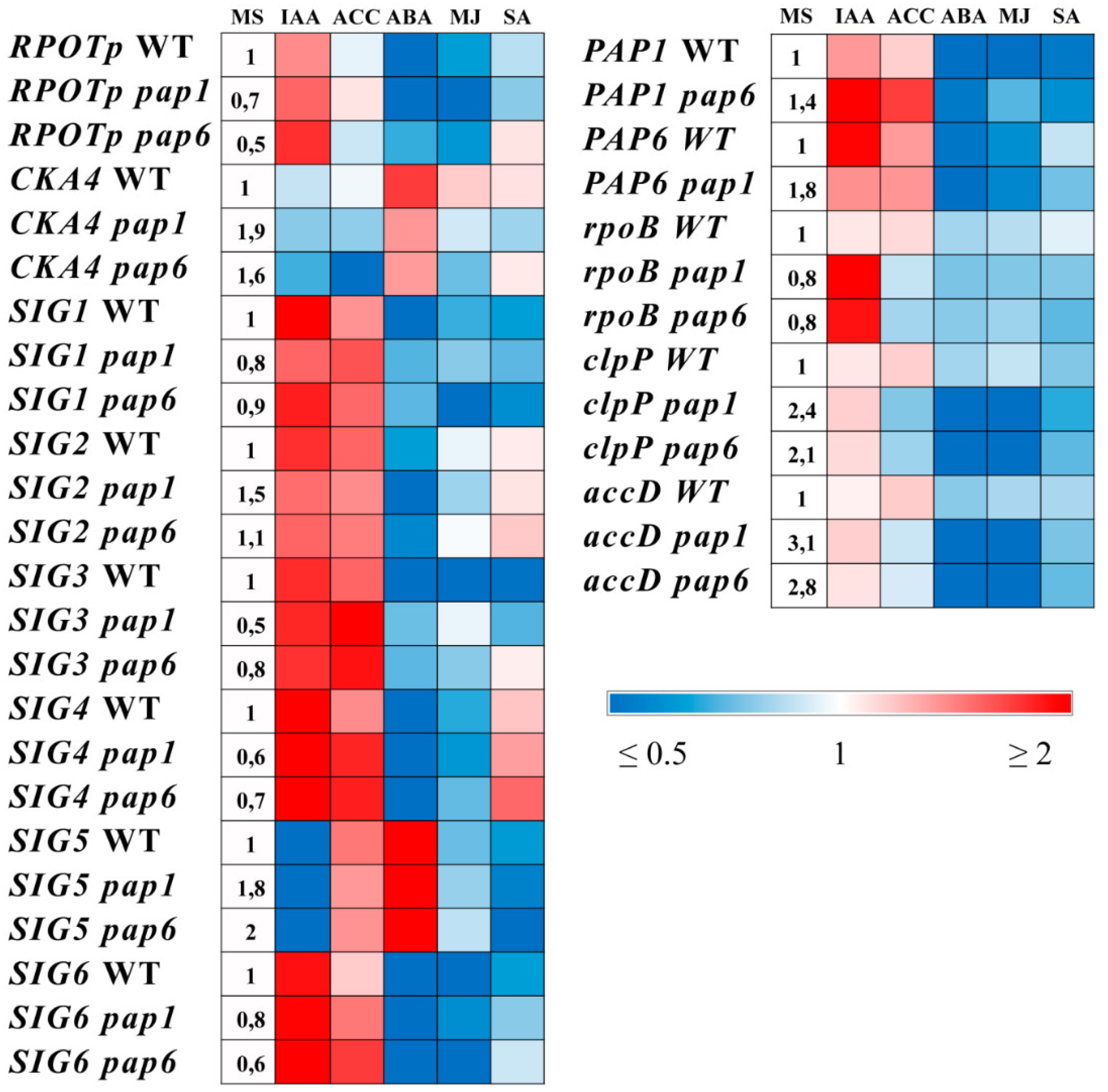
The expression of genes involved in hormone metabolism is altered in pap mutants
2.1.1. JA and SA
2.1.2. IAA
2.1.3. Ethylene and ABA
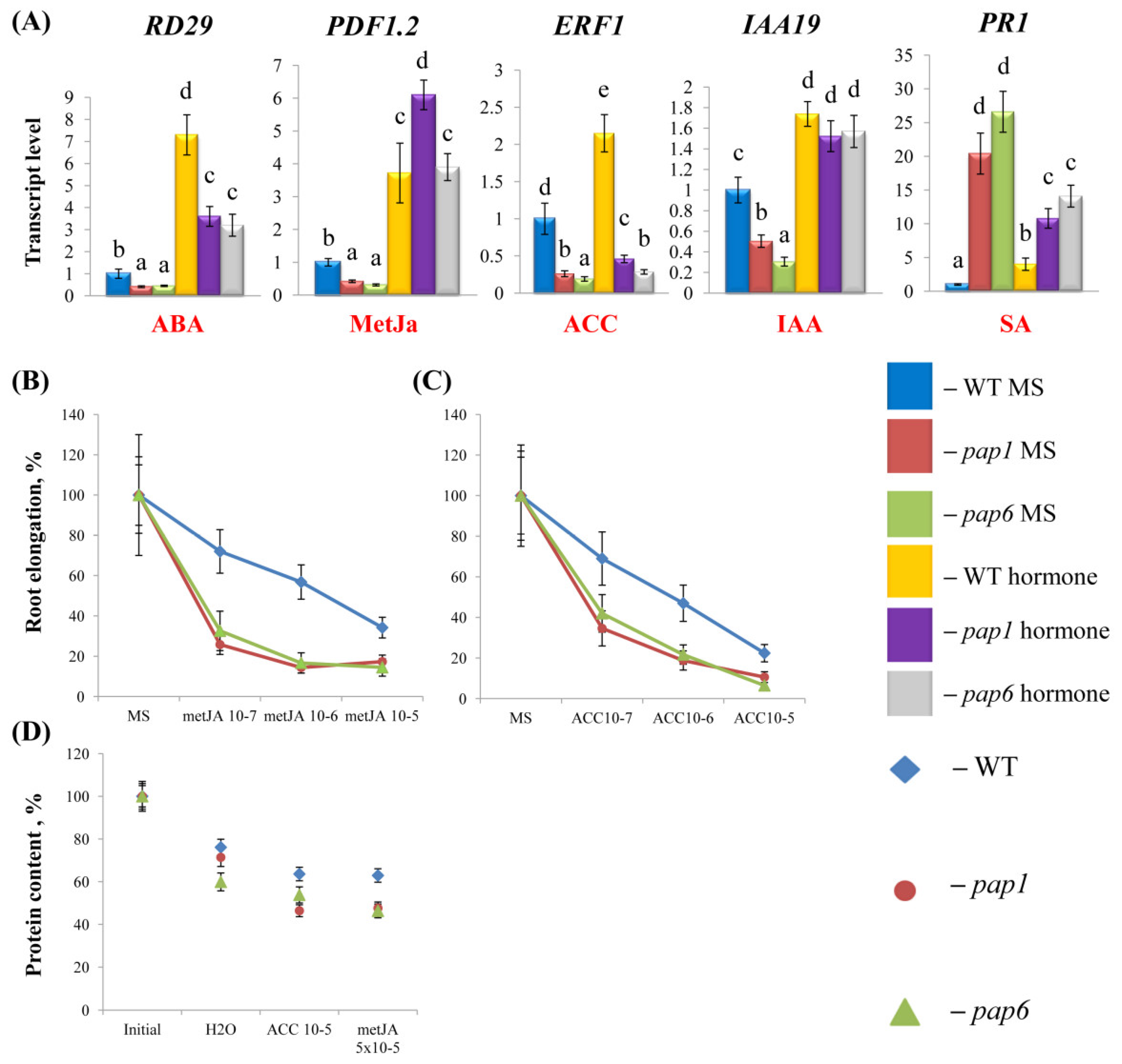
2.2. Pap mutants exhibited partially altered responses to hormone treatments
3. Discussion
4. Materials and methods
4.1. Plant material, growth conditions and hormone treatment
4.2. Hormone Extraction, Purification, and Determination
4.3. Quantitative RT‒PCR
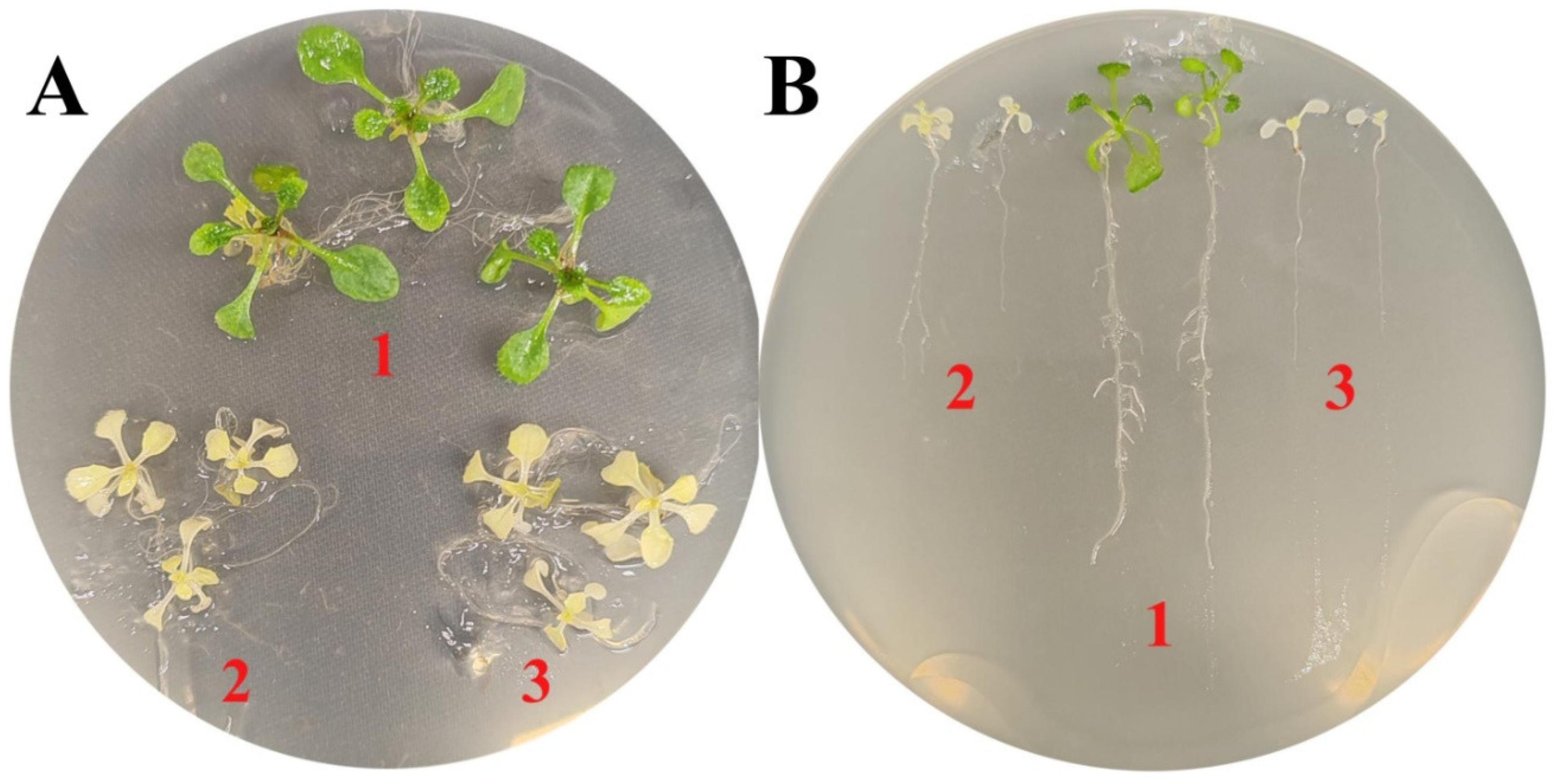
4.4. Growth sensitivity assays
4.5. Statistical Data Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Robles, P.; Quesada, V. Research Progress in the Molecular Functions of Plant mTERF Proteins. Cells. 2021, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Börner, T.; Aleynikova., A.Y.; Zubo, Y.O.; Kusnetsov, V.V. Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim. Biophys. Acta 2015, 1847, 761–769. [Google Scholar] [CrossRef]
- Liebers, M.; Pfannschmidt, T. Plastid RNA polymerases and nuclear-encoded proteins associated with them in Arabidopsis thaliana. Endocytobiosis and Cell Research 2017, 28, 20–32. [Google Scholar]
- Pfannschmidt, T.; Blanvillain, R.; Merendino, L.; Courtois, F.; Chevalier, F.; Liebers, M.; Lerbs-Mache, S. Plastid RNA polymerases: orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J. Exp. Bot. 2015, 66, 6957–6973. [Google Scholar] [CrossRef] [PubMed]
- Grübler, B.; Merendino, L.; Twardziok, S. O.; Mininno, M.; Allorent, G.; Chevalier, F.; Ravanel, S. Light and plastid signals regulate different sets of genes in the albino mutant pap7-1. Plant Physiol. 2017, 175, 1203–1219. [Google Scholar] [CrossRef]
- Grübler, B.; Cozzi, C.; Pfannschmidt, T. A Core Module of Nuclear Genes Regulated by Biogenic Retrograde Signals from Plastids. Plants 2021, 10, 296. [Google Scholar] [CrossRef] [PubMed]
