Submitted:
02 December 2023
Posted:
05 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Defining fibroblasts
3. Fibroblast activation and CAF markers
3.1. Origin
3.2. Fibroblast activation
3.3. Fibroblast markers
4. CAF subtypes in various cancer types
4.1. Breast cancer
4.2. Pancreatic and gastric cancer
4.3. Hepatocellular carcinoma and cholangiocarcinoma
5. CAF subtypes in oral cancer
| Subtype | Marker | Transcriptome | Pathways | Additional notes | Reference |
|---|---|---|---|---|---|
| myCAF | MHCII-/Ly6C- [9,58] | Acta2, contractile proteins (Tagln, Myl9, Tpm1, Tpm2, Mmp11, Postn, Hopx), Igfbp3, Thy1, Col12a1, Thbs2 [9] | Smooth muscle contraction, focal adhesion, ECM organization, collagen formation [9] | Activates through: - TGFβ [8] |
[8,9,10,55,56,58,59,63,65,66,68] |
| αSMA+ IL-6- [10] | Acta2, TGFβ response genes (Ctgf and Col1a1) [10] | Smooth muscle contraction, vascular wound healing [63] | |||
| αSMA [59] | Contractile proteins (Acta2, Tagln, Myl9, Tnc, Tpm1, Tpm2), growth factors (Tgfb1, Tgfb2, Ccn2/Ctgf, Pgf, Vegfa, Wnt5a, Pdgfrb), Igfbp3, Cnn2, Cnn3, Tmem119 [55] | ECM-receptor interaction, vascular smooth muscle contraction, focal adhesion [66] | |||
| Increased Acta2 and reduced Cxcl12, Des [56] | Contractile fiber, blood vessel development, tissue migration, ECM organization [68] | ||||
| Acta2, contractile proteins (Myh11, Mcam, Tagln, Mylk) [63] | |||||
| Pdpn, Col1a1 [65] | |||||
| Acta2, Mylk, Myl9 [66] | |||||
| Acta2, Tagln, Mmp11, Myl9, Postn, Tpm1, Tpm2 [68] | |||||
| iCAF | Ly6C+ [9,58] | Il6, Il8, chemokines (Cxcl1, Cxcl2, Ccl2, Cxcl12), Pdgfra, Cfd, Lmna, Dpt, Has1, Has2, Agtr1 [9] | Cytokine signaling, JAK/STAT signaling [8,10] | Activates through: - NF-κB signaling through IL-1 [8] - Cancer-derived EVs [25,59] - Hypoxia induced HIF1α [58] |
[8,9,10,13,25,55,56,58,59,63,65,66,68] |
| αSMA- IL-6+ [10] | Cytokines (Il6, Il11, Lif) and chemokines (Cxcl1, Cxcl2) [10] | IFNγ response, TNF/NF-κB, IL2/STAT5, IL6/JAK/STAT3, the complement pathway [9] | |||
| CXCL8 [59] | Fbln1, Igfi, Cxcl1, Igfbp6, Slpi, Saa1, and complement genes (C3, C7) and reduced Acta2 [13] | ECM, inflammatory response regulation, complement activation [13] | |||
| Ly6c1 and inflammatory cytokines (Il6, Cxcl1, Cxcl12, Mcp-1/Ccl2, Mcp-3/Ccl7, Il33), compliment pathway members (C3, C4b, C1s1, C1s2), Clec3b, Has1, Dpt, Col14a1, Ccl7, Ly6a/Sca-1, Saa3, Ogn, Prg4, Prelp, Efemp1, Htra3, Pdgfra [55] | Inflammation (iCAF) NFκB signaling pathway (iCAF-2) [63] | Function: - Recruit M2 macrophages by secreting CXCL12 [66] |
|||
