Submitted:
23 March 2024
Posted:
26 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and Method
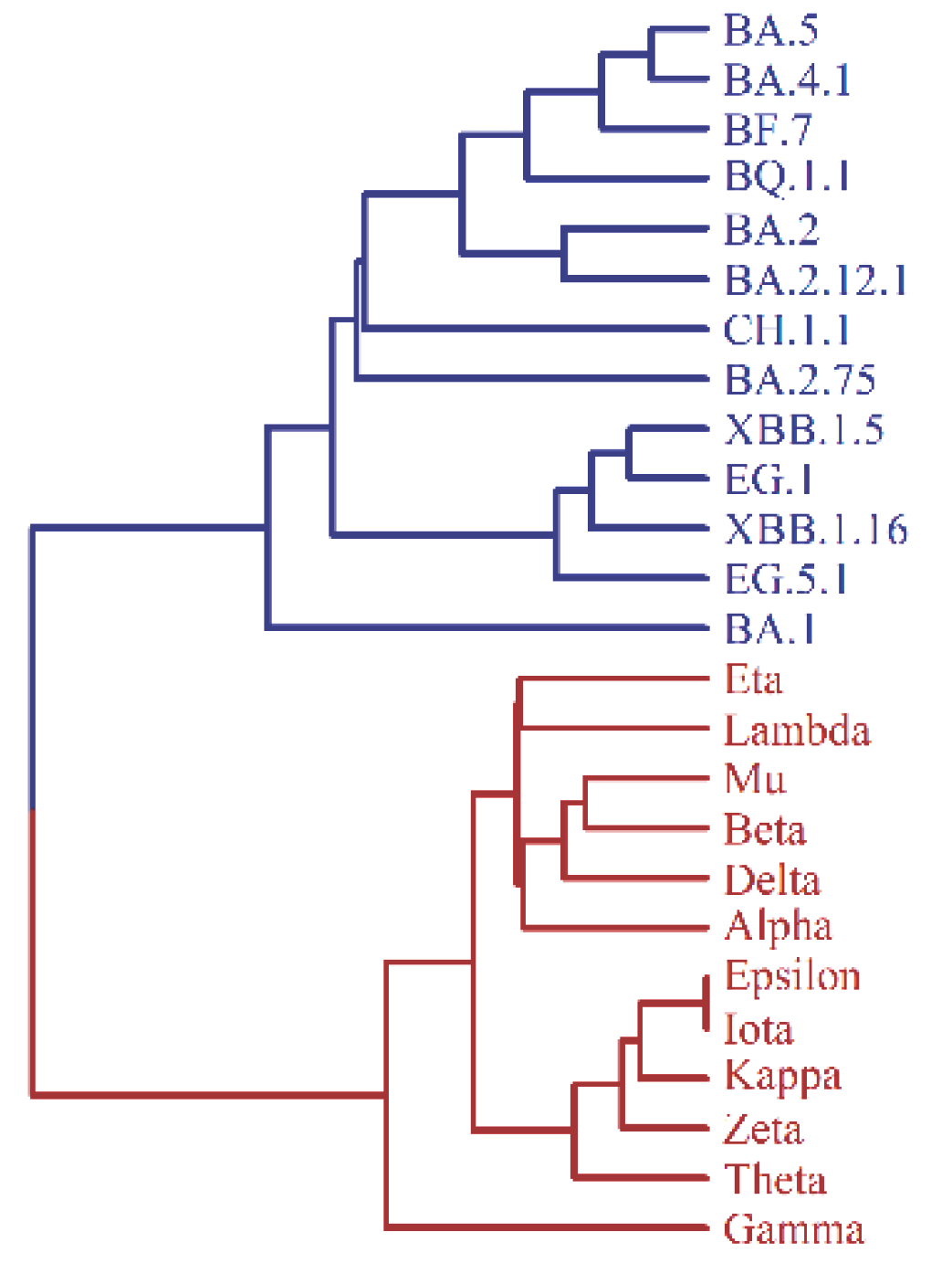
2.1. SARS-CoV-2 Variants
2.2. ACE2 Binding Affinity of a Single Mutation
2.3. The n-Distance between Two Sequences
2.4. The Construction Method of the Evolutionary Tree
3. Results
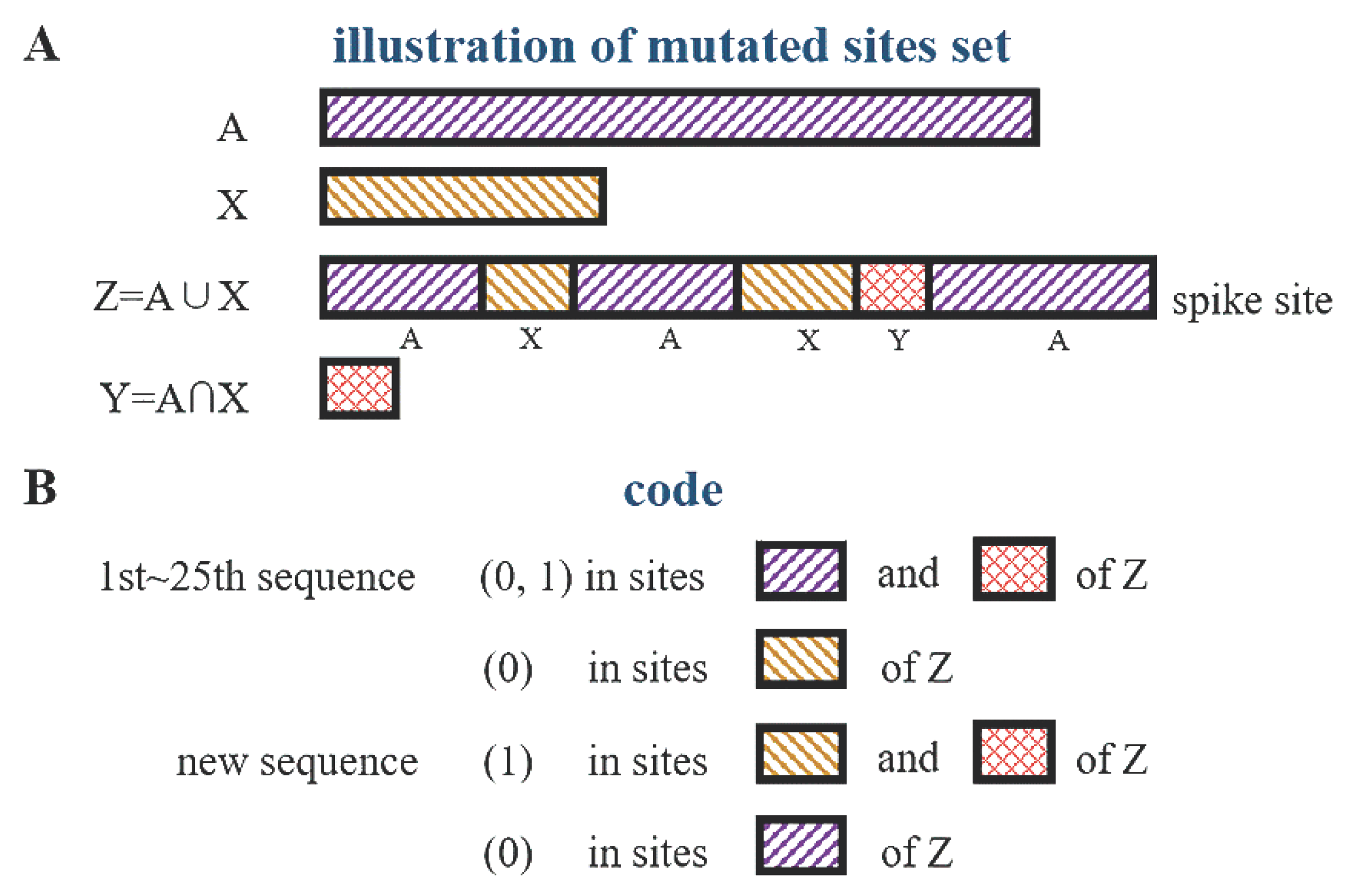
3.1. Four-Letter Representation of the Mutants
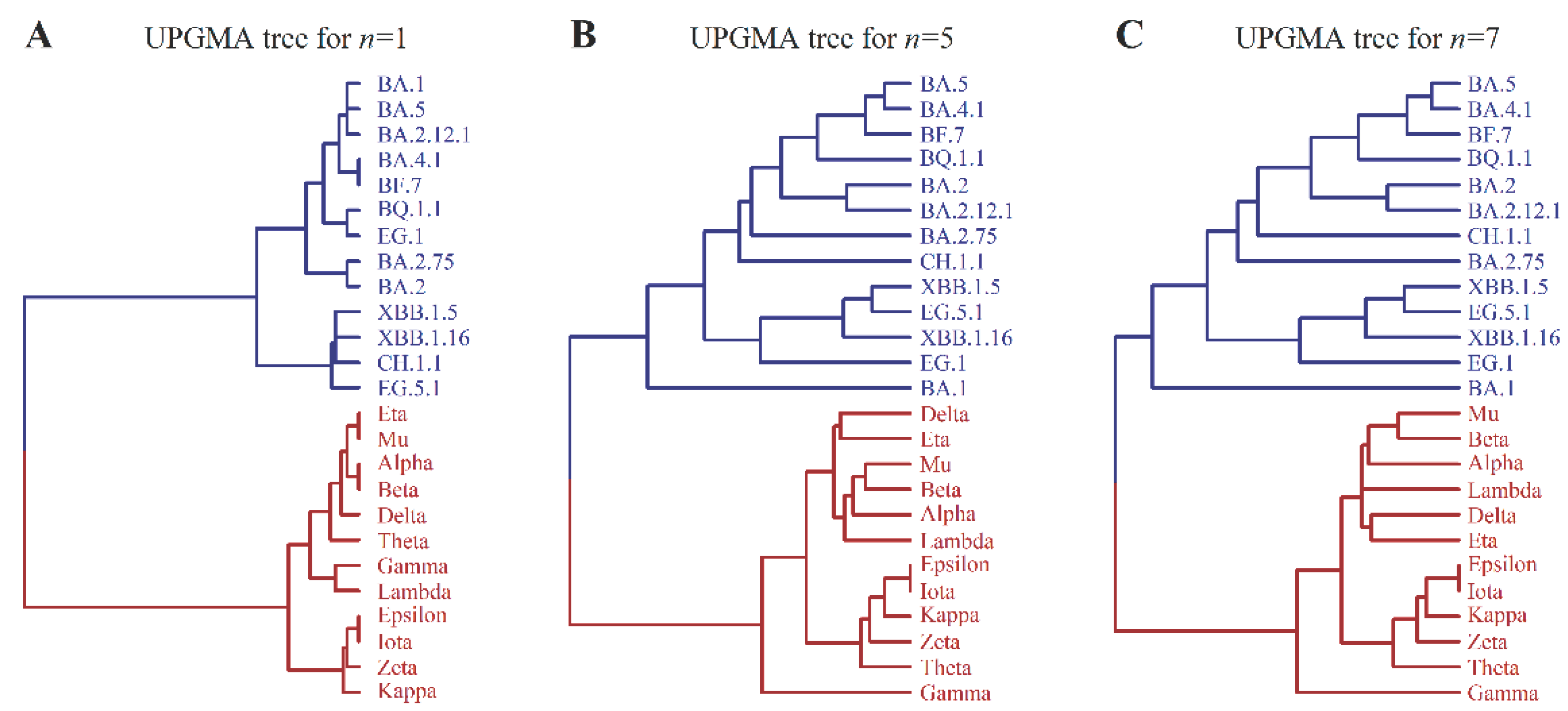
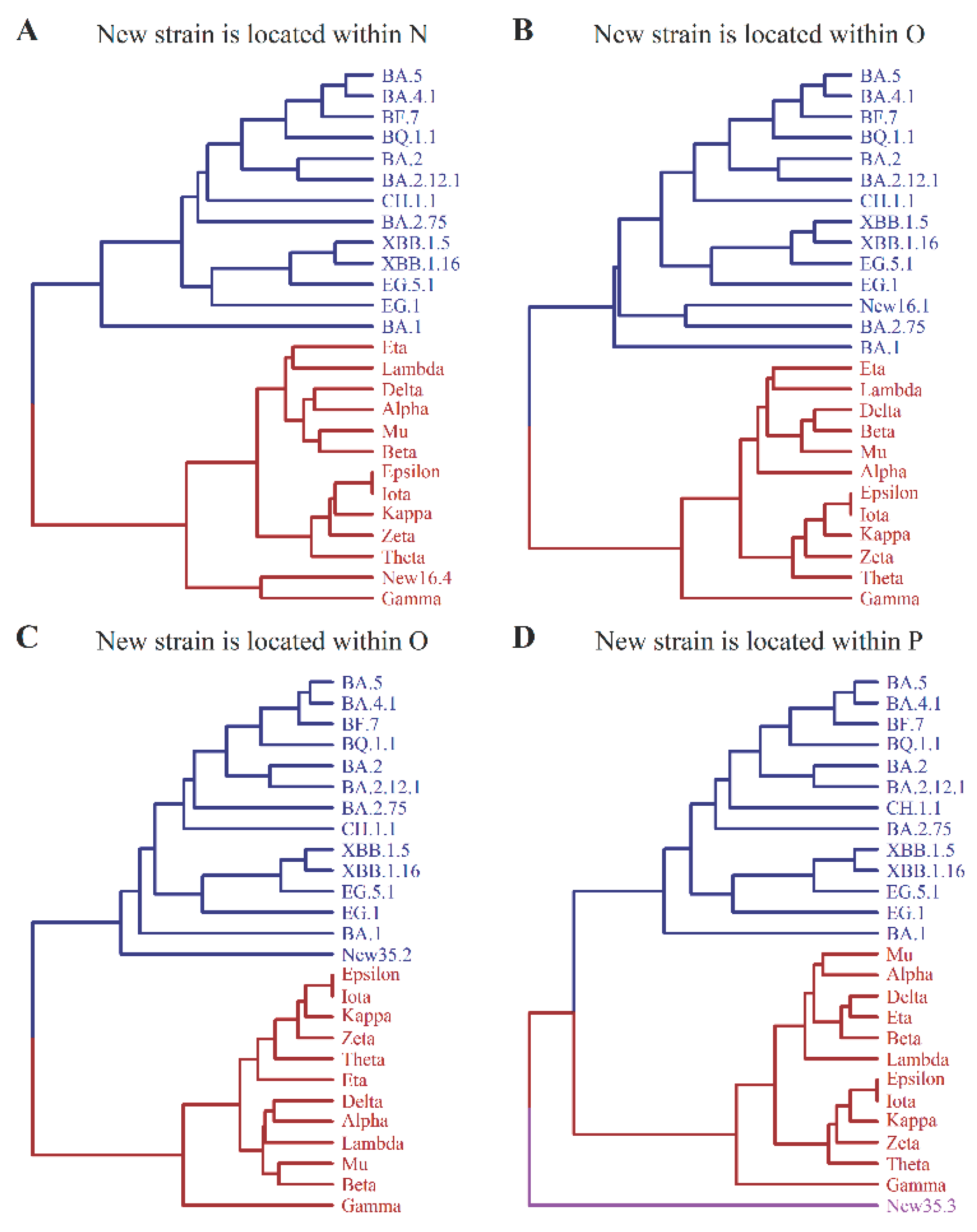
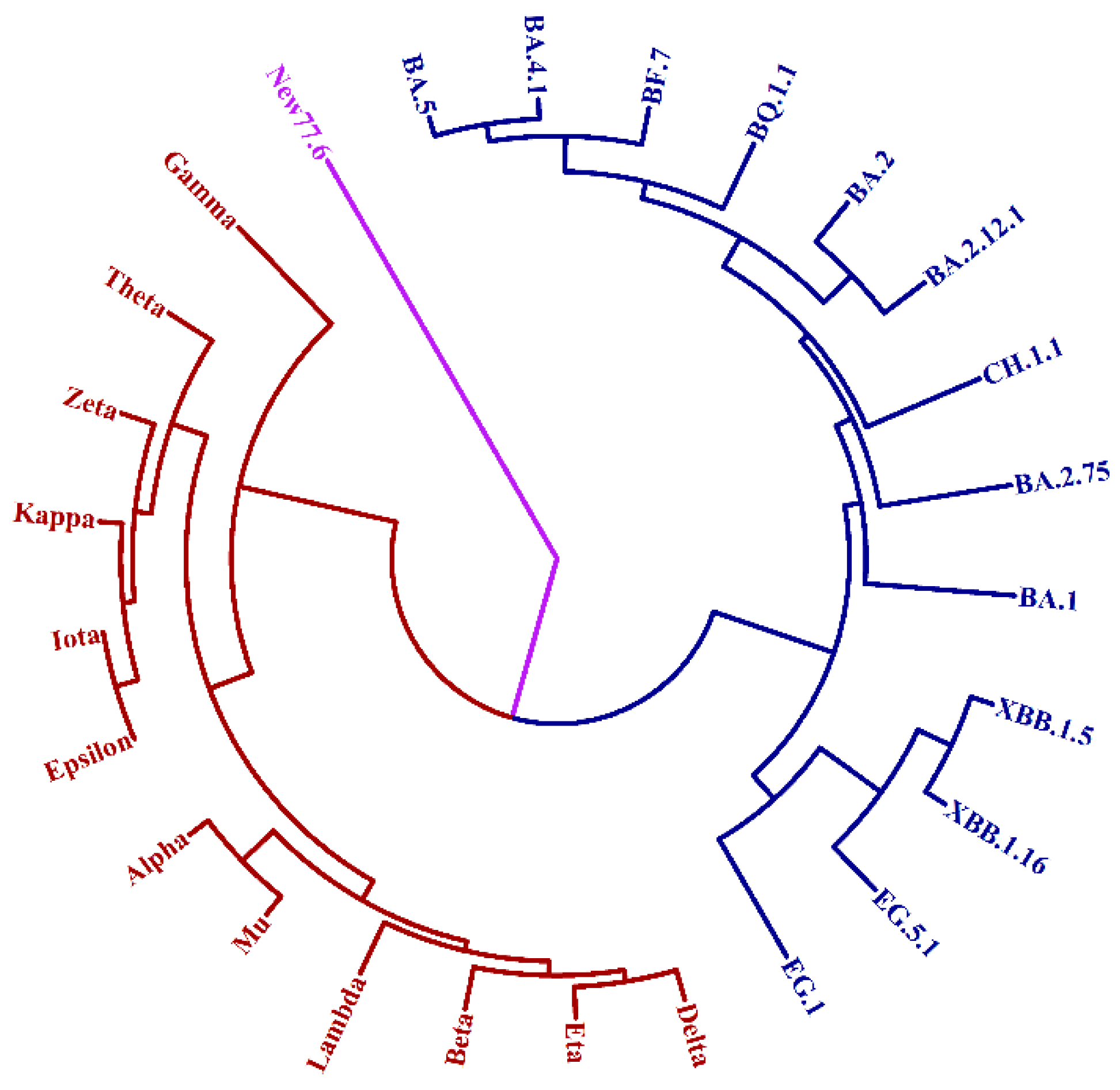
3.2. Evolutionary Tree of Virus Mutation Deduced by UPGMA Method
3.3. Generation of New Strain on the Tree by Stochastic Method
3.4. The Stochastic Property of the Intersection Set
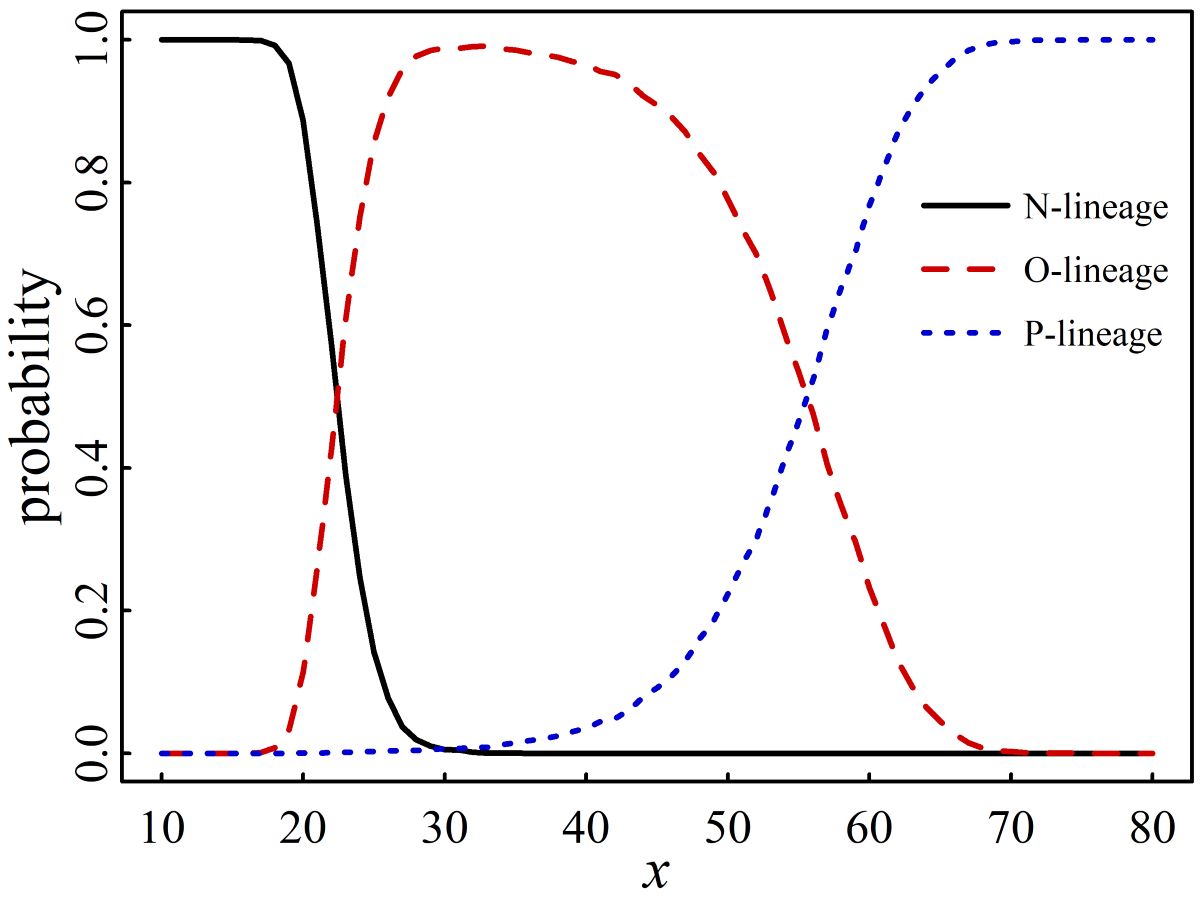
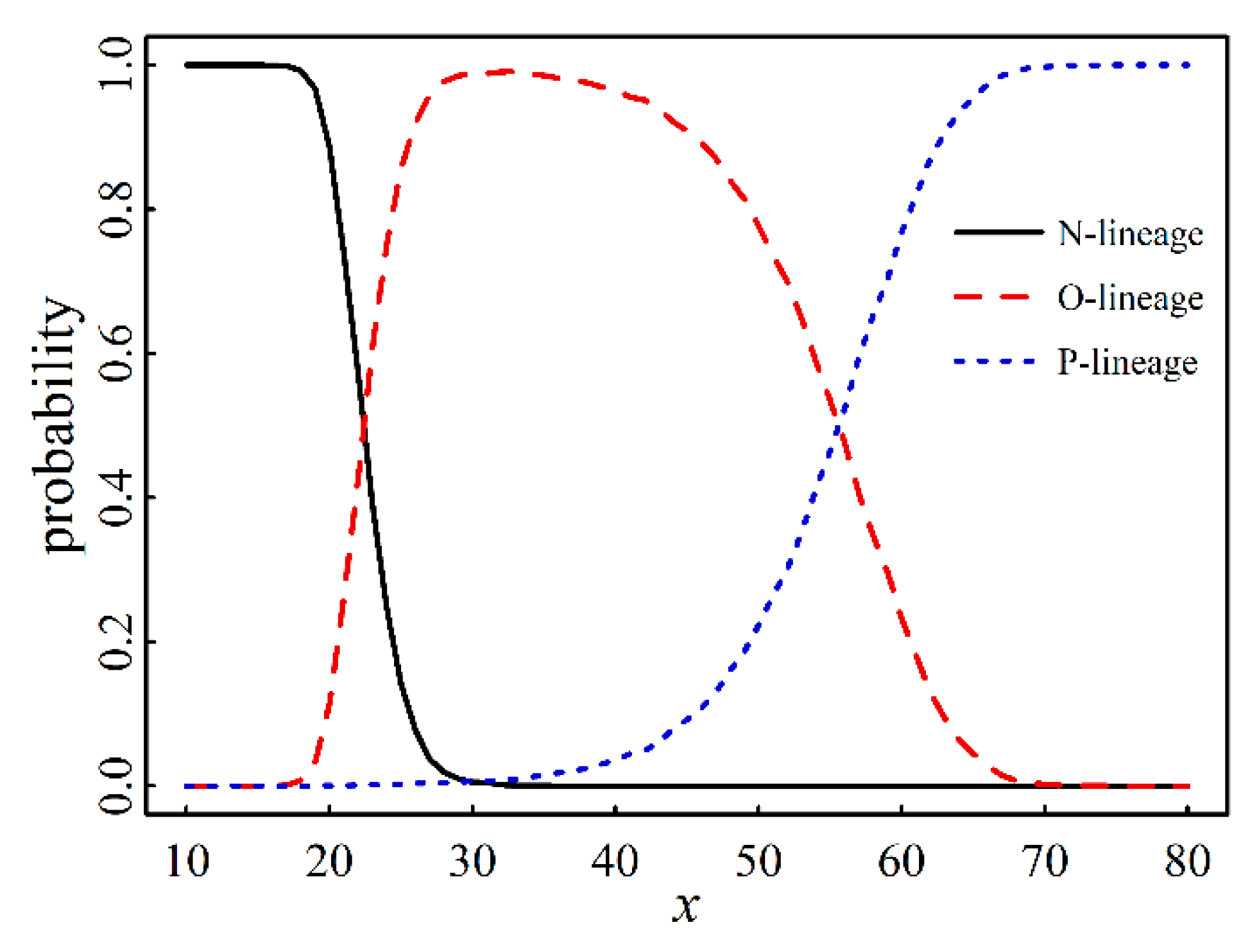
3.5. The Prediction of Macro-Lineage of SARS-CoV-2
4. Discussions
4.1. On Mutational Robustness
4.2. Electric Charge Dependence of Virus Mutation
4.3. Conclusions Drawn from the Current Study
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Božič, A.; Podgornik, R. Evolutionary changes in the number of dissociable amino acids on spike proteins and nucleoproteins of SARS-CoV-2 variants. Virus. Evol. 2023, 9, vead040. [Google Scholar] [CrossRef]
