Submitted:
21 December 2023
Posted:
21 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Application of Nucleic Acids Amplification Tests in Parasite Detection
3. CRISPR-Cas12a for POCT
3.1. Discovery of CRISPR
3.2. CRISPR-Cas12a Is More Suitable for Rapid On-Site Detection
3.3. CRISPR-Cas12a Has Been Applied to Parasite Detection
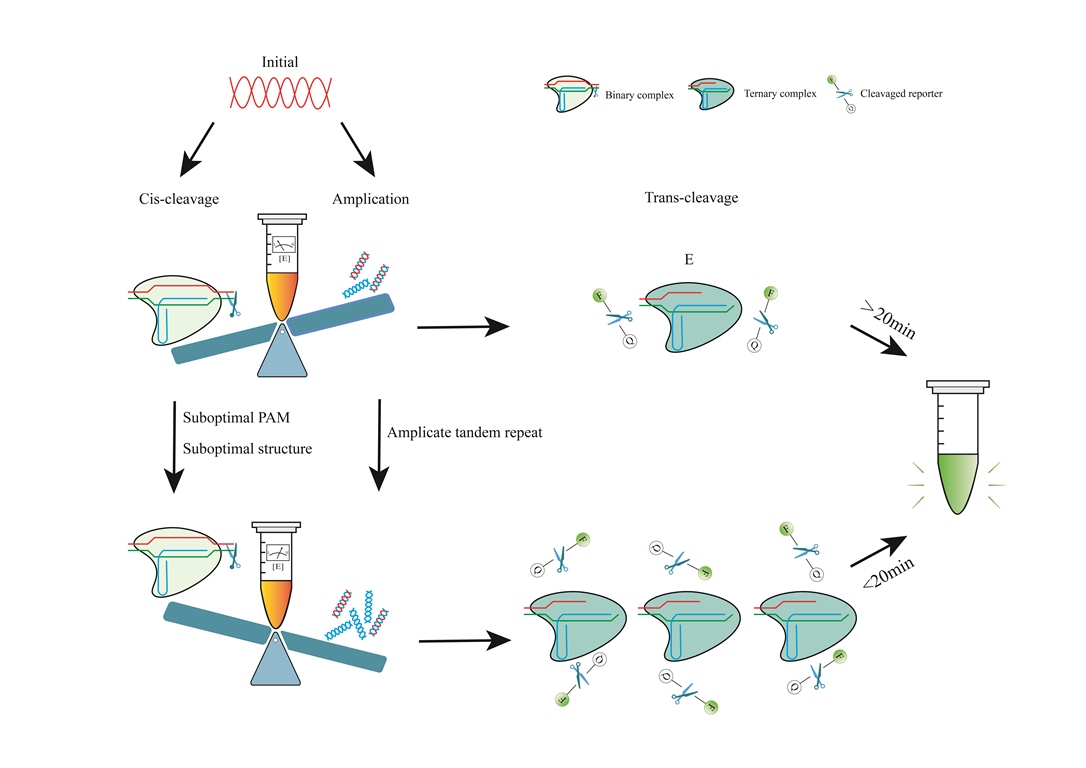
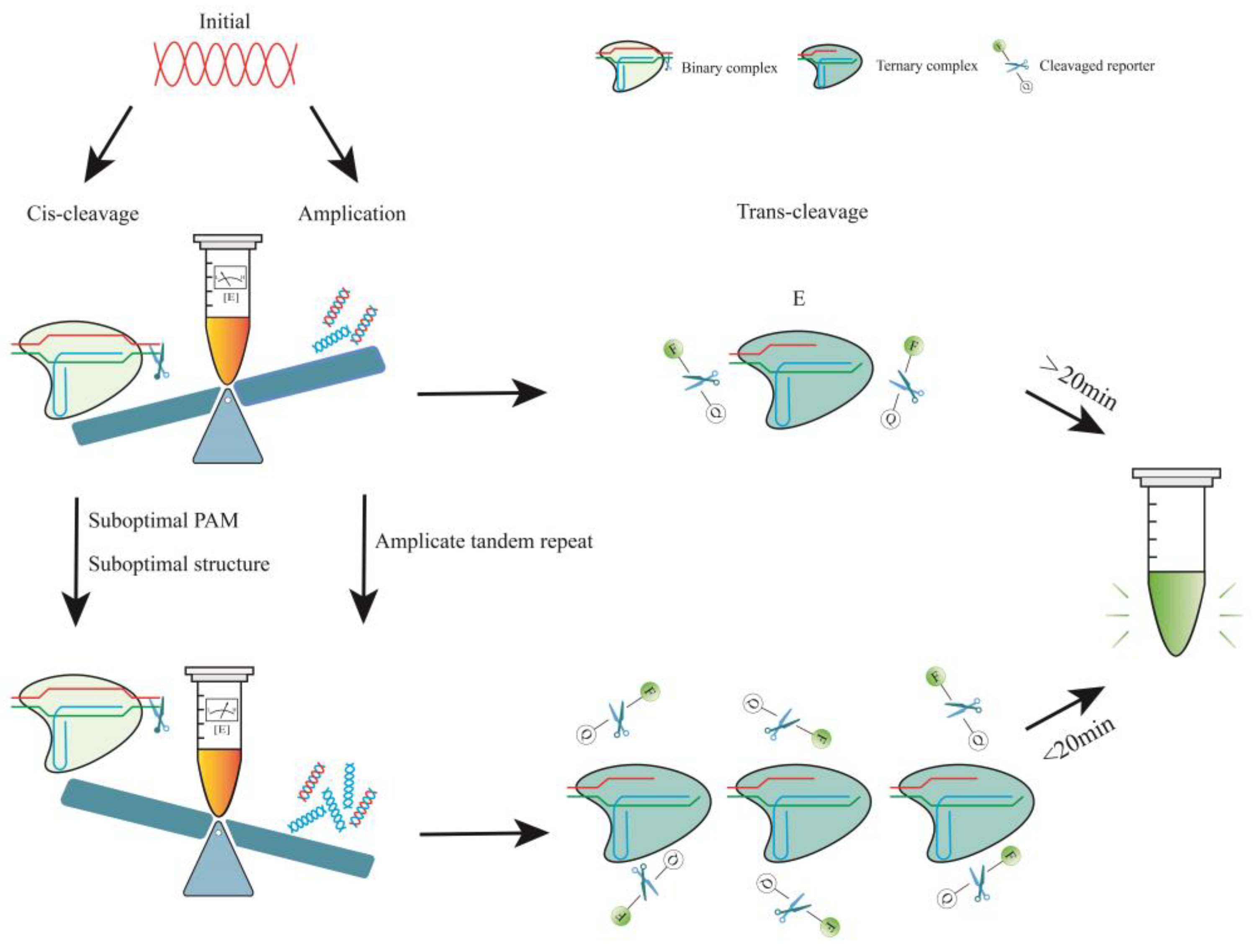
4. Optimization of the CRISPR-Cas12a One-Pot Detection Assay
4.1. One-Pot and One Step