| Increased Vim, Fap, Col3a1, Des, Il6, Cxcl12 and reduced Acta2 [56] | TNF signaling pathway, IL-17 signaling pathway, ECM-receptor interaction [66] | ||||
| iCAF: Cfd, C3, Cxcl14, Cxcl12, Il33; iCAF-2: inflammatory genes (Cxcl2, Tnfaip3), Icam1, Clu, Bdkrb1, Relb [63] | Blood vessel development, tissue migration, ECM organization [68] | ||||
| Cxcl12, Cxcl14, Igf1 [65] | |||||
| Cxcl12, Pdgfra, Thy1 [66] | |||||
| Chemokines (Pdgfra, Cxcl12, Cfd, Dpt, Lmna, Cxcl2, Ccl2) [68] | |||||
| apCAF | MHCII+ [9,58] | MHC-II genes (Cd74, H2-Aa, H2-Ab1), Saa3, Slpi [9] | Antigen presentation and processing, fatty-acid metabolism, MYC targets, MTORC1 signaling [9] | Function: - Activate CD4+ T cells [9] |
[9,13,55,57,58,65] |
| MHC-II (Cd74, Hla-Dra, Hla-Drb1) [13] | Leukocyte cell-cell adhesion, response to IFN-γ, antigen processing, and antigen presentation via MHCII [13] | ||||
| MHC-II (Cd74, H2-Aa, H2-Ab1, H2-Eb), Krt8, Krt18, Fsp1 [55] | Ribosome, antigen processing and presentation, phagosome [57] | ||||
| MHC-II (Cd74, Hla-Dra, Hla-Drb1, Hla-Dqb1, Hla-Drb5, Hla-Dpa1, Hla-Dpb1) [65] | |||||
| Matrix CAF (mCAF) [13] Cluster 3 [57] mCAF [62] dCAF [63] |
Fibulin-1 and PDGFRα [62] | High levels of ECM signatures (Col5a1, Col5a2, Col6a3), Postn, Fn1, Lum, Dcn, Vcan) and low levels of Acta2 [13] | ECM and collagen fibril organization [13] | [13,57,62,63] | |
| Acta2- [57] | ECM remodeling (protein processing in the endoplasmic reticulum, oxidative phosphorylation, proteoglycans in cancer, ECM-receptor interaction) [57] | ||||
| ECM-related genes such as glycoproteins (Dcn, Lum, and Vcan), structural proteins (Col14a1), matricellular proteins (Fbln1, Fbln2, and Smoc), and matrix-modifying enzymes (Lox and Loxl1) , Cxcl14 [62] | ECM signature [62] | ||||
| Col1a1, Col3a1, Stc1, Mmp1, Mmp11, Col10a1 [63] | ECM remodeling [63] | ||||
| S1 | CD29Med FAPHi αSMAHi FSP1Low-Hi PDGFRβMed-Hi CAV1Low [6,7,54] |
Function: - Promotes immunosuppression [7,54] - Increases migration properties of luminal BC cells [6] |
[6,7,54] | ||
| S4 | CD29Hi FAPLow αSMAHi FSP1Hi PDGFRβMed-Hi CAV1Neg-Low [6,7,54] |
[6,7,54] |
6. The prognostic role of CAF subtypes in oral cancer
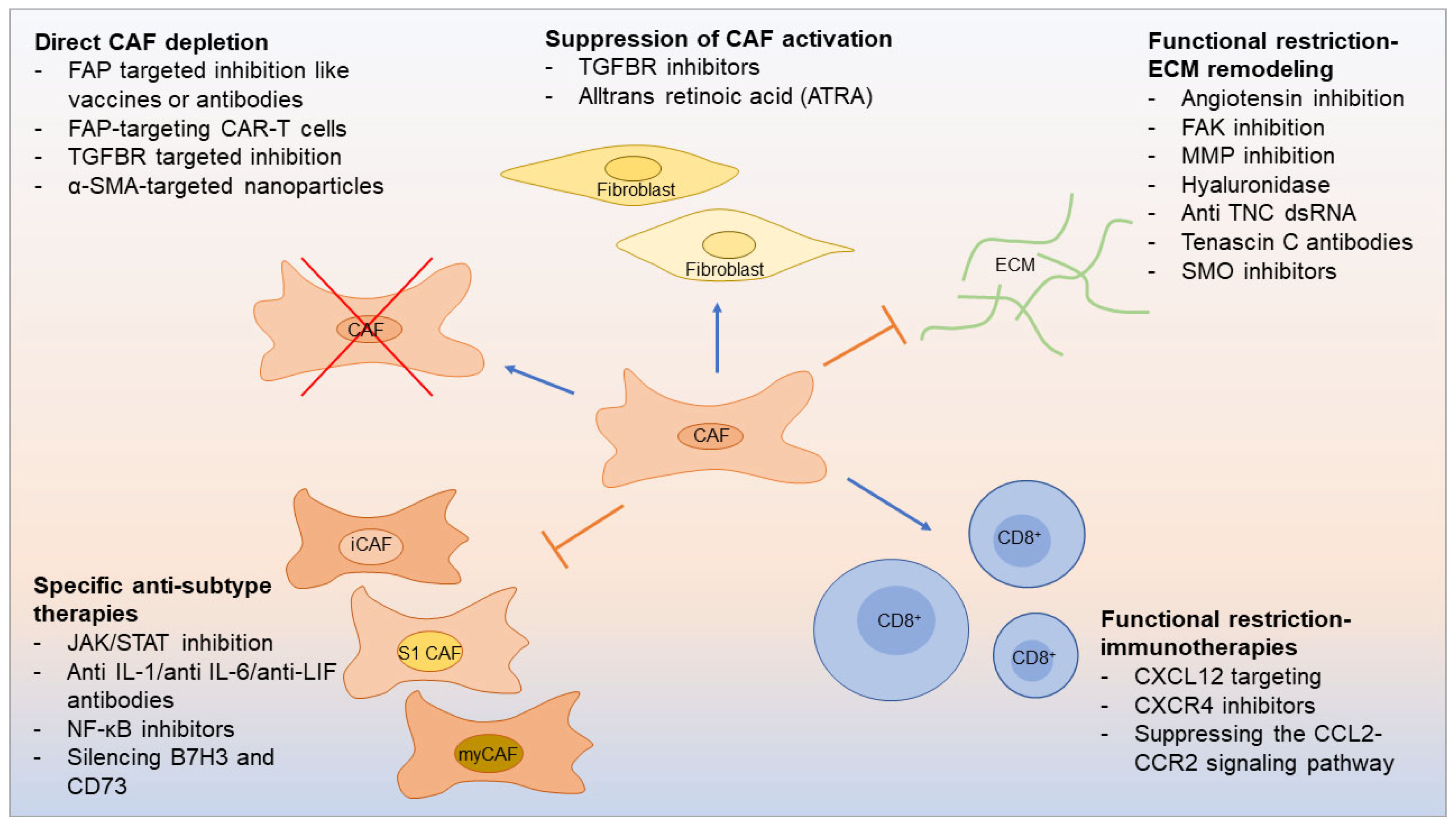
7. Therapeutic opportunities linked to CAF subtypes
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Suresh, G. M., R. Koppad, B. V. Prakash, K. S. Sabitha and P. S. Dhara. Prognostic indicators of oral squamous cell carcinoma. Ann Maxillofac Surg 2019, 9, 364–370. [CrossRef] [PubMed]
- American cancer society. Cancer facts & figures 2023; Atlanta, Ga: American Cancer Society, 2023.