- Callaway, E. Beyond Omicron: what’s next for COVID’s viral evolution. Nature 2021, 600, 204–207. [Google Scholar] [CrossRef]
- Cao, Y.; Jian, F.; Wang, J.; Yu, Y.; Song, W.; Yisimayi, A.; Wang, J.; An, R.; Chen, X.; Zhang, N.; Wang, Y.; Wang, P.; Zhao, L.; Sun, H.; Yu, L.; Yang, S.; Niu, X.; Xiao, T.; Gu, Q.; Shao, F.; Hao, X.; Xu, Y.; Jin, R.; Shen, Z.; Wang, Y.; Xie, X. S. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 2023, 614, 521–529. [Google Scholar] [CrossRef]
- Carabelli, A. M.; Peacock, T. P.; Thorne, L. G.; Harvey, W. T.; Hughes, J.; COVID-19 Genomics UK Consortium, Peacock, S. J.; Barclay, W. S.; de Silva, T. I.; Towers, G. J.; Robertson, D. L. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef]
- Cotten, M.; Phan, M. V. T. Evolution of increased positive charge on the SARS-CoV-2 spike protein may be adaptation to human transmission. iScience 2023, 26, 106230. [Google Scholar] [CrossRef]
- Gangavarapu, K.; Latif, A. A.; Mullen, J. L.; Alkuzweny, M.; Hufbauer, E.; Tsueng, G.; Haag, E.; Zeller, M.; Aceves, C. M.; Zaiets, K.; Cano, M.; Zhou, X.; Qian, Z.; Sattler, R.; Matteson, N. L.; Levy, J. I.; Lee, R. T. C.; Freitas, L.; Maurer-Stroh, S.; GISAID Core and Curation Team, Suchard, M. A.; Wu, C.; Su, A. I.; Andersen, K. G.; Hughes, L. D. Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Nat. methods 2023, 20, 512–522. [Google Scholar] [CrossRef]