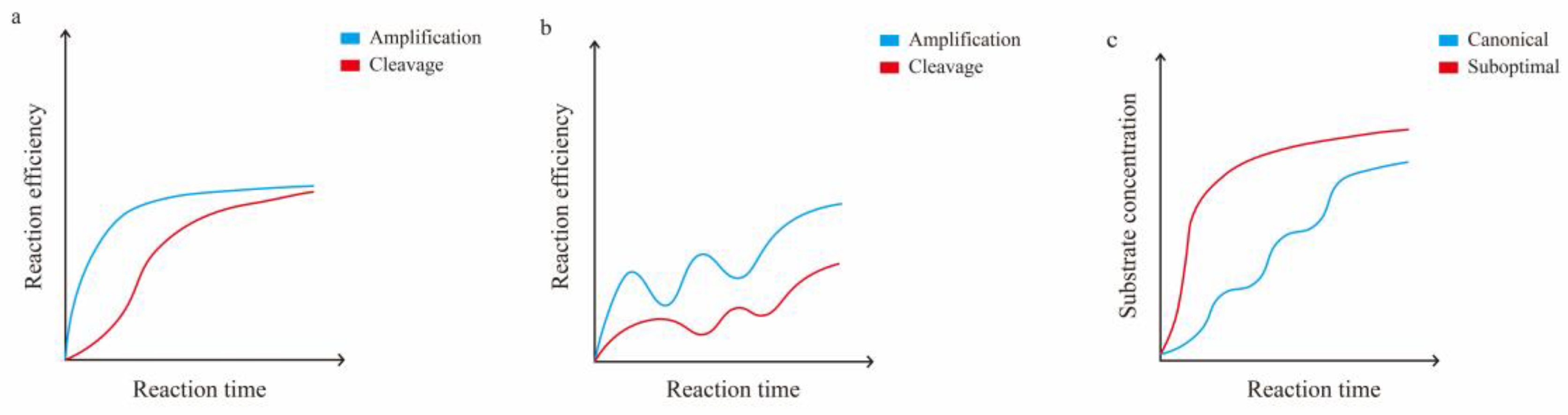
4.1.1. Determinants of Cas12a Enzyme Kinetics
4.1.2. Reduced crRNA Efficiency by PAM
4.1.3. Reduced crRNA Efficiency by Structure
4.2. One-Pot but Two Steps
4.2.1. Light-Activated crRNA to Initiate Cleavage
4.2.2. Physically Separate the Two Processes
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- WHO. Working to overcome the global impact of neglected tropical diseases: first WHO report on neglected tropical diseases. Available online: (accessed on 11 February).