- Zainab, H., A. Sultana and Shaimaa. Stromal desmoplasia as a possible prognostic indicator in different grades of oral squamous cell carcinoma. J Oral Maxillofac Pathol 2019, 23, 338–343. [CrossRef] [PubMed]
- Graizel, D., A. Zlotogorski-Hurvitz, I. Tsesis, E. Rosen, R. Kedem and M. Vered. Oral cancer-associated fibroblasts predict poor survival: Systematic review and meta-analysis. Oral Dis 2020, 26, 733–744. [CrossRef] [PubMed]
- Xiao, Z., L. Todd, L. Huang, E. Noguera-Ortega, Z. Lu, L. Huang, M. Kopp, Y. Li, N. Pattada, W. Zhong, et al. Desmoplastic stroma restricts t cell extravasation and mediates immune exclusion and immunosuppression in solid tumors. Nat Commun 2023, 14, 5110. [CrossRef] [PubMed]
- Bonneau, C., A. Eliès, Y. Kieffer, B. Bourachot, S. Ladoire, F. Pelon, D. Hequet, J. M. Guinebretière, C. Blanchet, A. Vincent-Salomon, et al. A subset of activated fibroblasts is associated with distant relapse in early luminal breast cancer. Breast Cancer Res 2020, 22, 76. [CrossRef] [PubMed]
- Costa, A., Y. Kieffer, A. Scholer-Dahirel, F. Pelon, B. Bourachot, M. Cardon, P. Sirven, I. Magagna, L. Fuhrmann, C. Bernard, et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell 2018, 33, 463–479.e10. [CrossRef] [PubMed]
- Biffi, G., T. E. Oni, B. Spielman, Y. Hao, E. Elyada, Y. Park, J. Preall and D. A. Tuveson. Il1-induced jak/stat signaling is antagonized by tgfβ to shape caf heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov 2019, 9, 282–301. [CrossRef]
- Elyada, E., M. Bolisetty, P. Laise, W. F. Flynn, E. T. Courtois, R. A. Burkhart, J. A. Teinor, P. Belleau, G. Biffi, M. S. Lucito, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov 2019, 9, 1102–1123. [CrossRef]
- Öhlund, D., A. Handly-Santana, G. Biffi, E. Elyada, A. S. Almeida, M. Ponz-Sarvise, V. Corbo, T. E. Oni, S. A. Hearn, E. J. Lee, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med 2017, 214, 579–596. [CrossRef]
- Chiavarina, B., R. Ronca, Y. Otaka, R. B. Sutton, S. Rezzola, T. Yokobori, P. Chiodelli, R. Souche, D. Pourquier, A. Maraver, et al. Fibroblast-derived prolargin is a tumor suppressor in hepatocellular carcinoma. Oncogene 2022, 41, 1410–1420. [CrossRef]
- Ying, F., M. S. M. Chan and T. K. W. Lee. Cancer-associated fibroblasts in hepatocellular carcinoma and cholangiocarcinoma. Cell Mol Gastroenterol Hepatol 2023, 15, 985–999. [CrossRef] [PubMed]
- Zhang, M., H. Yang, L. Wan, Z. Wang, H. Wang, C. Ge, Y. Liu, Y. Hao, D. Zhang, G. Shi, et al. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J Hepatol 2020, 73, 1118–1130. [CrossRef] [PubMed]
- Dhanda, J., A. Triantafyllou, T. Liloglou, H. Kalirai, B. Lloyd, R. Hanlon, R. J. Shaw, D. R. Sibson and J. M. Risk. Serpine1 and sma expression at the invasive front predict extracapsular spread and survival in oral squamous cell carcinoma. Br J Cancer 2014, 111, 2114–2121. [CrossRef] [PubMed]
- Kellermann, M. G., L. M. Sobral, S. D. da Silva, K. G. Zecchin, E. Graner, M. A. Lopes, I. Nishimoto, L. P. Kowalski and R. D. Coletta. Myofibroblasts in the stroma of oral squamous cell carcinoma are associated with poor prognosis. Histopathology 2007, 51, 849–853. [CrossRef] [PubMed]
- Vered, M., A. Dobriyan, D. Dayan, R. Yahalom, Y. P. Talmi, L. Bedrin, I. Barshack and S. Taicher. Tumor-host histopathologic variables, stromal myofibroblasts and risk score, are significantly associated with recurrent disease in tongue cancer. Cancer Sci 2010, 101, 274–280. [CrossRef] [PubMed]
- Vered, M., A. Shnaiderman-Shapiro, A. Zlotogorski-Hurvitz, T. Salo and R. Yahalom. Cancer-associated fibroblasts in the tumor microenvironment of tongue carcinoma is a heterogeneous cell population. Acta Histochem 2019, 121, 151446. [CrossRef]
- Marsh, D., K. Suchak, K. A. Moutasim, S. Vallath, C. Hopper, W. Jerjes, T. Upile, N. Kalavrezos, S. M. Violette, P. H. Weinreb, et al. Stromal features are predictive of disease mortality in oral cancer patients. J Pathol 2011, 223, 470–481. [CrossRef]
