Submitted:
27 December 2023
Posted:
28 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Experimental Procedures
2.1. Antibodies and reagents
2.2. Cell culture
2.3. Screening for E3 ubiquitin ligases that interact with tyrosinase
2.4. Plasmid construction and transfection, siRNA and transfection
2.5. Melanin content measurement
2.6. IP and Western blotting
2.7. Confocal immunofluorescence microscopy
2.8. Statistical analyses
3. Results
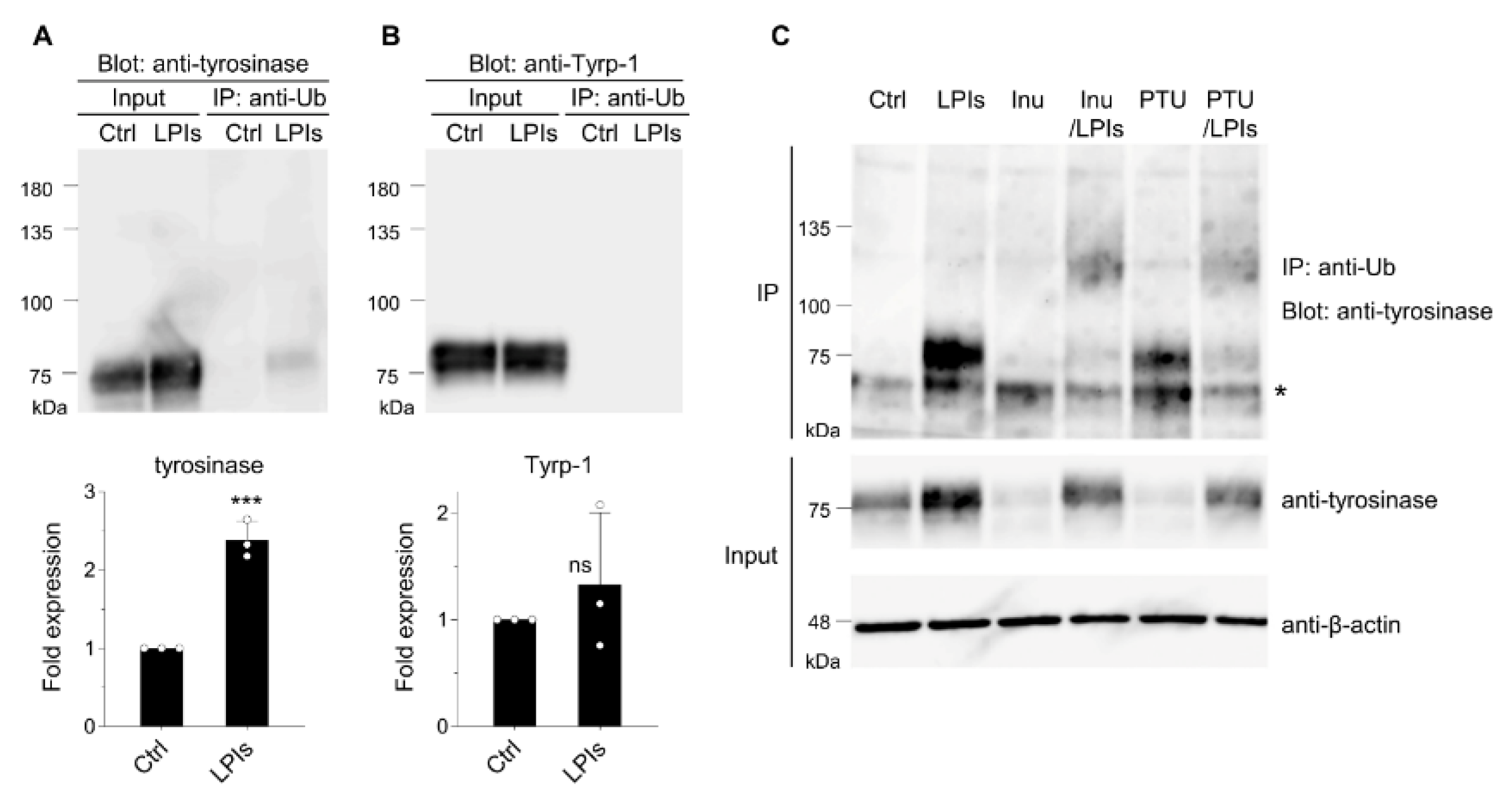
3.1. Tyrosinase ubiquitination in B16 melanoma cells
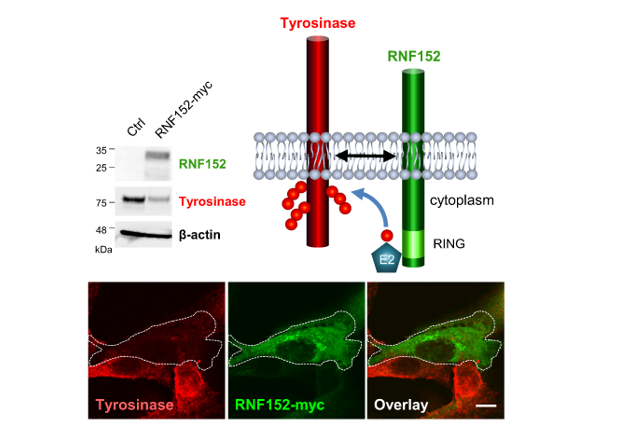
3.2. Identification of tyrosinase-specific E3 ubiquitin ligase
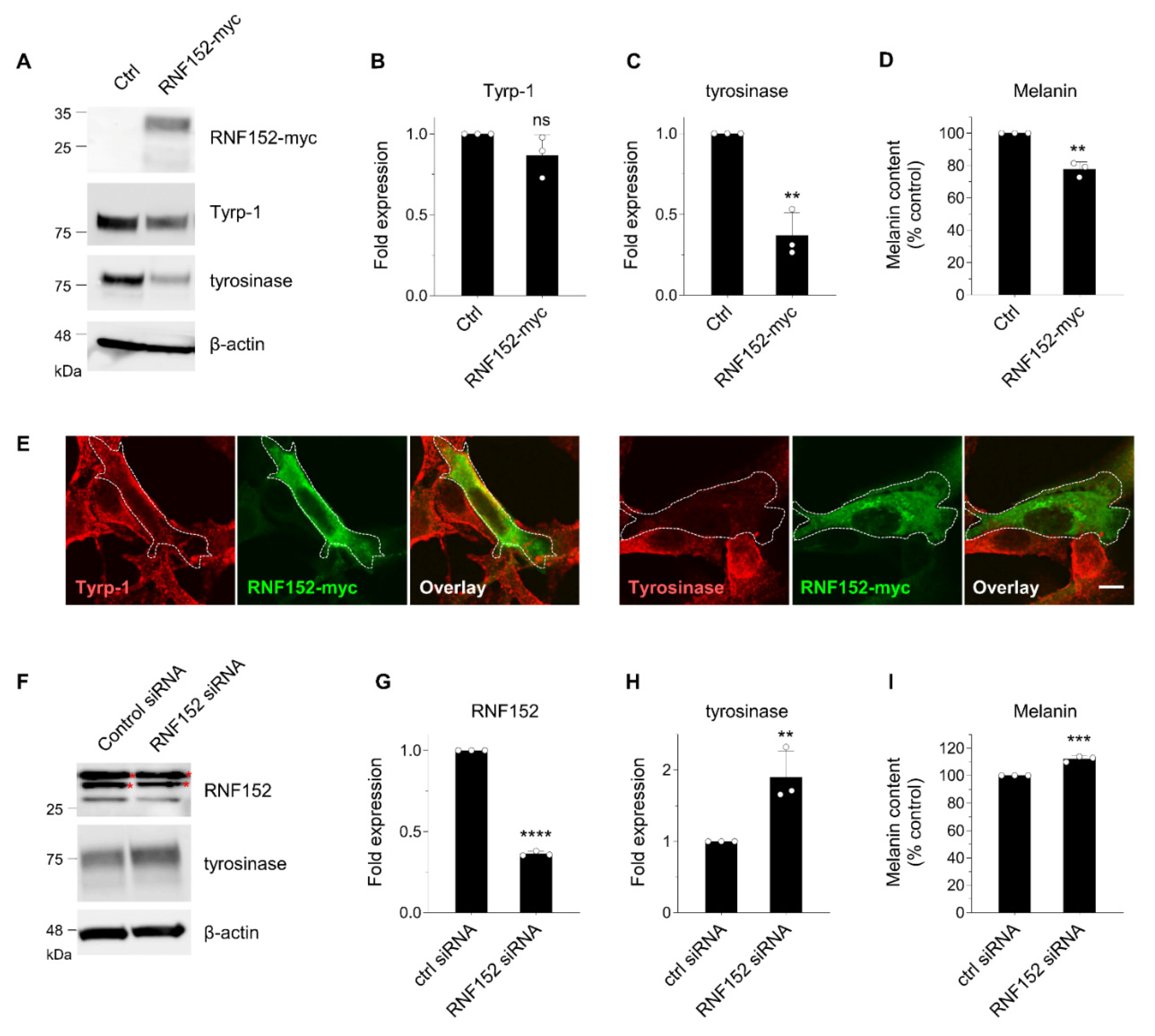
3.3. RNF152 regulates expression of tyrosinase in B16 cells.
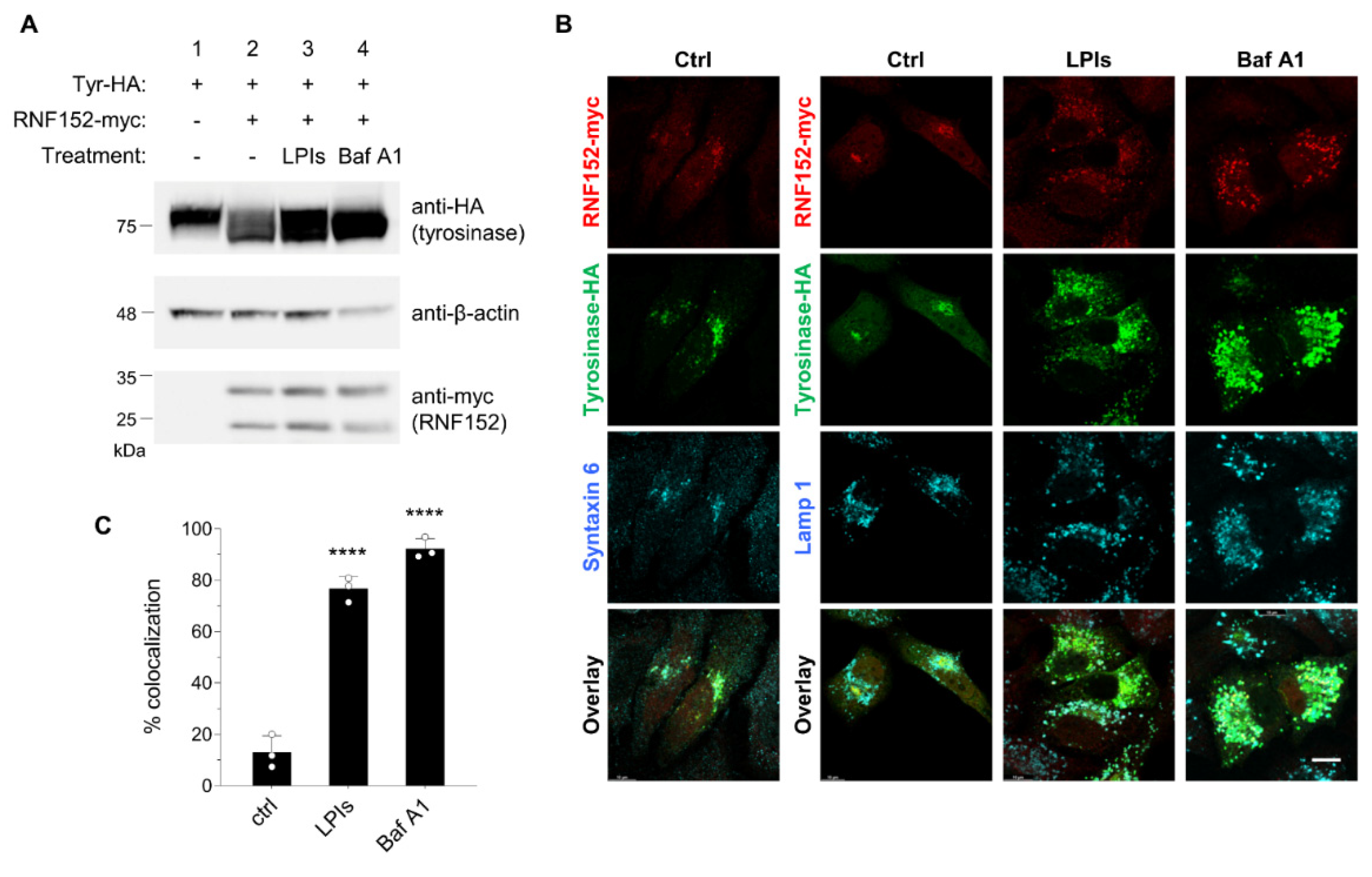
3.4. RNF152 co-localizes with tyrosinase in TGN and degrades it in lysosomes.
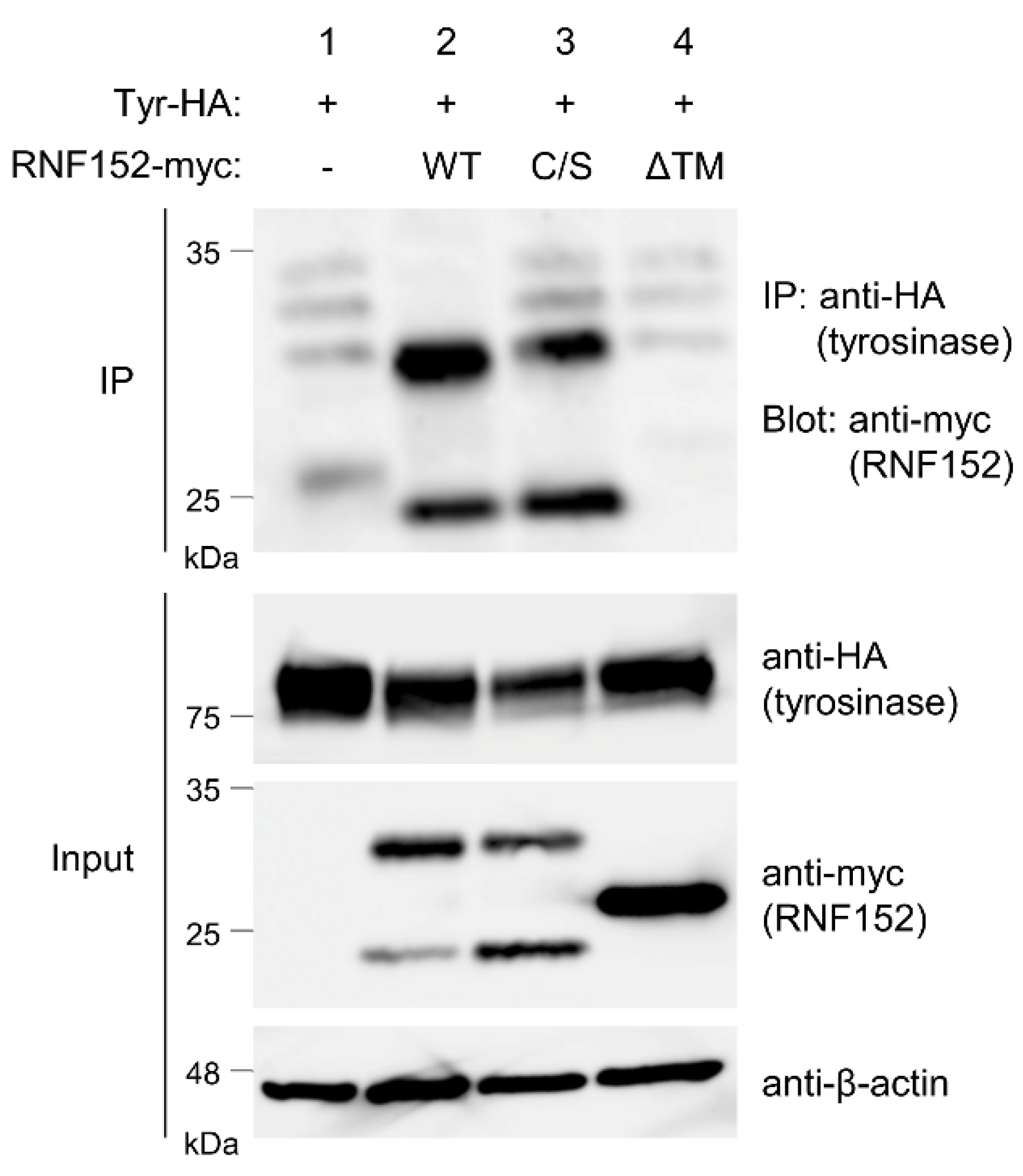
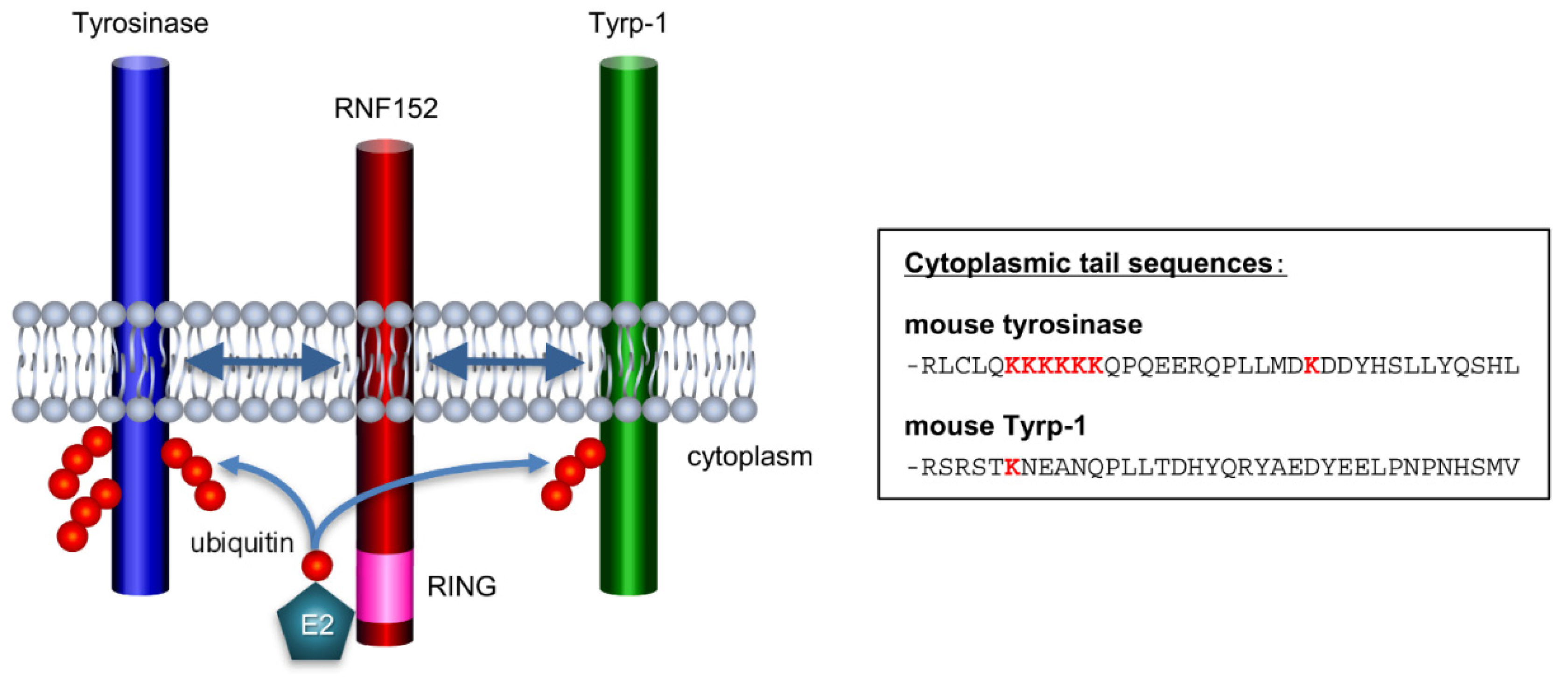
3.5. RNF152 is associated with tyrosinase through the transmembrane (TM) domain
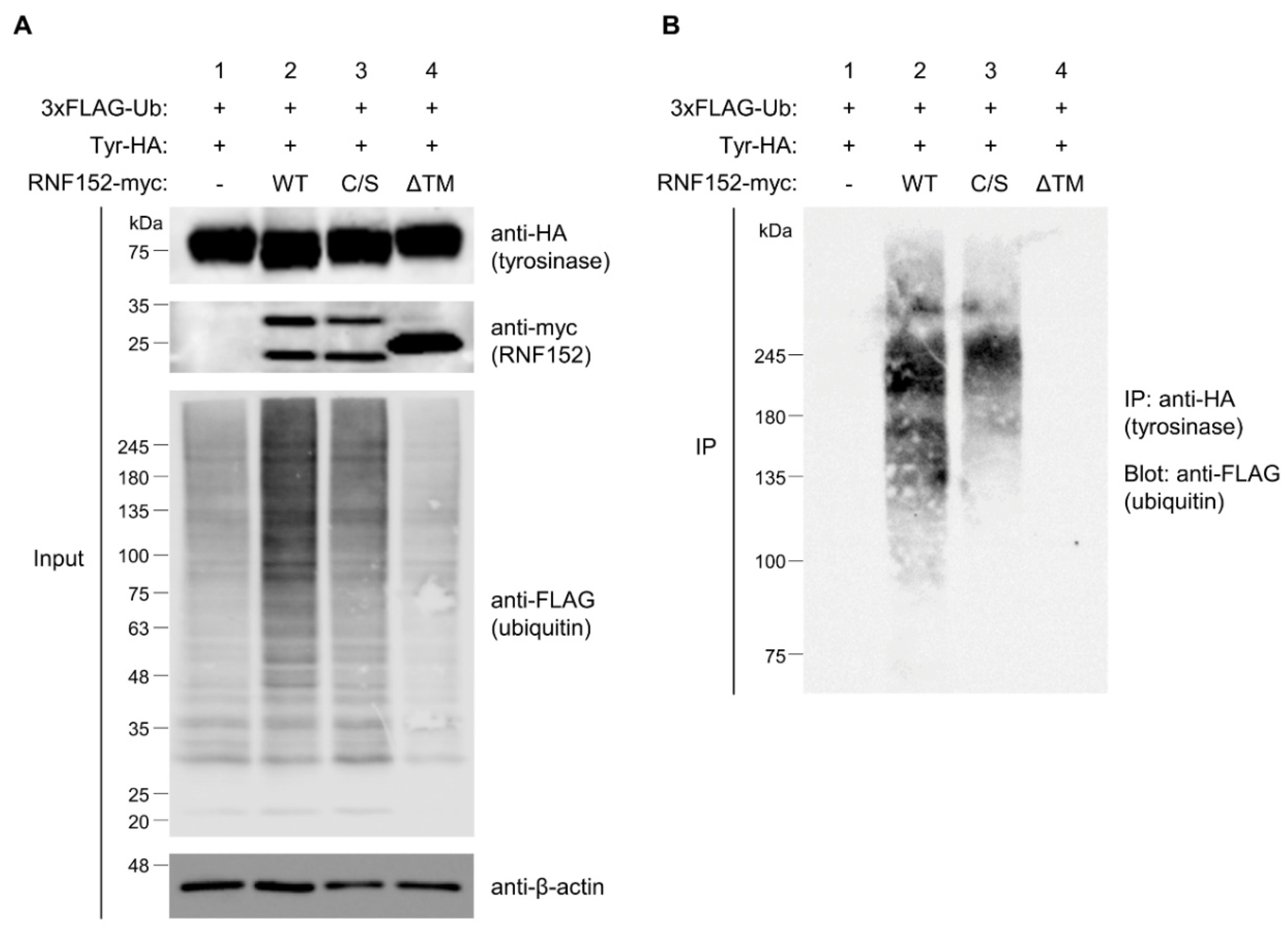
3.6. Both RING domain and TM domain of RNF152 are responsible for tyrosinase ubiquitination
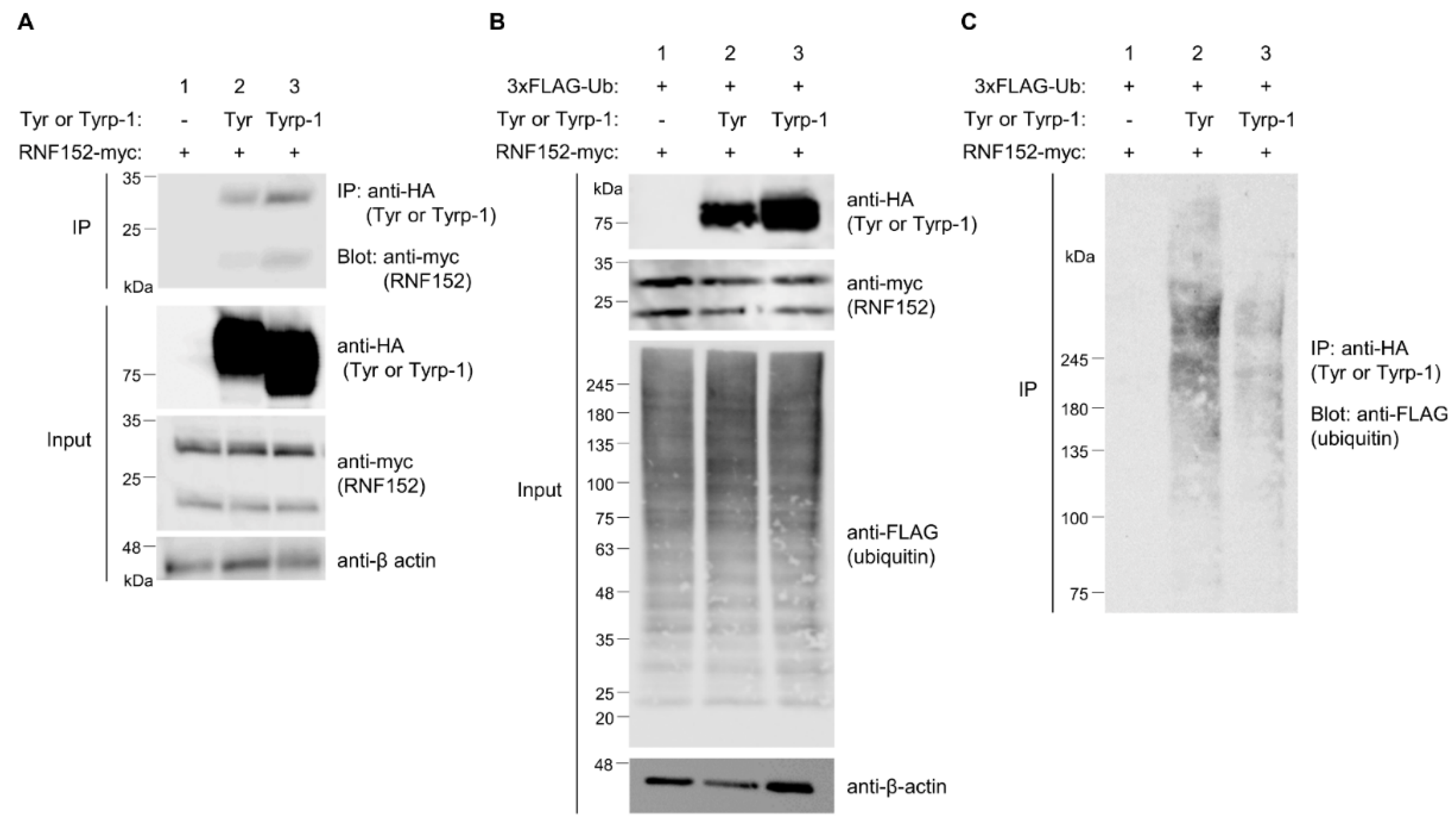
3.7. Tyrp-1 ubiquitination with RNF152
4. Discussion
Supplementary Materials
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Estadella, I.; Pedros-Gamez, O.; Colomer-Molera, M.; Bosch, M.; Sorkin, A.; Felipe, A. Endocytosis: A Turnover Mechanism Controlling Ion Channel Function. Cells 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Lecker, S.H.; Goldberg, A.L.; Mitch, W.E. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol 2006, 17, 1807–1819. [Google Scholar] [CrossRef] [PubMed]