- Torgerson, P.R.; Devleesschauwer, B.; Praet, N.; Speybroeck, N.; Willingham, A.L.; Kasuga, F.; Rokni, M.B.; Zhou, X.N.; Fevre, E.M.; Sripa, B.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 11 Foodborne Parasitic Diseases, 2010: A Data Synthesis. PLoS Med 2015, 12, e1001920. [Google Scholar] [CrossRef] [PubMed]
- WHO. World malaria report 2021. Available online: (accessed on 6 December).
- Theel, E.S.; Pritt, B.S. Parasites. Microbiol Spectr 2016, 4. [Google Scholar] [CrossRef]
- Meinel, T.R.; Gottstein, B.; Geib, V.; Keel, M.J.; Biral, R.; Mohaupt, M.; Brügger, J. Vertebral alveolar echinococcosis—a case report, systematic analysis, and review of the literature. The Lancet Infectious Diseases 2018, 18, e87–e98. [Google Scholar] [CrossRef]
- Daly, R.; Chiodini, P.L. Laboratory investigations and diagnosis of tropical diseases in travelers. Infectious disease clinics of North America 2012, 26, 803–818. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.S.; Fung, K.S.; Chau, S.; Poon, R.W.; Wong, S.C.; Yuen, K.Y. Molecular diagnosis in clinical parasitology: when and why? Exp Biol Med (Maywood) 2014, 239, 1443–1460. [Google Scholar] [CrossRef] [PubMed]
- Kettelhut, M.M.; Chiodini, P.L.; Edwards, H.; Moody, A. External quality assessment schemes raise standards: evidence from the UKNEQAS parasitology subschemes. Journal of clinical pathology 2003, 56, 927–932. [Google Scholar] [CrossRef]
- Rosenblatt, J.E. Laboratory diagnosis of infections due to blood and tissue parasites. Clin Infect Dis 2009, 49, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Kalogeropoulos, D.; Sakkas, H.; Mohammed, B.; Vartholomatos, G.; Malamos, K.; Sreekantam, S.; Kanavaros, P.; Kalogeropoulos, C. Ocular toxoplasmosis: a review of the current diagnostic and therapeutic approaches. International ophthalmology 2022, 42, 295–321. [Google Scholar] [CrossRef]
- Qian, W.; Yan, W.; Lv, C.; Bai, R.; Wang, T.; Wei, Z.; Zhang, M. Molecular Detection and Genotyping of Toxoplasma gondii and Neospora caninum in Slaughtered Goats in Central China. Foodborne pathogens and disease 2020, 17, 348–356. [Google Scholar] [CrossRef]
- Waitumbi, J.N.; Gerlach, J.; Afonina, I.; Anyona, S.B.; Koros, J.N.; Siangla, J.; Ankoudinova, I.; Singhal, M.; Watts, K.; Polhemus, M.E.; et al. Malaria prevalence defined by microscopy, antigen detection, DNA amplification and total nucleic acid amplification in a malaria-endemic region during the peak malaria transmission season. Trop Med Int Health 2011, 16, 786–793. [Google Scholar] [CrossRef]
- Alhassan, A.; Li, Z.; Poole, C.B.; Carlow, C.K. Expanding the MDx toolbox for filarial diagnosis and surveillance. Trends Parasitol 2015, 31, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Momcilovic, S.; Cantacessi, C.; Arsic-Arsenijevic, V.; Otranto, D.; Tasic-Otasevic, S. Rapid diagnosis of parasitic diseases: current scenario and future needs. Clin Microbiol Infect 2019, 25, 290–309. [Google Scholar] [CrossRef] [PubMed]
- Lalremruata, A.; Nguyen, T.T.; McCall, M.B.B.; Mombo-Ngoma, G.; Agnandji, S.T.; Adegnika, A.A.; Lell, B.; Ramharter, M.; Hoffman, S.L.; Kremsner, P.G.; et al. Recombinase Polymerase Amplification and Lateral Flow Assay for Ultrasensitive Detection of Low-Density Plasmodium falciparum Infection from Controlled Human Malaria Infection Studies and Naturally Acquired Infections. J Clin Microbiol 2020, 58. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.A.; Faisal, K.; Chowdhury, R.; Nath, R.; Ghosh, P.; Ghosh, D.; Hossain, F.; Abd El Wahed, A.; Mondal, D. Evaluation of molecular assays to detect Leishmania donovani in Phlebotomus argentipes fed on post-kala-azar dermal leishmaniasis patients. Parasit Vectors 2021, 14, 465. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Rong, R.; Zhang, H.Q.; Shi, C.J.; Zhu, X.Q.; Xia, C.M. Sensitive and rapid detection of Schistosoma japonicum DNA by loop-mediated isothermal amplification (LAMP). Int J Parasitol 2010, 40, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.A.; von Sternberg, R. Why repetitive DNA is essential to genome function. Biol Rev Camb Philos Soc 2005, 80, 227–250. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, W.; Zhang, L.; Zhang, Z.; Li, J.; Lu, G.; Zhu, Y.; Wang, Y.; Huang, Y.; Liu, J.; et al. The genome of the hydatid tapeworm Echinococcus granulosus. Nat Genet 2013, 45, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Prediger, E.; Huecas, M.E.; Nogueira, N.; Lizardi, P.M. Minichromosomal repetitive DNA in Trypanosoma cruzi: its use in a high-sensitivity parasite detection assay. Proc Natl Acad Sci U S A 1984, 81, 3356–3360. [Google Scholar] [CrossRef] [PubMed]