- Li, W. H. Molecular evolution; Sinauer Associates Inc.: Sunderland, MA, 1997. [Google Scholar]
- Luo, L.; Lv, J. Mathematical modelling of virus spreading in COVID-19. Viruses 2023, 15, 1788. [Google Scholar] [CrossRef]
- Ma, W.; Fu, H.; Jian, F.; Cao, Y.; Li, M. Immune evasion and ACE2 binding affinity contribute to SARS-CoV-2 evolution. Nat. Ecol. Evol. 2023, 7, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Mallapaty, S. Where did Omicron come from? Three key theories. Nature 2022, 602, 26–28. [Google Scholar] [CrossRef]
- Markov, P. V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N. I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Mohanty, V.; Greenbury, S. F.; Sarkany, T.; Narayanan, S.; Dingle, K.; Ahnert, S. E.; Louis, A. A. Maximum mutational robustness in genotype-phenotype maps follows a self-similar blancmange-like curve. J. R. Soc. Interface. 2023, 20, 20230169. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular evolution and phylogenetics; Oxford University Press: New York, 2000. [Google Scholar]
- Pawłowski, P. H. Additional Positive Electric Residues in the Crucial Spike Glycoprotein S Regions of the New SARS-CoV-2 Variants. Infect. Drug Resist. 2021, 14, 5099–5105. [Google Scholar] [CrossRef]