- Plikus, M. V., X. Wang, S. Sinha, E. Forte, S. M. Thompson, E. L. Herzog, R. R. Driskell, N. Rosenthal, J. Biernaskie and V. Horsley. Fibroblasts: Origins, definitions, and functions in health and disease. Cell 2021, 184, 3852–3872. [CrossRef]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef]
- Garcia, P. E., M. Adoumie, E. C. Kim, Y. Zhang, M. K. Scales, Y. S. El-Tawil, A. Z. Shaikh, H. J. Wen, F. Bednar, B. L. Allen, et al. Differential contribution of pancreatic fibroblast subsets to the pancreatic cancer stroma. Cell Mol Gastroenterol Hepatol 2020, 10, 581–599. [CrossRef]
- Helms, E. J., M. W. Berry, R. C. Chaw, C. C. DuFort, D. Sun, M. K. Onate, C. Oon, S. Bhattacharyya, H. Sanford-Crane, W. Horton, et al. Mesenchymal lineage heterogeneity underlies nonredundant functions of pancreatic cancer-associated fibroblasts. Cancer Discov 2022, 12, 484–501. [CrossRef] [PubMed]
- Hutton, C., F. Heider, A. Blanco-Gomez, A. Banyard, A. Kononov, X. Zhang, S. Karim, V. Paulus-Hock, D. Watt, N. Steele, et al. Single-cell analysis defines a pancreatic fibroblast lineage that supports anti-tumor immunity. Cancer Cell 2021, 39, 1227–1244.e20. [CrossRef] [PubMed]
- Lawson, J., C. Dickman, R. Towle, J. Jabalee, A. Javer and C. Garnis. Extracellular vesicle secretion of mir-142-3p from lung adenocarcinoma cells induces tumor promoting changes in the stroma through cell-cell communication. Mol Carcinog 2019, 58, 376–387. [CrossRef] [PubMed]
- Arebro, J., R. Towle, C. M. Lee, K. L. Bennewith and C. Garnis. Extracellular vesicles promote activation of pro-inflammatory cancer-associated fibroblasts in oral cancer. Front Cell Dev Biol 2023, 11, 1240159. [CrossRef] [PubMed]
- Yeon, J. H., H. E. Jeong, H. Seo, S. Cho, K. Kim, D. Na, S. Chung, J. Park, N. Choi and J. Y. Kang. Cancer-derived exosomes trigger endothelial to mesenchymal transition followed by the induction of cancer-associated fibroblasts. Acta Biomater 2018, 76, 146–153. [CrossRef] [PubMed]
- Krzysiek-Maczka, G., A. Targosz, U. Szczyrk, M. Strzalka, T. Brzozowski and A. Ptak-Belowska. Involvement of epithelial-mesenchymal transition-inducing transcription factors in the mechanism of helicobacter pylori-induced fibroblasts activation. J Physiol Pharmacol 2019, 70. [CrossRef] [PubMed]
- Tan, H. X., Z. B. Cao, T. T. He, T. Huang, C. L. Xiang and Y. Liu. Tgfβ1 is essential for mscs-cafs differentiation and promotes hct116 cells migration and invasion via jak/stat3 signaling. Onco Targets Ther 2019, 12, 5323–5334. [CrossRef] [PubMed]
- Abangan, R. S., Jr., C. R. Williams, M. Mehrotra, J. D. Duncan and A. C. Larue. Mcp1 directs trafficking of hematopoietic stem cell-derived fibroblast precursors in solid tumor. Am J Pathol 2010, 176, 1914–1926. [CrossRef] [PubMed]
- Nair, N., A. S. Calle, M. H. Zahra, M. Prieto-Vila, A. K. K. Oo, L. Hurley, A. Vaidyanath, A. Seno, J. Masuda, Y. Iwasaki, et al. A cancer stem cell model as the point of origin of cancer-associated fibroblasts in tumor microenvironment. Sci Rep 2017, 7, 6838. [CrossRef]
- Bochet, L., C. Lehuédé, S. Dauvillier, Y. Y. Wang, B. Dirat, V. Laurent, C. Dray, R. Guiet, I. Maridonneau-Parini, S. Le Gonidec, et al. Adipocyte-derived fibroblasts promote tumor progression and contribute to the desmoplastic reaction in breast cancer. Cancer Res 2013, 73, 5657–5668. [CrossRef]
- Ning, X., H. Zhang, C. Wang and X. Song. Exosomes released by gastric cancer cells induce transition of pericytes into cancer-associated fibroblasts. Med Sci Monit 2018, 24, 2350–2359. [CrossRef]
- Tan, H. X., W. Z. Gong, K. Zhou, Z. G. Xiao, F. T. Hou, T. Huang, L. Zhang, H. Y. Dong, W. L. Zhang, Y. Liu, et al. Cxcr4/tgf-β1 mediated hepatic stellate cells differentiation into carcinoma-associated fibroblasts and promoted liver metastasis of colon cancer. Cancer Biol Ther 2020, 21, 258–268. [CrossRef] [PubMed]