- Homan, W.L.; Vercammen, M.; De Braekeleer, J.; Verschueren, H. Identification of a 200- to 300-fold repetitive 529 bp DNA fragment in Toxoplasma gondii, and its use for diagnostic and quantitative PCR. Int J Parasitol 2000, 30, 69–75. [Google Scholar] [CrossRef]
- Demas, A.; Oberstaller, J.; DeBarry, J.; Lucchi, N.W.; Srinivasamoorthy, G.; Sumari, D.; Kabanywanyi, A.M.; Villegas, L.; Escalante, A.A.; Kachur, S.P.; et al. Applied genomics: data mining reveals species-specific malaria diagnostic targets more sensitive than 18S rRNA. J Clin Microbiol 2011, 49, 2411–2418. [Google Scholar] [CrossRef]
- Singh, R.; Singh, D.P.; Gupta, R.; Savargaonkar, D.; Singh, O.P.; Nanda, N.; Bhatt, R.M.; Valecha, N. Comparison of three PCR-based assays for the non-invasive diagnosis of malaria: detection of Plasmodium parasites in blood and saliva. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology 2014, 33, 1631–1639. [Google Scholar] [CrossRef]
- Azam, M.; Upmanyu, K.; Gupta, R.; Sruthy, K.S.; Matlani, M.; Savargaonkar, D.; Singh, R. Development of Two-Tube Loop-Mediated Isothermal Amplification Assay for Differential Diagnosis of Plasmodium falciparum and Plasmodium vivax and Its Comparison with Loopamp™ Malaria. Diagnostics 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Rosenzvit, M.C.; Canova, S.G.; Kamenetzky, L.; Ledesma, B.A.; Guarnera, E.A. Echinococcus granulosus: Cloning and characterization of a tandemly repeated DNA element. Exp. Parasitol. 1997, 87, 65–68. [Google Scholar] [CrossRef]
- Abbasi, I.; Hamburger, J.; Raoul, F.; Craig, P.S.; Campos-Ponce, M.; Branzburg, A.; Hafez, S.K.A. Copro-Diagnosis of Echinococcus Granulosus Infection in Dogs by Amplification of a Newly Identified Repeated DNA Sequence. The American Journal of Tropical Medicine and Hygiene 2003, 69, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Chapman, A.; Vallejo, V.; Mossie, K.G.; Ortiz, D.; Agabian, N.; Flisser, A. Isolation and characterization of species-specific DNA probes from Taenia solium and Taenia saginata and their use in an egg detection assay. J Clin Microbiol 1995, 33, 1283–1288. [Google Scholar] [CrossRef]
- González, L.M.; Montero, E.; Harrison, L.J.; Parkhouse, R.M.; Garate, T. Differential diagnosis of Taenia saginata and Taenia solium infection by PCR. J Clin Microbiol 2000, 38, 737–744. [Google Scholar] [CrossRef]
- Hamburger, J.; Turetski, T.; Kapeller, I.; Deresiewicz, R. Highly repeated short DNA sequences in the genome of Schistosoma mansoni recognized by a species-specific probe. Mol Biochem Parasitol 1991, 44, 73–80. [Google Scholar] [CrossRef]
- Mwangi, I.N.; Agola, E.L.; Mugambi, R.M.; Shiraho, E.A.; Mkoji, G.M. Development and Evaluation of a Loop-Mediated Isothermal Amplification Assay for Diagnosis of Schistosoma mansoni Infection in Faecal Samples. Journal of parasitology research 2018, 2018, 1267826. [Google Scholar] [CrossRef]
- Hamburger, J.; Xu, Y.X.; Ramzy, R.M.; Jourdane, J.; Ruppel, A. Development and laboratory evaluation of a polymerase chain reaction for monitoring Schistosoma mansoni infestation of water. Am J Trop Med Hyg 1998, 59, 468–473. [Google Scholar] [CrossRef]
- Hamburger, J.; He, N.; Abbasi, I.; Ramzy, R.M.; Jourdane, J.; Ruppel, A. Polymerase chain reaction assay based on a highly repeated sequence of Schistosoma haematobium: a potential tool for monitoring schistosome-infested water. Am J Trop Med Hyg 2001, 65, 907–911. [Google Scholar] [CrossRef]
- Hertel, J.; Hamburger, J.; Haberl, B.; Haas, W. Detection of bird schistosomes in lakes by PCR and filter-hybridization. Exp Parasitol 2002, 101, 57–63. [Google Scholar] [CrossRef]
- Lodh, N.; Caro, R.; Sofer, S.; Scott, A.; Krolewiecki, A.; Shiff, C. Diagnosis of Strongyloides stercoralis: Detection of parasite-derived DNA in urine. Acta tropica 2016, 163, 9–13. [Google Scholar] [CrossRef] [PubMed]
- McReynolds, L.A.; DeSimone, S.M.; Williams, S.A. Cloning and comparison of repeated DNA sequences from the human filarial parasite Brugia malayi and the animal parasite Brugia pahangi. Proc Natl Acad Sci U S A 1986, 83, 797–801. [Google Scholar] [CrossRef]