- Danilova, M.N.; Andreeva, A.A.; Doroshenko, A.S.; Kudryakova, N.V.; Kuznetsov, Vl. V.; Kusnetsov, V.V. Phytohormones Regulate the Expression of Nuclear Genes Encoding the Components of the Plastid Transcription Apparatus. Doklady Biochemistry and Biophysics 2018, 478, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, A.A.; Vankova, R.; Bychkov, I.A.; Kudryakova, N.V.; Danilova, M.N.; Lacek, J.; Pojidaeva, E.S.; Kusnetsov, V.V. Cytokinin-Regulated Expression of Arabidopsis thaliana PAP Genes and Its Implication for the Expression of Chloroplast-Encoded Genes. Biomolecules 2020, 10, 1658. [Google Scholar] [CrossRef] [PubMed]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef]
- Yagi, Y.; Ishizaki, Y.; Nakahira, Y.; Tozawa, Y.; Shiina, T. Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proc. Natl. Acad. Sci. USA 2012, 109, 7541–7546. [Google Scholar] [CrossRef]
- Arsova, B.; Hoja, U.; Wimmelbacher, M.; Greiner, E.; Üstün, Ş.; Melzer, M.; Börnke, F. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 2010, 22, 1498–1515. [Google Scholar] [CrossRef] [PubMed]
- Gilkerson, J.; Perez-Ruiz, J. M.; Chory, J.; Callis, J. The plastid-localized pfkB-type carbohydrate kinases FRUCTOKINASE-LIKE 1 and 2 are essential for growth and development of Arabidopsis thaliana. BMC Plant Biology 2012, 12, 1–17. [Google Scholar] [CrossRef]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Bandurski, R.S. Chemistry and Physiology of the bound auxin. Ann. Rev. Plant Physiol. 1982, 33, 403–430. [Google Scholar] [CrossRef]
- Leon-Reyes, A.; Van der Does, D.; De Lange, E.S.; Delker, C.; Wasternack, C.; Van Wees, S.C.; Ritsema, T.; Pieterse, C.M. Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 2010, 232, 1423–1432. [Google Scholar] [CrossRef]