- De Wever, O., Q. D. Nguyen, L. Van Hoorde, M. Bracke, E. Bruyneel, C. Gespach and M. Mareel. Tenascin-c and sf/hgf produced by myofibroblasts in vitro provide convergent pro-invasive signals to human colon cancer cells through rhoa and rac. Faseb j 2004, 18, 1016–1018. [CrossRef] [PubMed]
- Radhakrishnan, R., J. H. Ha, M. Jayaraman, J. Liu, K. M. Moxley, C. Isidoro, A. K. Sood, Y. S. Song and D. N. Dhanasekaran. Ovarian cancer cell-derived lysophosphatidic acid induces glycolytic shift and cancer-associated fibroblast-phenotype in normal and peritumoral fibroblasts. Cancer Lett 2019, 442, 464–474. [CrossRef]
- Su, Q., B. Zhang, L. Zhang, T. Dang, D. Rowley, M. Ittmann and L. Xin. Jagged1 upregulation in prostate epithelial cells promotes formation of reactive stroma in the pten null mouse model for prostate cancer. Oncogene 2017, 36, 618–627. [CrossRef] [PubMed]
- Strell, C., J. Paulsson, S. B. Jin, N. P. Tobin, A. Mezheyeuski, P. Roswall, C. Mutgan, N. Mitsios, H. Johansson, S. M. Wickberg, et al. Impact of epithelial-stromal interactions on peritumoral fibroblasts in ductal carcinoma in situ. J Natl Cancer Inst 2019, 111, 983–995. [CrossRef] [PubMed]
- Sanz-Moreno, V., C. Gaggioli, M. Yeo, J. Albrengues, F. Wallberg, A. Viros, S. Hooper, R. Mitter, C. C. Féral, M. Cook, et al. Rock and jak1 signaling cooperate to control actomyosin contractility in tumor cells and stroma. Cancer Cell 2011, 20, 229–245. [CrossRef]
- Erez, N., M. Truitt, P. Olson, S. T. Arron and D. Hanahan. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an nf-kappab-dependent manner. Cancer Cell 2010, 17, 135–147. [CrossRef]
- Amatangelo, M. D., D. E. Bassi, A. J. Klein-Szanto and E. Cukierman. Stroma-derived three-dimensional matrices are necessary and sufficient to promote desmoplastic differentiation of normal fibroblasts. Am J Pathol 2005, 167, 475–488. [CrossRef]
- Calvo, F., N. Ege, A. Grande-Garcia, S. Hooper, R. P. Jenkins, S. I. Chaudhry, K. Harrington, P. Williamson, E. Moeendarbary, G. Charras, et al. Mechanotransduction and yap-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol 2013, 15, 637–646. [CrossRef]
- Weinberg, F., N. Ramnath and D. Nagrath. Reactive oxygen species in the tumor microenvironment: An overview. Cancers (Basel) 2019, 11. [CrossRef] [PubMed]
- Fordyce, C., T. Fessenden, C. Pickering, J. Jung, V. Singla, H. Berman and T. Tlsty. DNA damage drives an activin a-dependent induction of cyclooxygenase-2 in premalignant cells and lesions. Cancer Prev Res (Phila) 2010, 3, 190–201. [CrossRef] [PubMed]
- Dai, G., X. Yao, Y. Zhang, J. Gu, Y. Geng, F. Xue and J. Zhang. Colorectal cancer cell-derived exosomes containing mir-10b regulate fibroblast cells via the pi3k/akt pathway. Bull Cancer 2018, 105, 336–349. [CrossRef] [PubMed]
- Ringuette Goulet, C., G. Bernard, S. Tremblay, S. Chabaud, S. Bolduc and F. Pouliot. Exosomes induce fibroblast differentiation into cancer-associated fibroblasts through tgfβ signaling. Mol Cancer Res 2018, 16, 1196–1204. [CrossRef] [PubMed]
- Ping, Q., R. Yan, X. Cheng, W. Wang, Y. Zhong, Z. Hou, Y. Shi, C. Wang and R. Li. Cancer-associated fibroblasts: Overview, progress, challenges, and directions. Cancer Gene Ther 2021, 28, 984–999. [CrossRef] [PubMed]
- Zhang, X., Y. Zhang, X. Qiu, J. Cai, Z. Yang and F. Song. Extracellular vesicles derived from lung cancer cells induce transformation of normal fibroblasts into lung cancer-associated fibroblasts and promote metastasis of lung cancer by delivering lncrna hotair. Stem Cells Int 2022, 2022, 3805013. [CrossRef] [PubMed]
- Shoucair, I., F. Weber Mello, J. Jabalee, S. Maleki and C. Garnis. The role of cancer-associated fibroblasts and extracellular vesicles in tumorigenesis. Int J Mol Sci 2020, 21, 103390/ijms21186837.