- Albers, A.; Sartono, E.; Wahyuni, S.; Yazdanbakhsh, M.; Maizels, R.M.; Klarmann-Schulz, U.; Pfarr, K.; Hoerauf, A. Real-time PCR detection of the HhaI tandem DNA repeat in pre- and post-patent Brugia malayi Infections: a study in Indonesian transmigrants. Parasit Vectors 2014, 7, 146. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.; McCarthy, J.; Bierwert, L.; Lizotte-Waniewski, M.; Chanteau, S.; Nutman, T.B.; Ottesen, E.A.; Williams, S.A. A polymerase chain reaction assay for detection of the parasite Wuchereria bancrofti in human blood samples. Am J Trop Med Hyg 1996, 54, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.U.; Atkinson, L.J.; Ramzy, R.M.; Helmy, H.; Farid, H.A.; Bockarie, M.J.; Susapu, M.; Laney, S.J.; Williams, S.A.; Weil, G.J. A real-time PCR-based assay for detection of Wuchereria bancrofti DNA in blood and mosquitoes. Am J Trop Med Hyg 2006, 74, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Saul, A.; Yeganeh, F.; Howard, R.J. Cloning and characterization of a novel multicopy, repetitive sequence of Plasmodium falciparum, REP51. Immunology and cell biology 1992, 70 ( Pt 5), 357-359. [CrossRef]
- Hotterbeekx, A.; Raimon, S.; Abd-Elfarag, G.; Carter, J.Y.; Sebit, W.; Suliman, A.; Siewe Fodjo, J.N.; De Witte, P.; Logora, M.Y.; Colebunders, R.; et al. Onchocerca volvulus is not detected in the cerebrospinal fluid of persons with onchocerciasis-associated epilepsy. Int J Infect Dis 2020, 91, 119–123. [Google Scholar] [CrossRef]
- Macfarlane, C.L.; Quek, S.; Pionnier, N.; Turner, J.D.; Wanji, S.; Wagstaff, S.C.; Taylor, M.J. The insufficiency of circulating miRNA and DNA as diagnostic tools or as biomarkers of treatment efficacy for Onchocerca volvulus. Scientific reports 2020, 10, 6672. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.M.; Fessler, M.K.; Bloomfield, R.A.; Sandke, W.D.; Malekshahi, C.R.; Keroack, C.D.; Duignan, P.J.; Torquato, S.D.; Williams, S.A. A novel quantitative real-time PCR diagnostic assay for fecal and nasal swab detection of an otariid lungworm, Parafilaroides decorus. International journal for parasitology. Parasites and wildlife 2020, 12, 85–92. [Google Scholar] [CrossRef]
- van Dongen, J.E.; Berendsen, J.T.W.; Steenbergen, R.D.M.; Wolthuis, R.M.F.; Eijkel, J.C.T.; Segerink, L.I. Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosens Bioelectron 2020, 166, 112445. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of bacteriology 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.; Embden, J.D.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 2002, 43, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Mohanraju, P.; Makarova, K.S.; Zetsche, B.; Zhang, F.; Koonin, E.V.; van der Oost, J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 2016, 353, aad5147. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Ma, E.B.; Harrington, L.B.; Da Costa, M.; Tian, X.R.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nature reviews. Microbiology 2017, 15, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell 2015, 60, 385–397. [Google Scholar] [CrossRef]
- Swarts, D.C.; Jinek, M. Mechanistic Insights into the cis- and trans-Acting DNase Activities of Cas12a. Mol Cell 2019, 73, 589–600 e584. [Google Scholar] [CrossRef]
- Strohkendl, I.; Saifuddin, F.A.; Rybarski, J.R.; Finkelstein, I.J.; Russell, R. Kinetic Basis for DNA Target Specificity of CRISPR-Cas12a. Mol Cell 2018, 71, 816–824 e813. [Google Scholar] [CrossRef]
- Wang, R.; Qian, C.; Pang, Y.; Li, M.; Yang, Y.; Ma, H.; Zhao, M.; Qian, F.; Yu, H.; Liu, Z.; et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens Bioelectron 2021, 172, 112766. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhong, M.; Liu, Y.; Ma, P.; Dang, L.; Meng, Q.; Wan, W.; Ma, X.; Liu, J.; Yang, G.; et al. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci Bull (Beijing) 2020, 65, 1436–1439. [Google Scholar] [CrossRef]
- Lee, R.A.; Puig, H.; Nguyen, P.Q.; Angenent-Mari, N.M.; Donghia, N.M.; McGee, J.P.; Dvorin, J.D.; Klapperich, C.M.; Pollock, N.R.; Collins, J.J. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc Natl Acad Sci U S A 2020, 117, 25722–25731. [Google Scholar] [CrossRef]
- WHO. Malaria rapid diagnostic test performance: summary results of WHO product testing of malaria RDTs: Round 1-8 (2008–2018). Available online: (accessed on 23 April).