- Starr, T. N.; Greaney, A. J.; Hilton, S. K.; Ellis, D.; Crawford, K. H. D.; Dingens, A. S.; Navarro, M. J.; Bowen, J. E.; Tortorici, M. A.; Walls, A. C.; King, N. P.; Veesler, D.; Bloom, J. D. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Iketani, S.; Li, Z.; Liu, L.; Guo, Y.; Huang, Y.; Bowen, A. D.; Liu, M.; Wang, M.; Yu, J.; Valdez, R.; Lauring, A. S.; Sheng, Z.; Wang, H. H.; Gordon, A.; Liu, L.; Ho, D. D. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 2023, 186, 279–286.e8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H. X.; Pang, X. Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Chem. Rev. 2018, 118, 1691–1741. [Google Scholar] [CrossRef] [PubMed]
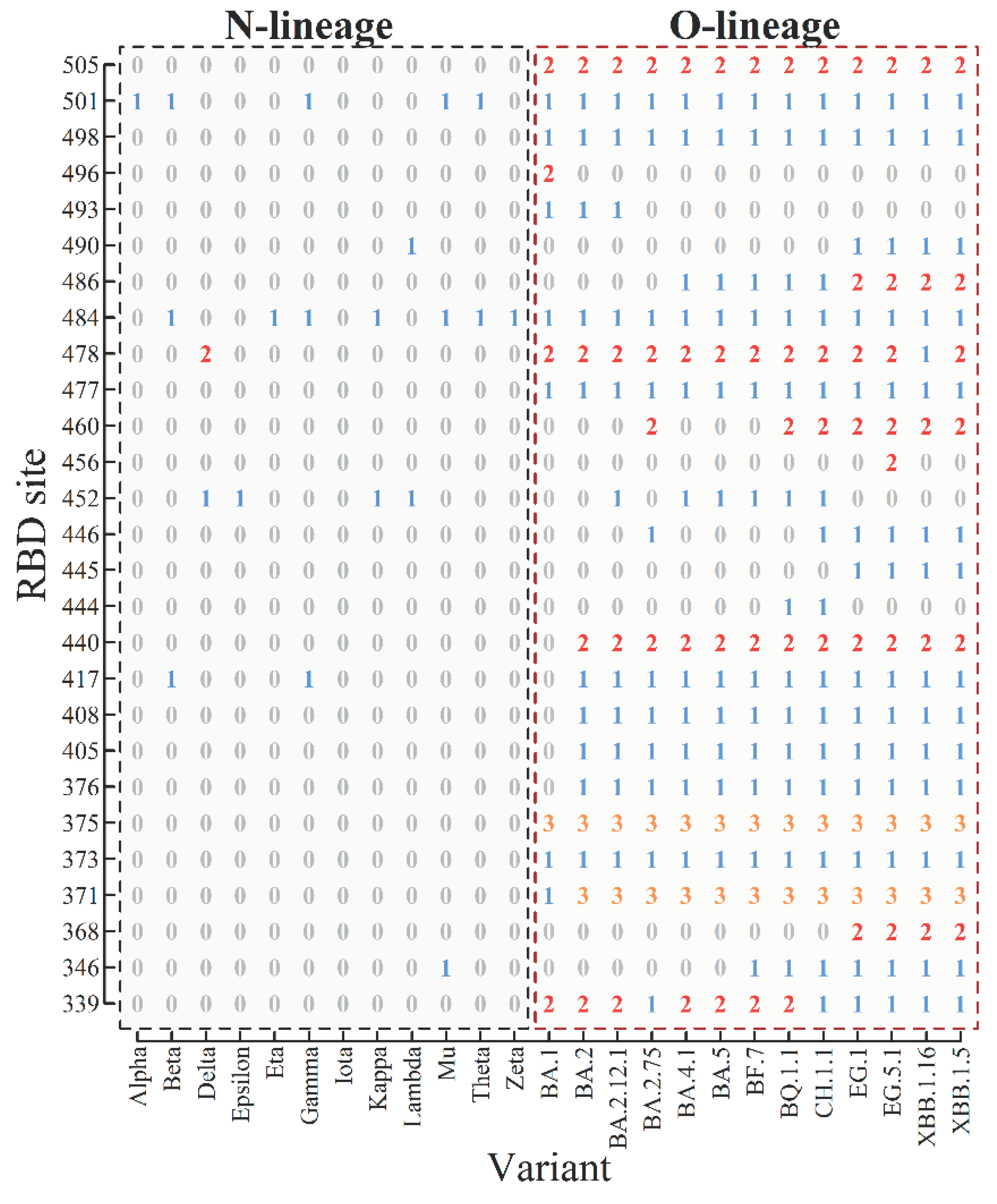
| macro-lineage | variant | number of mutated sites | mutated sites 1) |
|---|---|---|---|
| N-lineage | Alpha | 10 | 69, 70, 144, 501, 570, 614, 681, 716, 982, 1118 |
| Beta | 10 | 80, 215, 241, 242, 243, 417, 484, 501, 614, 701 | |
| Gamma | 12 | 18, 20, 26, 138, 190, 417, 484, 501, 614, 655, 1027, 1176 | |
| Delta | 9 | 19, 156, 157, 158, 452, 478, 614, 681, 950 | |
| Epsilon | 4 | 13, 152, 452, 614 | |
| Zeta | 3 | 484, 614, 1176 | |
| Eta | 9 | 52, 67, 69, 70, 144, 484, 614, 677, 888 | |