- Öhlund, D., E. Elyada and D. Tuveson. Fibroblast heterogeneity in the cancer wound. J Exp Med 2014, 211, 1503–1523. [CrossRef] [PubMed]
- Purcell, J. W., S. G. Tanlimco, J. Hickson, M. Fox, M. Sho, L. Durkin, T. Uziel, R. Powers, K. Foster, T. McGonigal, et al. Lrrc15 is a novel mesenchymal protein and stromal target for antibody-drug conjugates. Cancer Res 2018, 78, 4059–4072. [CrossRef]
- Dominguez, C. X., S. Müller, S. Keerthivasan, H. Koeppen, J. Hung, S. Gierke, B. Breart, O. Foreman, T. W. Bainbridge, A. Castiglioni, et al. Single-cell rna sequencing reveals stromal evolution into lrrc15(+) myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov 2020, 10, 232–253. [CrossRef]
- Available online: https://fibroxplorer.com/.
- Buechler, M. B., R. N. Pradhan, A. T. Krishnamurty, C. Cox, A. K. Calviello, A. W. Wang, Y. A. Yang, L. Tam, R. Caothien, M. Roose-Girma, et al. Cross-tissue organization of the fibroblast lineage. Nature 2021, 593, 575–579. [CrossRef]
- Givel, A. M., Y. Kieffer, A. Scholer-Dahirel, P. Sirven, M. Cardon, F. Pelon, I. Magagna, G. Gentric, A. Costa, C. Bonneau, et al. Mir200-regulated cxcl12β promotes fibroblast heterogeneity and immunosuppression in ovarian cancers. Nat Commun 2018, 9, 1056. [CrossRef]
- Sebastian, A., N. R. Hum, K. A. Martin, S. F. Gilmore, I. Peran, S. W. Byers, E. K. Wheeler, M. A. Coleman and G. G. Loots. Single-cell transcriptomic analysis of tumor-derived fibroblasts and normal tissue-resident fibroblasts reveals fibroblast heterogeneity in breast cancer. Cancers (Basel) 2020, 12, 103390/cancers12051307.
- Bernard, V., A. Semaan, J. Huang, F. A. San Lucas, F. C. Mulu, B. M. Stephens, P. A. Guerrero, Y. Huang, J. Zhao, N. Kamyabi, et al. Single-cell transcriptomics of pancreatic cancer precursors demonstrates epithelial and microenvironmental heterogeneity as an early event in neoplastic progression. Clin Cancer Res 2019, 25, 2194–2205. [CrossRef]
- Zhang, Q., Y. Wang, C. Xia, L. Ding, Y. Pu, X. Hu, H. Cai and Q. Hu. Integrated analysis of single-cell rna-seq and bulk rna-seq reveals distinct cancer-associated fibroblasts in head and neck squamous cell carcinoma. Ann Transl Med 2021, 9, 1017. [CrossRef]
- Schwörer, S., F. V. Cimino, M. Ros, K. M. Tsanov, C. Ng, S. W. Lowe, C. Carmona-Fontaine and C. B. Thompson. Hypoxia potentiates the inflammatory fibroblast phenotype promoted by pancreatic cancer cell-derived cytokines. Cancer Res 2023, 83, 1596–1610. [CrossRef]
- Naito, Y., Y. Yamamoto, N. Sakamoto, I. Shimomura, A. Kogure, M. Kumazaki, A. Yokoi, M. Yashiro, T. Kiyono, K. Yanagihara, et al. Cancer extracellular vesicles contribute to stromal heterogeneity by inducing chemokines in cancer-associated fibroblasts. Oncogene 2019, 38, 5566–5579. [CrossRef]
- Mizutani, Y., H. Kobayashi, T. Iida, N. Asai, A. Masamune, A. Hara, N. Esaki, K. Ushida, S. Mii, Y. Shiraki, et al. Meflin-positive cancer-associated fibroblasts inhibit pancreatic carcinogenesis. Cancer Res 2019, 79, 5367–5381. [CrossRef]
- Li, H., E. T. Courtois, D. Sengupta, Y. Tan, K. H. Chen, J. J. L. Goh, S. L. Kong, C. Chua, L. K. Hon, W. S. Tan, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet 2017, 49, 708–718. [CrossRef] [PubMed]
- Bartoschek, M., N. Oskolkov, M. Bocci, J. Lövrot, C. Larsson, M. Sommarin, C. D. Madsen, D. Lindgren, G. Pekar, G. Karlsson, et al. Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell rna sequencing. Nat Commun 2018, 9, 5150. [CrossRef] [PubMed]
- albo, P. M., Jr., X. Zang and D. Zheng. Molecular features of cancer-associated fibroblast subtypes and their implication on cancer pathogenesis, prognosis, and immunotherapy resistance. Clin Cancer Res 2021, 27, 2636–2647. [CrossRef] [PubMed]
- Hu, C., Y. Zhang, C. Wu and Q. Huang. Heterogeneity of cancer-associated fibroblasts in head and neck squamous cell carcinoma: Opportunities and challenges. Cell Death Discov 2023, 9, 124. [CrossRef] [PubMed]
- Song, H., C. Lou, J. Ma, Q. Gong, Z. Tian, Y. You, G. Ren, W. Guo, Y. Wang, K. He, et al. Single-cell transcriptome analysis reveals changes of tumor immune microenvironment in oral squamous cell carcinoma after chemotherapy. Front Cell Dev Biol 2022, 10, 914120. [CrossRef] [PubMed]