- Lei, R.; Li, L.; Wu, P.; Fei, X.; Zhang, Y.; Wang, J.; Zhang, D.; Zhang, Q.; Yang, N.; Wang, X. RPA/CRISPR/Cas12a-Based On-Site and Rapid Nucleic Acid Detection of Toxoplasma gondii in the Environment. ACS Synth Biol 2022, 11, 1772–1781. [Google Scholar] [CrossRef]
- Galvani, A.T.; Christ, A.P.G.; Padula, J.A.; Barbosa, M.R.F.; de Araújo, R.S.; Sato, M.I.Z.; Razzolini, M.T.P. Real-time PCR detection of Toxoplasma gondii in surface water samples in São Paulo, Brazil. Parasitol Res 2019, 118, 631–640. [Google Scholar] [CrossRef]
- Yu, F.; Zhang, K.; Wang, Y.; Li, D.; Cui, Z.; Huang, J.; Zhang, S.; Li, X.; Zhang, L. CRISPR/Cas12a-based on-site diagnostics of Cryptosporidium parvum IId-subtype-family from human and cattle fecal samples. Parasit Vectors 2021, 14, 208. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Deng, F.; Hall, T.; Vesey, G.; Goldys, E.M. CRISPR/Cas12a-powered immunosensor suitable for ultra-sensitive whole Cryptosporidium oocyst detection from water samples using a plate reader. Water research 2021, 203, 117553. [Google Scholar] [CrossRef]
- Kanitchinda, S.; Srisala, J.; Suebsing, R.; Prachumwat, A.; Chaijarasphong, T. CRISPR-Cas fluorescent cleavage assay coupled with recombinase polymerase amplification for sensitive and specific detection of Enterocytozoon hepatopenaei. Biotechnology reports (Amsterdam, Netherlands) 2020, 27, e00485. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Peng, D.; Jiang, C.; Zhao, W.; Li, G.; Huang, W.; Kong, L.; Gao, H.; Zheng, J.; Peng, H. Rapid and Visual Detection of Heterodera schachtii Using Recombinase Polymerase Amplification Combined with Cas12a-Mediated Technology. International journal of molecular sciences 2021, 22. [Google Scholar] [CrossRef]
- Fleming, K.A.; Horton, S.; Wilson, M.L.; Atun, R.; DeStigter, K.; Flanigan, J.; Sayed, S.; Adam, P.; Aguilar, B.; Andronikou, S.; et al. The Lancet Commission on diagnostics: transforming access to diagnostics. Lancet 2021, 398, 1997–2050. [Google Scholar] [CrossRef]
- Kosack, C.S.; Page, A.L.; Klatser, P.R. A guide to aid the selection of diagnostic tests. Bull World Health Organ 2017, 95, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Huyke, D.A.; Ramachandran, A.; Bashkirov, V.I.; Kotseroglou, E.K.; Kotseroglou, T.; Santiago, J.G. Enzyme Kinetics and Detector Sensitivity Determine Limits of Detection of Amplification-Free CRISPR-Cas12 and CRISPR-Cas13 Diagnostics. Anal Chem 2022, 94, 9826–9834. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, A.; Santiago, J.G. CRISPR Enzyme Kinetics for Molecular Diagnostics. Anal Chem 2021, 93, 7456–7464. [Google Scholar] [CrossRef] [PubMed]
- Garneau, J.E.; Dupuis, M.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadán, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Ren, K.; Qiu, X.; Zheng, J.; Guo, M.; Guan, X.; Liu, H.; Li, N.; Zhang, B.; Yang, D.; et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 2016, 532, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Yang, H.; Rajashankar, K.R.; Huang, Z.; Patel, D.J. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell research 2016, 26, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Yamano, T.; Nishimasu, H.; Zetsche, B.; Hirano, H.; Slaymaker, I.M.; Li, Y.; Fedorova, I.; Nakane, T.; Makarova, K.S.; Koonin, E.V.; et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell 2016, 165, 949–962. [Google Scholar] [CrossRef] [PubMed]
- Yamano, T.; Zetsche, B.; Ishitani, R.; Zhang, F.; Nishimasu, H.; Nureki, O. Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1. Mol Cell 2017, 67, 633–645 e633. [Google Scholar] [CrossRef]
- Kim, H.K.; Song, M.; Lee, J.; Menon, A.V.; Jung, S.; Kang, Y.M.; Choi, J.W.; Woo, E.; Koh, H.C.; Nam, J.W.; et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat Methods 2017, 14, 153–159. [Google Scholar] [CrossRef]
- Lu, S.; Tong, X.; Han, Y.; Zhang, K.; Zhang, Y.; Chen, Q.; Duan, J.; Lei, X.; Huang, M.; Qiu, Y.; et al. Fast and sensitive detection of SARS-CoV-2 RNA using suboptimal protospacer adjacent motifs for Cas12a. Nat Biomed Eng 2022, 6, 286–297. [Google Scholar] [CrossRef]