| Theta | 7 | 484, 501, 614, 681, 1092, 1101, 1176 | |
| Iota | 4 | 5, 95, 253, 614 | |
| Kappa | 5 | 452, 484, 614, 681, 1071 | |
| Lambda | 14 | 75, 76, 246, 247, 248, 249, 250, 251, 252, 253, 452, 490, 614, 859 | |
| Mu | 9 | 95, 144, 145, 346, 484, 501, 614, 681, 950 | |
| O-lineage | BA.1 | 33 | 67, 69, 70, 95, 142, 143, 144, 145, 211, 212, 339, 371, 373, 375, 477, 478, 484, 493, 496, 498, 501, 505, 547, 614, 655, 679, 681, 764, 796, 856, 954, 969, 981 |
| BA.2 | 31 | 19, 24, 25, 26, 27, 142, 213, 339, 371, 373, 375, 376, 405, 408, 417, 440, 477, 478, 484, 493, 498, 501, 505, 614, 655, 679, 681, 764, 796, 954, 969 | |
| BA.2.12.1 | 33 | 19, 24, 25, 26, 27, 142, 213, 339, 371, 373, 375, 376, 405, 408, 417, 440, 452, 477, 478, 484, 493, 498, 501, 505, 614, 655, 679, 681, 704, 764, 796, 954, 969 | |
| BA.2.75 | 30 | 19, 24, 210, 213, 257, 339, 371, 373, 375, 376, 405, 408, 417, 440, 446, 460, 477, 478, 484, 498, 501, 505, 614, 655, 679, 681, 764, 796, 954, 969 | |
| BA.4.1 | 35 | 3, 19, 24, 25, 26, 27, 69, 70, 142, 213, 339, 371, 373, 375, 376, 405, 408, 417, 440, 452, 477, 478, 484, 486, 498, 501, 505, 614, 655, 679, 681, 764, 796, 954, 969 | |
| BA.5 | 34 | 19, 24, 25, 26, 27, 69, 70, 142, 213, 339, 371, 373, 375, 376, 405, 408, 417, 440, 452, 477, 478, 484, 486, 498, 501, 505, 614, 655, 679, 681, 764, 796, 954, 969 | |