- Wang, Z., H. Zhang, Y. Zhai, F. Li, X. Shi and M. Ying. Single-cell profiling reveals heterogeneity of primary and lymph node metastatic tumors and immune cell populations and discovers important prognostic significance of ccdc43 in oral squamous cell carcinoma. Front Immunol 2022, 13, 843322. [CrossRef] [PubMed]
- Costea, D. E., A. Hills, A. H. Osman, J. Thurlow, G. Kalna, X. Huang, C. Pena Murillo, H. Parajuli, S. Suliman, K. K. Kulasekara, et al. Identification of two distinct carcinoma-associated fibroblast subtypes with differential tumor-promoting abilities in oral squamous cell carcinoma. Cancer Res 2013, 73, 3888–3901. [CrossRef] [PubMed]
- Yang, W., S. Zhang, T. Li, Z. Zhou and J. Pan. Single-cell analysis reveals that cancer-associated fibroblasts stimulate oral squamous cell carcinoma invasion via the tgf-β/smad pathway. Acta Biochim Biophys Sin (Shanghai) 2022, 55, 262–273. [CrossRef] [PubMed]
- Patel, A. K., K. Vipparthi, V. Thatikonda, I. Arun, S. Bhattacharjee, R. Sharan, P. Arun and S. Singh. A subtype of cancer-associated fibroblasts with lower expression of alpha-smooth muscle actin suppresses stemness through bmp4 in oral carcinoma. Oncogenesis 2018, 7, 78. [CrossRef]
- Bienkowska, K. J., C. J. Hanley and G. J. Thomas. Cancer-associated fibroblasts in oral cancer: A current perspective on function and potential for therapeutic targeting. Front Oral Health 2021, 2, 686337. [CrossRef]
- Almangush, A., I. Heikkinen, N. Bakhti, L. K. Mäkinen, J. H. Kauppila, M. Pukkila, J. Hagström, J. Laranne, Y. Soini, L. P. Kowalski, et al. Prognostic impact of tumour-stroma ratio in early-stage oral tongue cancers. Histopathology 2018, 72, 1128–1135. [CrossRef] [PubMed]
- Hendry, S., R. Salgado, T. Gevaert, P. A. Russell, T. John, B. Thapa, M. Christie, K. van de Vijver, M. V. Estrada, P. I. Gonzalez-Ericsson, et al. Assessing tumor-infiltrating lymphocytes in solid tumors: A practical review for pathologists and proposal for a standardized method from the international immuno-oncology biomarkers working group: Part 2: Tils in melanoma, gastrointestinal tract carcinomas, non-small cell lung carcinoma and mesothelioma, endometrial and ovarian carcinomas, squamous cell carcinoma of the head and neck, genitourinary carcinomas, and primary brain tumors. Adv Anat Pathol 2017, 24, 311–335. [CrossRef]
- Froeling, F. E., C. Feig, C. Chelala, R. Dobson, C. E. Mein, D. A. Tuveson, H. Clevers, I. R. Hart and H. M. Kocher. Retinoic acid-induced pancreatic stellate cell quiescence reduces paracrine wnt-β-catenin signaling to slow tumor progression. Gastroenterology 2011, 141, 1486–97, 97.e1-14. [CrossRef] [PubMed]
- Li, Z. L., H. L. Zhang, Y. Huang, J. H. Huang, P. Sun, N. N. Zhou, Y. H. Chen, J. Mai, Y. Wang, Y. Yu, et al. Autophagy deficiency promotes triple-negative breast cancer resistance to t cell-mediated cytotoxicity by blocking tenascin-c degradation. Nat Commun 2020, 11, 3806. [CrossRef] [PubMed]
- Cazet, A. S., M. N. Hui, B. L. Elsworth, S. Z. Wu, D. Roden, C. L. Chan, J. N. Skhinas, R. Collot, J. Yang, K. Harvey, et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat Commun 2018, 9, 2897. [CrossRef] [PubMed]
- Mao, X., J. Xu, W. Wang, C. Liang, J. Hua, J. Liu, B. Zhang, Q. Meng, X. Yu and S. Shi. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: New findings and future perspectives. Mol Cancer 2021, 20, 131. [CrossRef] [PubMed]
- Özdemir, B. C., T. Pentcheva-Hoang, J. L. Carstens, X. Zheng, C. C. Wu, T. R. Simpson, H. Laklai, H. Sugimoto, C. Kahlert, S. V. Novitskiy, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell 2015, 28, 831–833. [CrossRef] [PubMed]
- Rhim, A. D., P. E. Oberstein, D. H. Thomas, E. T. Mirek, C. F. Palermo, S. A. Sastra, E. N. Dekleva, T. Saunders, C. P. Becerra, I. W. Tattersall, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell 2014, 25, 735–747. [CrossRef]
- Lee, J. J., R. M. Perera, H. Wang, D. C. Wu, X. S. Liu, S. Han, J. Fitamant, P. D. Jones, K. S. Ghanta, S. Kawano, et al. Stromal response to hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A 2014, 111, E3091–100. [CrossRef] [PubMed]