- Apaja, P.M.; Lukacs, G.L. Protein homeostasis at the plasma membrane. Physiology (Bethesda) 2014, 29, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Rapoport, T.A. Mechanistic insights into ER-associated protein degradation. Curr Opin Cell Biol 2018, 53, 22–28. [Google Scholar] [CrossRef]
- Piper, R.C.; Luzio, J.P. Ubiquitin-dependent sorting of integral membrane proteins for degradation in lysosomes. Curr Opin Cell Biol 2007, 19, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Piper, R.C.; Dikic, I.; Lukacs, G.L. Ubiquitin-dependent sorting in endocytosis. Cold Spring Harb Perspect Biol 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- d'Azzo, A.; Bongiovanni, A.; Nastasi, T. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic (Copenhagen, Denmark) 2005, 6, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Bartee, E.; Mansouri, M.; Hovey Nerenberg, B.T.; Gouveia, K.; Fruh, K. Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. Journal of virology 2004, 78, 1109–1120. [Google Scholar] [CrossRef]
- Fujita, H.; Iwabu, Y.; Tokunaga, K.; Tanaka, Y. Membrane-associated RING-CH (MARCH) 8 mediates the ubiquitination and lysosomal degradation of the transferrin receptor. Journal of cell science 2013, 126, 2798–2809. [Google Scholar] [CrossRef]
- Fukuda, H.; Nakamura, N.; Hirose, S. MARCH-III Is a novel component of endosomes with properties similar to those of MARCH-II. Journal of biochemistry 2006, 139, 137–145. [Google Scholar] [CrossRef]
- Tada, T.; Zhang, Y.; Fujita, H.; Tokunaga, K. MARCH8: the tie that binds to viruses. FEBS J 2022, 289, 3642–3654. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wu, W.; Wu, Y.; Zheng, J.; Suo, T.; Tang, H.; Tang, J. RNF152, a novel lysosome localized E3 ligase with pro-apoptotic activities. Protein & cell 2010, 1, 656–663. [Google Scholar] [CrossRef]