- Gawroński, P.; Górecka, M.; Bederska, M.; Rusaczonek, A.; Slesak, I.; Kruk, J.; Karpiński, S. Isochorismate synthase 1 is required for thylakoid organization, optimal plastoquinone redox status, and state transitions in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 3669–3679. [Google Scholar] [CrossRef]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L-J. Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: DoWe Understand What They Are Whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y; Zhang, D.; Yan Chu, J.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Despre´ s, C. The Arabidopsis NPR1 Protein Is a Receptor for the Plant Defense Hormone Salicylic Acid. Cell Reports 2012, 1, 639–647. [Google Scholar] [CrossRef]
- Zhong, Q.; Hu, H.; Fan, B.; Zhu, C.; Chen, Z. Biosynthesis and Roles of Salicylic Acid in Balancing Stress Response and Growth in Plants. Int. J. Mol. Sci. 2021, 22, 11672. [Google Scholar] [CrossRef]
- Wang, D.; Pajerowska-Mukhtar, K.; Culler, A.H.; Dong, X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 2007, 17, 1784–1790. [Google Scholar] [CrossRef]
- Kong, W.; Li, Y.; Zhang, M.; Jin, F.; Li, J. A Novel Arabidopsis MicroRNA Promotes IAA Biosynthesis via the Indole-3-acetaldoxime Pathway by Suppressing SUPERROOT1. Plant Cell Physiol. 2015, 56, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Hu, Y.; Aoi, Y.; Hira, H.; Ge, C.; Dai, X.; Kasahara, H.; Zhao, Y. Local conjugation of auxin by the GH3 amido synthetases is required for normal development of roots and flowers in Arabidopsis. Biochem. Biophys. Res. Commun. 2022, 589, 16e22. [Google Scholar] [CrossRef] [PubMed]
- Pattyn, J.; Vaughan-Hirsch, J.; Van de Poel, B. The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytol. 2021, 229, 770–782. [Google Scholar] [CrossRef]