- Biffi, G. and D. A. Tuveson. Diversity and biology of cancer-associated fibroblasts. Physiol Rev 2021, 101, 147–176. [CrossRef] [PubMed]
- Feig, C., J. O. Jones, M. Kraman, R. J. Wells, A. Deonarine, D. S. Chan, C. M. Connell, E. W. Roberts, Q. Zhao, O. L. Caballero, et al. Targeting cxcl12 from fap-expressing carcinoma-associated fibroblasts synergizes with anti-pd-l1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci U S A 2013, 110, 20212–20217. [CrossRef]
- Kraman, M., P. J. Bambrough, J. N. Arnold, E. W. Roberts, L. Magiera, J. O. Jones, A. Gopinathan, D. A. Tuveson and D. T. Fearon. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science 2010, 330, 827–830. [CrossRef]
- Lo, A., C. P. Li, E. L. Buza, R. Blomberg, P. Govindaraju, D. Avery, J. Monslow, M. Hsiao and E. Puré. Fibroblast activation protein augments progression and metastasis of pancreatic ductal adenocarcinoma. JCI Insight 2017, 2. [CrossRef] [PubMed]
- Peltier, A., R. D. Seban, I. Buvat, F. C. Bidard and F. Mechta-Grigoriou. Fibroblast heterogeneity in solid tumors: From single cell analysis to whole-body imaging. Semin Cancer Biol 2022, 86, 262–272. [CrossRef] [PubMed]
- Fridman, W. H., F. Pagès, C. Sautès-Fridman and J. Galon. The immune contexture in human tumours: Impact on clinical outcome. Nat Rev Cancer 2012, 12, 298–306. [CrossRef] [PubMed]
- Jimenez, D. G., A. Sobti, D. Askmyr, C. Sakellariou, S. C. Santos, S. Swoboda, O. Forslund, L. Greiff and M. Lindstedt. Tonsillar cancer with high cd8(+) t-cell infiltration features increased levels of dendritic cells and transcriptional regulation associated with an inflamed tumor microenvironment. Cancers 2021, 13. [CrossRef] [PubMed]
- Almangush, A., I. O. Bello, I. Heikkinen, J. Hagström, C. Haglund, L. P. Kowalski, P. Nieminen, R. D. Coletta, A. A. Mäkitie, T. Salo, et al. Stromal categorization in early oral tongue cancer. Virchows Arch 2021, 478, 925–932. [CrossRef] [PubMed]
- Harper, J. and R. C. Sainson. Regulation of the anti-tumour immune response by cancer-associated fibroblasts. Semin Cancer Biol 2014, 25, 69–77. [CrossRef] [PubMed]
- Liu, T., C. Han, S. Wang, P. Fang, Z. Ma, L. Xu and R. Yin. Cancer-associated fibroblasts: An emerging target of anti-cancer immunotherapy. J Hematol Oncol 2019, 12, 86. [CrossRef] [PubMed]
- Karakasheva, T. A., E. W. Lin, Q. Tang, E. Qiao, T. J. Waldron, M. Soni, A. J. Klein-Szanto, V. Sahu, D. Basu, S. Ohashi, et al. Il-6 mediates cross-talk between tumor cells and activated fibroblasts in the tumor microenvironment. Cancer Res 2018, 78, 4957–4970. [CrossRef]
| Cancer form | CAF subtype | Subtyping method | Reference | Year |
|---|---|---|---|---|
| Breast | CAF-S1, -S2, -S3, -S4 | Flow cytometry and IHC | [6,7] | 2018, 2020 |
| myCAF, iCAF, apCAF. Six clusters | ScRNA-seq | [55] | 2020 | |
| vCAF, mCAF, dCAF | ScRNA-seq, IHC | [62] | 2018 | |
| Ovarian | CAF-S1, -S2, -S3, -S4 | Flow cytometry and IHC | [54] | 2018 |
| PDAC * | myCAF, iCAF | IF, IHC, flow cytometry, RNA in situ hybridization, RNA-seq | [10] | 2017 |
| myCAF, iCAF, apCAF | RNA-seq, RNA in situ hybridization, flow cytometry, IHC | [9] | 2019 | |
| myCAF, iCAF | ScRNA-seq | [8] | 2019 | |
| myCAF, iCAF | Single-cell digital microdissection | [56] | 2019 | |
| myCAF, iCAF | Flow cytometry | [58] | 2023 | |
| DGC * | myCAF, iCAF | IF | [59] | 2019 |
| ICC * | vCAF, mCAF, iCAF, apCAF, eCAF, lipofibroblast | ScRNA-seq | [13] | 2020 |
| HCC * | CAF_Port, CAF_HSC, CAF_VSMC | Proteomics, scRNA-seq | [11] | 2022 |
| CRC * | CAF-A, CAF-B (myofibroblast markers) | ScRNA-seq | [61] | 2017 |
| HNSCC * | CAF cluster 0-8 (cluster 4: apCAF) | ScRNA-seq | [57] | 2021 |
| OSCC * | iCAF, apCAF, myCAF | ScRNA-seq | [65] | 2022 |
| iCAF, myo-CAF (mCAF) | ScRNA-seq | [66] | 2022 | |
| CAF-N, CAF-D | RT PCR | [67] | 2013 | |
| Myofibroblast (mCAF), iCAF | ScRNA-seq | [68] | 2022 | |
| Melanoma, HNSCC, LC * | myCAF, dCAF, iCAF, iCAF-2, nCAF, pCAF | ScRNA-seq | [63] | 2021 |
| Gingivobuccal OC * | C1-type CAF, C2-type CAF | Gene expression microarray | [69] | 2018 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