- Deng, L.; Jiang, C.; Chen, L.; Jin, J.; Wei, J.; Zhao, L.; Chen, M.; Pan, W.; Xu, Y.; Chu, H.; et al. The ubiquitination of rag A GTPase by RNF152 negatively regulates mTORC1 activation. Molecular cell 2015, 58, 804–818. [Google Scholar] [CrossRef] [PubMed]
- Xiong, M.G.; Xu, Z.S.; Li, Y.H.; Wang, S.Y.; Wang, Y.Y.; Ran, Y. RNF152 positively regulates TLR/IL-1R signaling by enhancing MyD88 oligomerization. EMBO Rep 2020, 21, e48860. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Liu, S.; Sun, W.; Yu, H.; Tang, W.; Liu, W.; Ji, J.; Liu, B. Ring finger protein 152-dependent degradation of TSPAN12 suppresses hepatocellular carcinoma progression. Cancer Cell Int 2021, 21, 122. [Google Scholar] [CrossRef]
- Okamoto, T.; Imaizumi, K.; Kaneko, M. The Role of Tissue-Specific Ubiquitin Ligases, RNF183, RNF186, RNF182 and RNF152, in Disease and Biological Function. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Jimbow, K.; Gomez, P.F.; Toyofuku, K.; Chang, D.; Miura, S.; Tsujiya, H.; Park, J.S. Biological role of tyrosinase related protein and its biosynthesis and transport from TGN to stage I melanosome, late endosome, through gene transfection study. Pigment cell research 1997, 10, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Marks, M.S.; Seabra, M.C. The melanosome: membrane dynamics in black and white. Nature reviews. Molecular cell biology 2001, 2, 738–748. [Google Scholar] [CrossRef]
- Orlow, S.J.; Boissy, R.E.; Moran, D.J.; Pifko-Hirst, S. Subcellular distribution of tyrosinase and tyrosinase-related protein-1: implications for melanosomal biogenesis. The Journal of investigative dermatology 1993, 100, 55–64. [Google Scholar] [CrossRef]