- Oñate-Sánchez, L.; Anderson, J.P.; Young, J.; Singh., K.B. AtERF14, a Member of the ERF Family of Transcription Factors, Plays a Nonredundant Role in Plant Defense. Plant Physiol. 2007, 143, 400–409. [Google Scholar] [CrossRef]
- Cheng, M-C.; Liao, P-M.; Kuo, W‒W.; Lin, T-P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 Regulates Abiotic Stress-Responsive Gene Expression by Binding to Different cis-Acting Elements in Response to Different Stress Signals. Plant Physiol. 2013, 162, 1566–1582. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Yu, Y.; Li, S.; Wang, J.; Tang, S.; Huang, R. Abscisic Acid Antagonizes Ethylene Production through the ABI4-Mediated Transcriptional Repression of ACS4 and ACS8 in Arabidopsis. Mol. Plant. 2016, 9, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Roychoudhury, A. Abscisic acid in plants under abiotic stress: crosstalk with major phytohormones. Plant Cell Rep. 2023, 42, 961–974. [Google Scholar] [CrossRef]
- Danilova, M.N.; Kudryakova, N.V.; Doroshenko, A.S.; Zabrodin, D.A.; Rakhmankulova, Z.F.; Oelmüller, R.; Kusnetsov, V.V. Opposite roles of the Arabidopsis cytokinin receptors AHK2 and AHK3 in the expression of plastid genes and genes for the plastid transcriptional machinery during senescence. Plant Mol. Biol. 2017, 93, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chang, H.P.; Hu, S.; Lu, X.T.; Yuan, C.Y.; Zhang, C.; Wang, P.; Xiao, W.J.; Xiao, L.T.; Xue, G.P.; Guo, X. Plastid casein kinase 2 knockout reduces abscisic acid (ABA) sensitivity, thermotolerance, and expression of ABA and heat stress responsive nuclear genes. J. Exp. Bot. 2014, 65, 4159–4175. [Google Scholar] [CrossRef]
- Yamburenko, M.V.; Zubo, Y.O.; Börner, T. Abscisic acid affects transcription of chloroplast genes via protein phosphatase 2C-dependent activation of nuclear genes: repression by guanosine-3′-5′-bisdiphosphate and activation by sigma factor 5. Plant J. 2015, 82, 1030–1041. [Google Scholar] [CrossRef]