| BF.7 | 35 | 19, 24, 25, 26, 27, 69, 70, 142, 213, 339, 346, 371, 373, 375, 376, 405, 408, 417, 440, 452, 477, 478, 484, 486, 498, 501, 505, 614, 655, 679, 681, 764, 796, 954, 969 | |
| BQ.1.1 | 37 | 19, 24, 25, 26, 27, 69, 70, 142, 213, 339, 346, 371, 373, 375, 376, 405, 408, 417, 440, 444, 452, 460, 477, 478, 484, 486, 498, 501, 505, 614, 655, 679, 681, 764, 796, 954, 969 | |
| CH.1.1 | 41 | 19, 24, 25, 26, 27, 142, 147, 152, 157, 210, 213, 257, 339, 346, 371, 373, 375, 376, 405, 408, 417, 440, 444, 446, 452, 460, 477, 478, 484, 486, 498, 501, 505, 614, 655, 679, 681, 764, 796, 954, 969 | |
| XBB.1.5 | 42 | 19, 24, 25, 26, 27, 83, 142, 144, 146, 183, 213, 252, 339, 346, 368, 371, 373, 375, 376, 405, 408, 417, 440, 445, 446, 460, 477, 478, 484, 486, 490, 498, 501, 505, 614, 655, 679, 681, 764, 796, 954, 969 | |
| XBB.1.16 | 43 | 19, 24, 25, 26, 27, 83, 142, 144, 146, 180, 183, 213, 252, 339, 346, 368, 371, 373, 375, 376, 405, 408, 417, 440, 445, 446, 460, 477, 478, 484, 486, 490, 498, 501, 505, 614, 655, 679, 681, 764, 796, 954, 969 | |
| EG.1 | 37 | 19, 24, 25, 26, 27, 83, 142, 144, 146, 183, 213, 252, 339, 346, 368, 371, 373, 375, 376, 405, 408, 417, 440, 445, 446, 460, 477, 478, 484, 486, 490, 498, 501, 505, 613, 614, 655 | |