- Raposo, G.; Marks, M.S. The dark side of lysosome-related organelles: specialization of the endocytic pathway for melanosome biogenesis. Traffic (Copenhagen, Denmark) 2002, 3, 237–248. [Google Scholar] [CrossRef]
- Raposo, G.; Tenza, D.; Murphy, D.M.; Berson, J.F.; Marks, M.S. Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. The Journal of cell biology 2001, 152, 809–824. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Hebert, D.N. Tyrosinase maturation through the mammalian secretory pathway: bringing color to life. Pigment cell research 2006, 19, 3–18. [Google Scholar] [CrossRef]
- Ando, H.; Kondoh, H.; Ichihashi, M.; Hearing, V.J. Approaches to identify inhibitors of melanin biosynthesis via the quality control of tyrosinase. The Journal of investigative dermatology 2007, 127, 751–761. [Google Scholar] [CrossRef]
- Niki, Y.; Adachi, N.; Fukata, M.; Fukata, Y.; Oku, S.; Makino-Okamura, C.; Takeuchi, S.; Wakamatsu, K.; Ito, S.; Declercq, L.; et al. S-Palmitoylation of Tyrosinase at Cysteine(500) Regulates Melanogenesis. The Journal of investigative dermatology 2023, 143, 317–327 e316. [Google Scholar] [CrossRef]
- Solano, F.; Briganti, S.; Picardo, M.; Ghanem, G. Hypopigmenting agents: an updated review on biological, chemical and clinical aspects. Pigment cell research 2006, 19, 550–571. [Google Scholar] [CrossRef]
- Ando, H.; Watabe, H.; Valencia, J.C.; Yasumoto, K.; Furumura, M.; Funasaka, Y.; Oka, M.; Ichihashi, M.; Hearing, V.J. Fatty acids regulate pigmentation via proteasomal degradation of tyrosinase: a new aspect of ubiquitin-proteasome function. The Journal of biological chemistry 2004, 279, 15427–15433. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Wen, Z.M.; Kim, H.Y.; Valencia, J.C.; Costin, G.E.; Watabe, H.; Yasumoto, K.; Niki, Y.; Kondoh, H.; Ichihashi, M.; et al. Intracellular composition of fatty acid affects the processing and function of tyrosinase through the ubiquitin-proteasome pathway. The Biochemical journal 2006, 394, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.M.; Orlow, S.J. Degradation of tyrosinase induced by phenylthiourea occurs following Golgi maturation. Pigment cell research 2005, 18, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Fujita, H.; Motokawa, T.; Katagiri, T.; Yokota, S.; Yamamoto, A.; Himeno, M.; Tanaka, Y. Inulavosin, a melanogenesis inhibitor, leads to mistargeting of tyrosinase to lysosomes and accelerates its degradation. The Journal of investigative dermatology 2009, 129, 1489–1499. [Google Scholar] [CrossRef]
- Fujita, H.; Menezes, J.C.; Santos, S.M.; Yokota, S.; Kamat, S.P.; Cavaleiro, J.A.; Motokawa, T.; Kato, T.; Mochizuki, M.; Fujiwara, T.; et al. Inulavosin and its benzo-derivatives, melanogenesis inhibitors, target the copper loading mechanism to the active site of tyrosinase. Pigment cell & melanoma research 2014, 27, 376–386. [Google Scholar] [CrossRef]
- Chen, X.K.; Kwan, J.S.; Chang, R.C.; Ma, A.C. 1-phenyl 2-thiourea (PTU) activates autophagy in zebrafish embryos. Autophagy 2021, 17, 1222–1231. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.A.; Park, N.H.; Na, Y.J.; Lee, H.K.; Lee, J.H.; Kim, Y.J.; Lee, C.S. Coumestrol Down-Regulates Melanin Production in Melan-a Murine Melanocytes through Degradation of Tyrosinase. Biological & pharmaceutical bulletin 2017, 40, 535–539. [Google Scholar] [CrossRef]