- Corsi, G.I.; Qu, K.; Alkan, F.; Pan, X.; Luo, Y.; Gorodkin, J. CRISPR/Cas9 gRNA activity depends on free energy changes and on the target PAM context. Nat Commun 2022, 13, 3006. [Google Scholar] [CrossRef]
- Zhuo, C.; Zhang, J.; Lee, J.-H.; Jiao, J.; Cheng, D.; Liu, L.; Kim, H.-W.; Tao, Y.; Li, M. Spatiotemporal control of CRISPR/Cas9 gene editing. Signal Transduction and Targeted Therapy 2021, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Brown, W.; Bardhan, A.; Delaney, M.; Ilk, A.S.; Rauen, R.R.; Kahn, S.I.; Tsang, M.; Deiters, A. Spatiotemporal Control of CRISPR/Cas9 Function in Cells and Zebrafish using Light-Activated Guide RNA. Angewandte Chemie International Edition 2020, 59, 8998–9003. [Google Scholar] [CrossRef]
- Jain, P.K.; Ramanan, V.; Schepers, A.G.; Dalvie, N.S.; Panda, A.; Fleming, H.E.; Bhatia, S.N. Development of Light-Activated CRISPR Using Guide RNAs with Photocleavable Protectors. Angewandte Chemie (International ed. in English) 2016, 55, 12440–12444. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Qiu, Z.; Bi, Z.; Tian, T.; Jiang, Y.; Zhou, X. Photocontrolled crRNA activation enables robust CRISPR-Cas12a diagnostics. Proc Natl Acad Sci U S A 2022, 119, e2202034119. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Liu, R.; Qiu, Z.; Cao, F.; Tian, T.; Lu, Y.; Jiang, Y.; Zhou, X. Light-Start CRISPR-Cas12a Reaction with Caged crRNA Enables Rapid and Sensitive Nucleic Acid Detection. Angewandte Chemie International Edition 2023, 62. [Google Scholar] [CrossRef]
- Pang, B.; Xu, J.; Liu, Y.; Peng, H.; Feng, W.; Cao, Y.; Wu, J.; Xiao, H.; Pabbaraju, K.; Tipples, G.; et al. Isothermal Amplification and Ambient Visualization in a Single Tube for the Detection of SARS-CoV-2 Using Loop-Mediated Amplification and CRISPR Technology. Anal Chem 2020, 92, 16204–16212. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shi, Y.; Chen, Y.; Yang, Z.; Wu, H.; Zhou, Z.; Li, J.; Ping, J.; He, L.; Shen, H.; et al. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in the point-of-care detection. Biosens Bioelectron 2020, 169, 112642. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Liu, Y.; Yang, M.; Zheng, J.; Liu, C.; Ye, W.; Song, S.; Bai, T.; Song, C.; Wang, M.; et al. The engineered CRISPR-Mb2Cas12a variant enables sensitive and fast nucleic acid-based pathogens diagnostics in the field. Plant Biotechnology Journal 2023, 21, 1465–1478. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, B.; Yang, L.; Zhao, C.; Wang, Y.; Tang, Y.; Yang, G.; Wang, P.; Gao, S. CRISPR/Cas12a combined with recombinase polymerase amplification for rapid and sensitive detection of <italic>Vibrio vulnificus</italic> in one tube. Acta Biochimica et Biophysica Sinica 2023, 55, 322–326. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Smith, B.M.; Jain, P.K. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat Commun 2020, 11, 4906. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhou, X. CRISPR-Based Biosensing Strategies: Technical Development and Application Prospects. Annual Review of Analytical Chemistry 2023, 16, 311–332. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhao, W.; Ma, S.; Li, Z.; Yao, Y.; Fei, T. A chemical-enhanced system for CRISPR-Based nucleic acid detection. Biosens Bioelectron 2021, 192, 113493. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, M.; Merlo, R.; Bagheri, N.; Moscone, D.; Valenti, A.; Saha, A.; Arantes, P.R.; Ippodrino, R.; Ricci, F.; Treglia, I.; et al. Enhancement of CRISPR/Cas12a trans-cleavage activity using hairpin DNA reporters. Nucleic Acids Res 2022, 50, 8377–8391. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Hu, R.; Xia, J.; Xu, Z.; Chen, D.; Xi, J.; Liu, B.F.; Zhu, J.; Li, Y.; Yang, Y.; et al. G-triplex: A new type of CRISPR-Cas12a reporter enabling highly sensitive nucleic acid detection. Biosens Bioelectron 2021, 187, 113292. [Google Scholar] [CrossRef]