| EG.5.1 | 44 | 19, 24, 25, 26, 27, 52, 83, 142, 144, 146, 183, 213, 252, 339, 346, 368, 371, 373, 375, 376, 405, 408, 417, 440, 445, 446, 456, 460, 477, 478, 484, 486, 490, 498, 501, 505, 614, 655, 679, 681, 764, 796, 954, 969 | |
| total | 25 variants | 104 mutation sits | 3, 5, 13, 18, 19, 20, 24, 25, 26, 27, 52, 67, 69, 70, 75, 76, 80, 83, 95, 138, 142, 143, 144, 145, 146, 147, 152, 156, 157, 158, 180, 183, 190, 210, 211, 212, 213, 215, 241, 242, 243, 246, 247, 248, 249, 250, 251, 252, 253, 257, 339, 346, 368, 371, 373, 375, 376, 405, 408, 417, 440, 444, 445, 446, 452, 456, 460, 477, 478, 484, 486, 490, 493, 496, 498, 501, 505, 547, 570, 613, 614, 655, 677, 679, 681, 701, 704, 716, 764, 796, 856, 859, 888, 950, 954, 969, 981, 982, 1027, 1071, 1092, 1101, 1118, 1176 |
| x | distribution of y 1) | |||||
| mean | sd | median | max | skew | kurtosis | |
| 13 | 1.06 | 0.98 | 1 | 7 | 0.84 | 0.54 |
| 13 | 1.06 | 0.98 | 1 | 7 | 0.84 | 0.55 |
| 13 | 1.06 | 0.98 | 1 | 7 | 0.83 | 0.52 |
| 13 | 1.06 | 0.98 | 1 | 7 | 0.82 | 0.46 |
| 13 | 1.06 | 0.98 | 1 | 7 | 0.83 | 0.51 |
| 13 | 1.07 | 0.99 | 1 | 7 | 0.84 | 0.53 |
| 16 | 1.31 | 1.09 | 1 | 7 | 0.75 | 0.42 |
| 16 | 1.31 | 1.09 | 1 | 8 | 0.77 | 0.49 |
| 16 | 1.30 | 1.09 | 1 | 9 | 0.75 | 0.41 |