- Cho, Y.H.; Park, J.E.; Lim, D.S.; Lee, J.S. Tranexamic acid inhibits melanogenesis by activating the autophagy system in cultured melanoma cells. J Dermatol Sci 2017, 88, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Isogawa, K.; Asano, M.; Hayazaki, M.; Koga, K.; Watanabe, M.; Suzuki, K.; Kobayashi, T.; Kawaguchi, K.; Ishizuka, A.; Kato, S.; et al. Thioxothiazolidin derivative, 4-OST, inhibits melanogenesis by enhancing the specific recruitment of tyrosinase-containing vesicles to lysosome. J Cell Biochem 2021, 122, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Kawaguchi, K.; Nakamura, Y.; Furuta, K.; Takemori, H. GIF-2209, an Oxindole Derivative, Accelerates Melanogenesis and Melanosome Secretion via the Modification of Lysosomes in B16F10 Mouse Melanoma Cells. Molecules 2021, 27. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Guo, Y.; Wang, H.; Xiao, Y.; Liu, W.; Lyu, L. The ubiquitin-proteasome system in melanin metabolism. J Cosmet Dermatol 2022, 21, 6661–6668. [Google Scholar] [CrossRef] [PubMed]
- Sawasaki, T.; Ogasawara, T.; Morishita, R.; Endo, Y. A cell-free protein synthesis system for high-throughput proteomics. Proc Natl Acad Sci U S A 2002, 99, 14652–14657. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Uematsu, A.; Yamanaka, S.; Imamura, M.; Nakajima, T.; Doi, K.; Yasuoka, S.; Takahashi, C.; Takeda, H.; Sawasaki, T. Establishment of a Wheat Cell-Free Synthesized Protein Array Containing 250 Human and Mouse E3 Ubiquitin Ligases to Identify Novel Interaction between E3 Ligases and Substrate Proteins. PloS one 2016, 11, e0156718. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Kim, S.Y.; Park, S.H.; Choi, Y.G.; Kwon, S.B.; Kim, M.K.; Na, J.I.; Youn, S.W.; Park, K.C. Inhibitory effects of 4-n-butylresorcinol on tyrosinase activity and melanin synthesis. Biological & pharmaceutical bulletin 2005, 28, 2216–2219. [Google Scholar] [CrossRef]