- Aluru, M.R.; Zola, J.; Foudree, A.; Rodermel, S.R. Chloroplast Photooxidation-Induced Transcriptome Reprogramming in Arabidopsis immutans White Leaf Sectors. Plant Physiol. 2009, 150, 904–923. [Google Scholar] [CrossRef]
- Pogorelko, G.V.; Kambakam, S.; Nolan, T.; Foudree, A.; Zabotina, O.A.; Rodermel, S.R. Impaired Chloroplast Biogenesis in Immutans, an Arabidopsis Variegation Mutant, Modifies Developmental Programming, Cell Wall Composition and Resistance to Pseudomonas syringae. PLoS ONE 2016, 1, e0150983. [Google Scholar] [CrossRef] [PubMed]
- Hirosawa, Y; Tada, A.; Matsuura, T.; Mori, I.C.; Ogura, Y.; Hayashi, T.; Uehara, S.; Ito-Inaba, Y.; Inaba, T. Salicylic Acid Acts Antagonistically to Plastid Retrograde Signaling by Promoting the Accumulation of Photosynthesis-associated Proteins in Arabidopsis. Plant Cell Physiol. 2021, 62, 1728–1744. [Google Scholar] [CrossRef]
- Kurepin, L.V.; Ivanov, A.G.; Zaman, M.; Pharis, R.P.; Allakhverdiev, S.I.; Hurry, V.; Hüner, P.A. Stress-related hormones and glycinebetaine interplayin protection of photosynthesis under abiotic stress conditions. Photosyn. Res. 2015, 126, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Q.; Hu, H.; Fan, B.; Zhu, C.; Chen, Z. Biosynthesis and Roles of Salicylic Acid in Balancing Stress Response and Growth in Plants. Int. J. Mol. Sci. 2021, 22, 11672. [Google Scholar] [CrossRef]
- Choi, J.; Huh, S.U.; Mikiko Kojima, M.; Sakakibara, H.; Paek, K-H.; Hwang, I. The Cytokinin-Activated Transcription Factor ARR2 Promotes Plant Immunity via TGA3/NPR1-Dependent Salicylic Acid Signaling in Arabidopsis Developmental. Cell 2010, 19, 284–295. [Google Scholar] [CrossRef]
- Mothes, K.; Engelbrecht, L.; Schűtte, H.R. Ǘber den Akkumulationvon α- minoisobuttersaure im Blatgewebe unter dem Einfluss von Kinetin. Physiol Plantarum 1961, 14, 72–76. [Google Scholar] [CrossRef]