| 16 | 1.30 | 1.09 | 1 | 8 | 0.74 | 0.42 |
| 16 | 1.31 | 1.10 | 1 | 7 | 0.77 | 0.45 |
| 16 | 1.31 | 1.09 | 1 | 8 | 0.75 | 0.40 |
| 39 | 3.19 | 1.68 | 3 | 13 | 0.46 | 0.12 |
| 39 | 3.20 | 1.69 | 3 | 13 | 0.48 | 0.17 |
| 39 | 3.19 | 1.69 | 3 | 14 | 0.47 | 0.18 |
| 39 | 3.18 | 1.69 | 3 | 13 | 0.47 | 0.19 |
| 39 | 3.18 | 1.68 | 3 | 13 | 0.46 | 0.15 |
| 39 | 3.18 | 1.69 | 3 | 12 | 0.47 | 0.16 |
| 76 | 6.22 | 2.32 | 6 | 18 | 0.32 | 0.03 |
| 76 | 6.21 | 2.32 | 6 | 17 | 0.32 | 0.07 |
| 76 | 6.21 | 2.31 | 6 | 19 | 0.33 | 0.08 |
| 76 | 6.20 | 2.33 | 6 | 17 | 0.31 | 0.05 |
| 76 | 6.21 | 2.32 | 6 | 18 | 0.32 | 0.06 |
| 76 | 6.21 | 2.31 | 6 | 18 | 0.31 | 0.03 |
| Macro-lineages | Mutant | Number of AA sites | Positively charged | Negatively charged | Neutrally charged |
|---|---|---|---|---|---|
| N-lineage | Lambda | 14 | 3 | 1 | 10 |
| Gamma | 12 | 3 | 5 | 6 | |
| Beta | 10 | 5 | 2 | 4 | |
| Alpha | 10 | 5 | 3 | 3 | |
| Mu | 9 | 7 | 1 | 2 | |
| Eta | 9 | 6 | 1 | 3 | |
| Delta | 9 | 7 | 1 | 1 | |
| Theta | 7 | 6 | 3 | 1 | |
| Kappa | 5 | 5 | 0 | 0 | |
| Iota | 4 | 2 | 0 | 2 | |
| Epsilon | 4 | 2 | 0 | 2 | |
| Zeta | 3 | 3 | 0 | 1 | |
| O-lineage | BA.1 | 33 | 16 | 6 | 13 |
| BA.5 | 34 | 14 | 8 | 14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