- Tachiyama, R.; Ishikawa, D.; Matsumoto, M.; Nakayama, K.I.; Yoshimori, T.; Yokota, S.; Himeno, M.; Tanaka, Y.; Fujita, H. Proteome of ubiquitin/MVB pathway: possible involvement of iron-induced ubiquitylation of transferrin receptor in lysosomal degradation. Genes to cells : devoted to molecular & cellular mechanisms 2011, 16, 448–466. [Google Scholar] [CrossRef]
- Yurkow, E.J.; Laskin, J.D. Purification of tyrosinase to homogeneity based on its resistance to sodium dodecyl sulfate-proteinase K digestion. Arch Biochem Biophys 1989, 275, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Allouche, J.; Rachmin, I.; Adhikari, K.; Pardo, L.M.; Lee, J.H.; McConnell, A.M.; Kato, S.; Fan, S.; Kawakami, A.; Suita, Y.; et al. NNT mediates redox-dependent pigmentation via a UVB- and MITF-independent mechanism. Cell 2021, 184, 4268–4283 e4220. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Hearing, V.J. Direct interaction of tyrosinase with Tyrp1 to form heterodimeric complexes in vivo. Journal of cell science 2007, 120, 4261–4268. [Google Scholar] [CrossRef] [PubMed]
- Halaban, R.; Cheng, E.; Hebert, D.N. Coexpression of wild-type tyrosinase enhances maturation of temperature-sensitive tyrosinase mutants. The Journal of investigative dermatology 2002, 119, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Winder, A.J.; Wittbjer, A.; Rosengren, E.; Rorsman, H. The mouse brown (b) locus protein has dopachrome tautomerase activity and is located in lysosomes in transfected fibroblasts. Journal of cell science 1993, 106 Pt 1, 153–166. [Google Scholar] [CrossRef]