- Lobato, I.M.; O'Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. Trends Analyt Chem 2018, 98, 19–35. [Google Scholar] [CrossRef]
| Discipline | Strength | Weakness |
| Morphology | ·Accuracy (gold standard) ·Can detect multiple species at the same time |
·Lower sensitivity ·Difficulty distinguishing parasite-like egg ·High demand for professional skills |
| Immunology | ·Strong specificity ·Strong sensitivity |
·High cost and time consuming ·False positives for cross-reactivity ·False negatives in immunocompromised patients ·Inability to differentiate between ongoing and past infections |
| Molecular biology | ·Strong specificity ·Strong sensitivity ·Strong repeatability |
·High cost ·Limitations related to sample preparation and equipment ·Logistics systems requiring fresh sample analysis (e.g. cryogenic) |
| Parasite | Repeat Sequence Name | Length(bp) | Quantity | GenBank Accession | Refs |
|---|---|---|---|---|---|
| Protozoa | |||||
| Trypanosoma cruzi | TCNRE | 195 | 12% of the total genome | K01772 | [20] |
| Toxoplasma gondii | / | 529 | 200-300 copies per genome | AF146527 | [21] |
| Plasmodium falciparum | Pfr364 | 716 | 41 copies per genome | / | [22,23] |
| Plasmodium vivax | Pvr47 | 333 | 14 copies per genome | / | [22,23,24] |
| Cestodes | |||||
| Echinococcus granulosus | EgG1 Hae III repeat | 269 | 6900 copies per haploid genome (1% of E. granulosus genomic DNA) |
DQ157697 | [25,26] |
| Taenia solium | Tsol-9 | 158 | None Related Description | U45987 | [27] |
| Taenia saginata | HDP1 | 1272 | 0.4% of the T. saginata DNA | AJ133764 | [28] |
| Trematodes | |||||
| Schistosoma mansoni | Sml-7(DraI) | 121 | 12% of the total genome | M61098 | [29,30,31] |
| Schistosoma haematobium | DraI | 121 | over 15% of the S. haematobium genome | DQ157698 | [32] |
| Trichobilharzia ocellata | ToSau3A | 396 | 10,000 copies per haploid genome (1.5% of the T. ocellata genome) |
AF442689 | [33] |
| Nematodes | |||||
| Strongyloides stercoralis | / | 765 | None Related Description | AY028262 | [34] |
| Brugia malayi | HhaI repeat | 320 | several thousand copies per haploid genome (about 12% of the genome) |
M12691 | [35,36] |
| Wuchereria bancrofti | SspI | 195 | 300 copies per haploid genome | L20344 | [37] |
| LDR | 1674 | None Related Description | AY297458 | [38] | |
| Onchocerca volvulus | O-150 | 149 | 4500 copies per haploid genome | J04659 | [39,40,41] |
| Parafilaroides decorus | Pd65 | 689 | None Related Description | MT053285 | [42] |
| Technology | Device dependency | Specificity | Reaction time (min) |
Number of primers | Quantification | Cost | Results View Method | POCT potential |
| PCR | Moderate | Strong | 60-180 | 2 | No | High | Gel electrophoresis | Moderate |
| q-PCR (qRT-PCR) |
High | Strong | >60 | 2 | Yes | Extremely high | Fluorescent and computer system | LOW |
| d-PCR | High | Strong | >60 | 2 | Yes | Extremely high | Fluorescent and computer system | LOW |
| LAMP | Low | Strong | <60 | 4-6 | No | Low | ·Gel electrophoresis ·Color ·Turbidity |
High |
| RPA | Low | Moderate | 20-60 | 2 | No | Low | ·Gel electrophoresis ·Fluorescent ·Lateral flow |
High |
| Cas12a | Low | Strong | 20-60 | 2 | No | Low | ·fluorescent ·Lateral flow |
High |
| Species | Method | Time (min) | LOD | Specificity | Refs |
|---|---|---|---|---|---|
| Plasmodium falciparum | Cas12a-RPA | 30 (+10) a | 0.36 parasites/μL | 100% | [56] |
| Plasmodium vivax | Cas12a-RPA | 30 (+10) a | 1.2 parasites/μL | 100% | |
| Toxoplasma gondii | Cas12a-RPA | 35 (+20) a | 99~115 copies/μL b | 100% | [58] |
| Cryptosporidium parvum | Cas12a-RPA (two steps) | 30 + 60 (+20) a | 10 oocysts | 100% | [60] |
| Cas12a-RPA | 90 | 1 oocyst | 100% | [61] | |
| Enterocytozoon hepatopenaei | Cas12a-RPA | 60 | 50 copies/μL b | 100% | [62] |
| Heterodera schachtii | Cas12a-RPA | 60 | 10-4 single cysts | 100% | [63] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