- Mothes, K.; Engelbrecht, L. On the activity of a kinetin-like root factor. Life Sci. 1963, 11, 852–857. [Google Scholar] [CrossRef]
- Dobrev, P. I.; Kamınek, M. Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J. Chromatogr. A 2002, 950, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Dobrev, P. I.; Vankova, R. Quantification of abscisic acid, cytokinin, and auxin content in salt-stressed plant tissues. Methods Mol. Biol. 2012, 913, 251–261. [Google Scholar] [CrossRef]
- Bychkov, I.A.; Andreeva, A.A.; Kudryakova, N.V.; Pojidaeva, E.S.; Kusnetsov, V.V. The role of PAP4/FSD3 and PAP9/FSD2 in heat stress responses of chloroplast genes. Plant Sci. 2022, 322, 111359. [Google Scholar] [CrossRef] [PubMed]
| Phytohormones | WT | pap1 | pap6 |
|---|---|---|---|
| ABA | 358.91±72.24 | 178.43±9.84* | 168.73±20.68* |
| PA | 84.39±12.01 | 14.64±2.62** | 6.73±2.16** |
| 9OH-ABA | 15.01±2.8546 | 2.70±0.46** | 1.14±1.14** |
| DPA | 31.22±7.04 | 29.49±7.02 | 18.76±3.91 |
| BzA | 655.04±61.87 | 1092.02±146.66* | 1160.01±90.71* |
| SA | 882.35±182.37 | 2588.49. ±498.01 | 3264.72±897.05* |
| JA | 151.58±29.71 | 145.89±30.01 | 140.03±13.77 |
| JA-Ileu | 16.31±3.27 | 9.3225±3.03 | 14.17±3.50 |
| cisOPDA | 137.52±7.42 | 94.85±24.90 | 58.77±13.50* |
| ACC | 338.17±86.45 | 11.00±2.63** | 34.28±7.05** |
| IAA | 67.44±2.89 | 64.63±6.54 | 66.74±4.22 |
| OxIAA-GE | 922.21±129.18 | 1461.79±151.29* | 1232.99±119.36 |
| PAA | 98.06±8.44 | 133.42±21.69 | 162.57±18.38 |
| IAA-Asp | 56.51±17.94 | 5.80±1.12* | 12.76±2.35* |
| IAA-GE | 20.11±1.91 | 33.34±5.95 | 32.02±5.82 |
| OxIAA | 248.51±38.26 | 312.15±47.86 | 326.04±26.21 |
| IAN | 4284.60 ±1411.27 | 5577.52±833.42 | 6208.17±2741.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