- Calvo, P.A.; Frank, D.W.; Bieler, B.M.; Berson, J.F.; Marks, M.S. A cytoplasmic sequence in human tyrosinase defines a second class of di-leucine-based sorting signals for late endosomal and lysosomal delivery. The Journal of biological chemistry 1999, 274, 12780–12789. [Google Scholar] [CrossRef] [PubMed]
- Simmen, T.; Schmidt, A.; Hunziker, W.; Beermann, F. The tyrosinase tail mediates sorting to the lysosomal compartment in MDCK cells via a di-leucine and a tyrosine-based signal. Journal of cell science 1999, 112 Pt 1, 45–53. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, X.; Chen, L.; Liu, Y.Y.; Venkatarangan, V.; Reist, L.; Hanson, P.; Xu, H.; Wang, Y.; Li, M. A conserved ubiquitin- and ESCRT-dependent pathway internalizes human lysosomal membrane proteins for degradation. PLoS Biol 2021, 19, e3001361. [Google Scholar] [CrossRef]
- Goto, E.; Ishido, S.; Sato, Y.; Ohgimoto, S.; Ohgimoto, K.; Nagano-Fujii, M.; Hotta, H. c-MIR, a human E3 ubiquitin ligase, is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar activity. The Journal of biological chemistry 2003, 278, 14657–14668. [Google Scholar] [CrossRef]
| E3 binding assay result to V5-Tyrosinase | predicted localization* | predicted number of transmembrane domain* | ||||
| Sample/Mock | Inulavosin/DMSO | |||||
| Symbol | rank | DMSO | Inulavosin | |||
| RNF152 | 1 | 16.65 | 22.13 | 1.33 | Lysosome | 1 |
| VPS41 | 2 | 17.99 | 20.97 | 1.17 | Cytosol, lysosome, Golgi, Endosome | 0 |
| RNF41 | 3 | 17.84 | 20.55 | 1.15 | Cytosol | 0 |
| ZNF598 | 4 | 16.43 | 18.52 | 1.13 | Cytosol | 0 |
| TRIM21 | 5 | 19.82 | 21.73 | 1.10 | Cytosol, Nucleus | 0 |
| *Predictions were obtained from the following sites; GeneCard: https://www.genecards.org Uniprote: https://www.uniprot.org |
||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





