Submitted:
24 November 2023
Posted:
27 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Overview
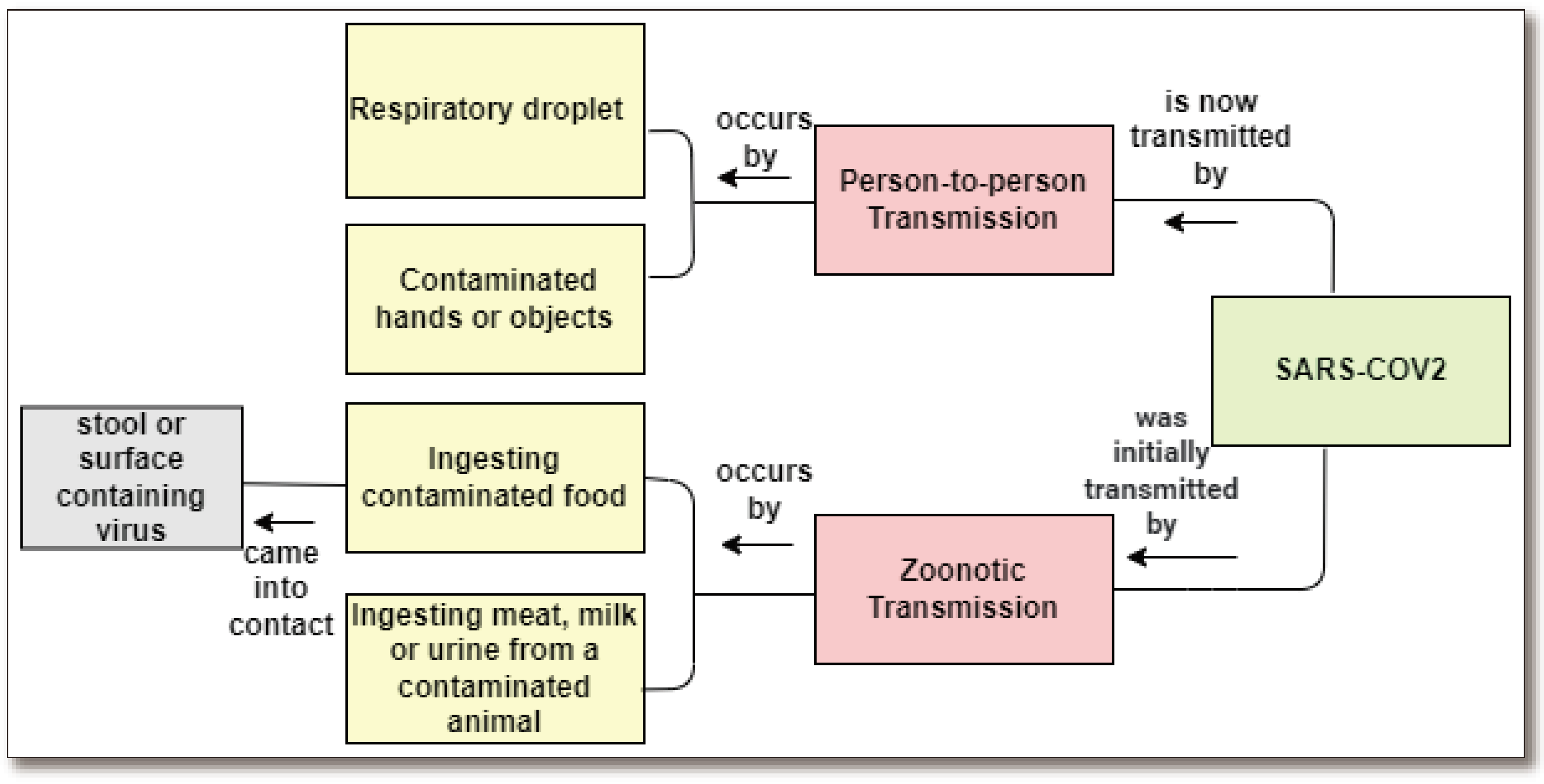
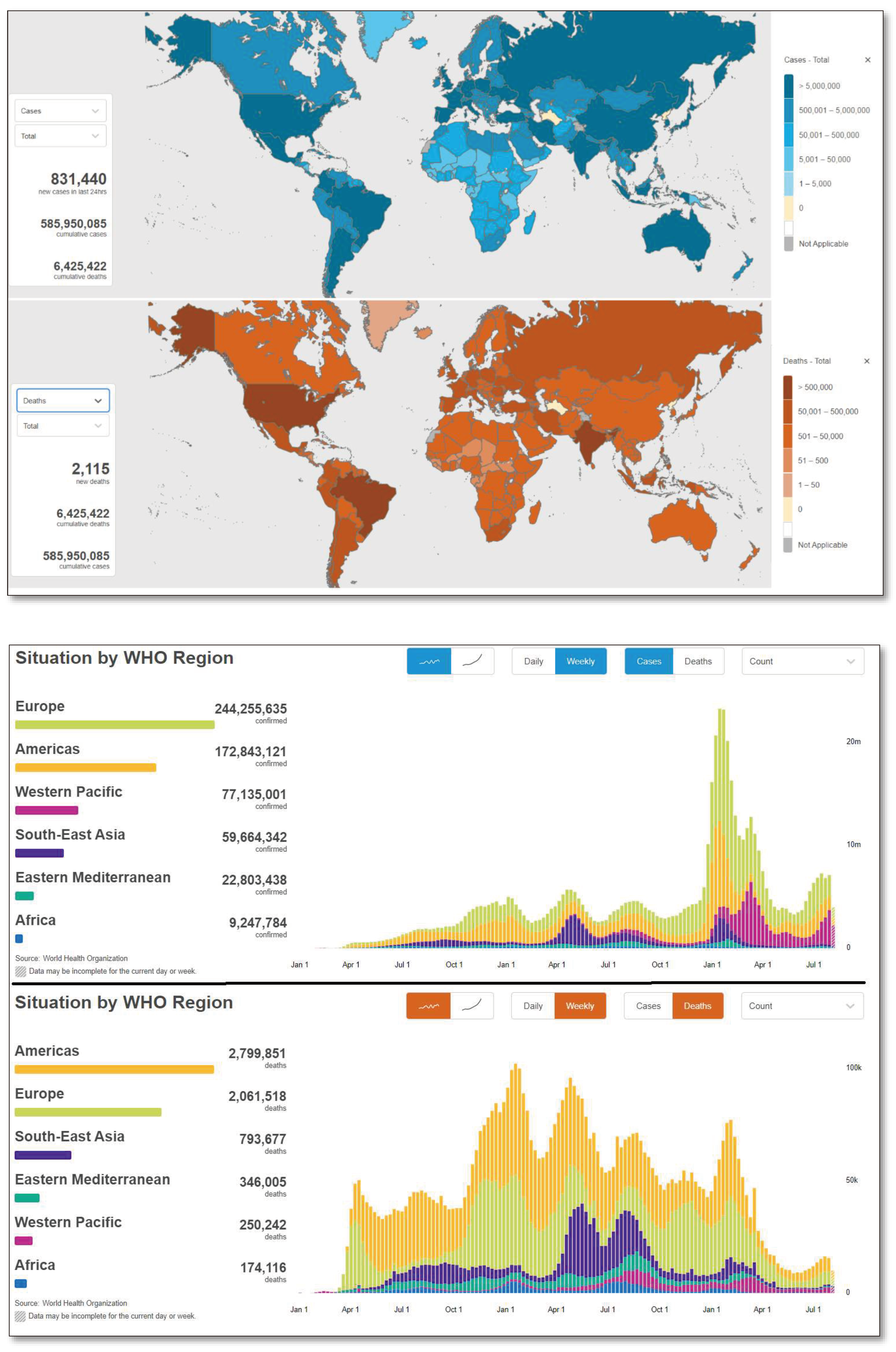
1.2. Global Scenario
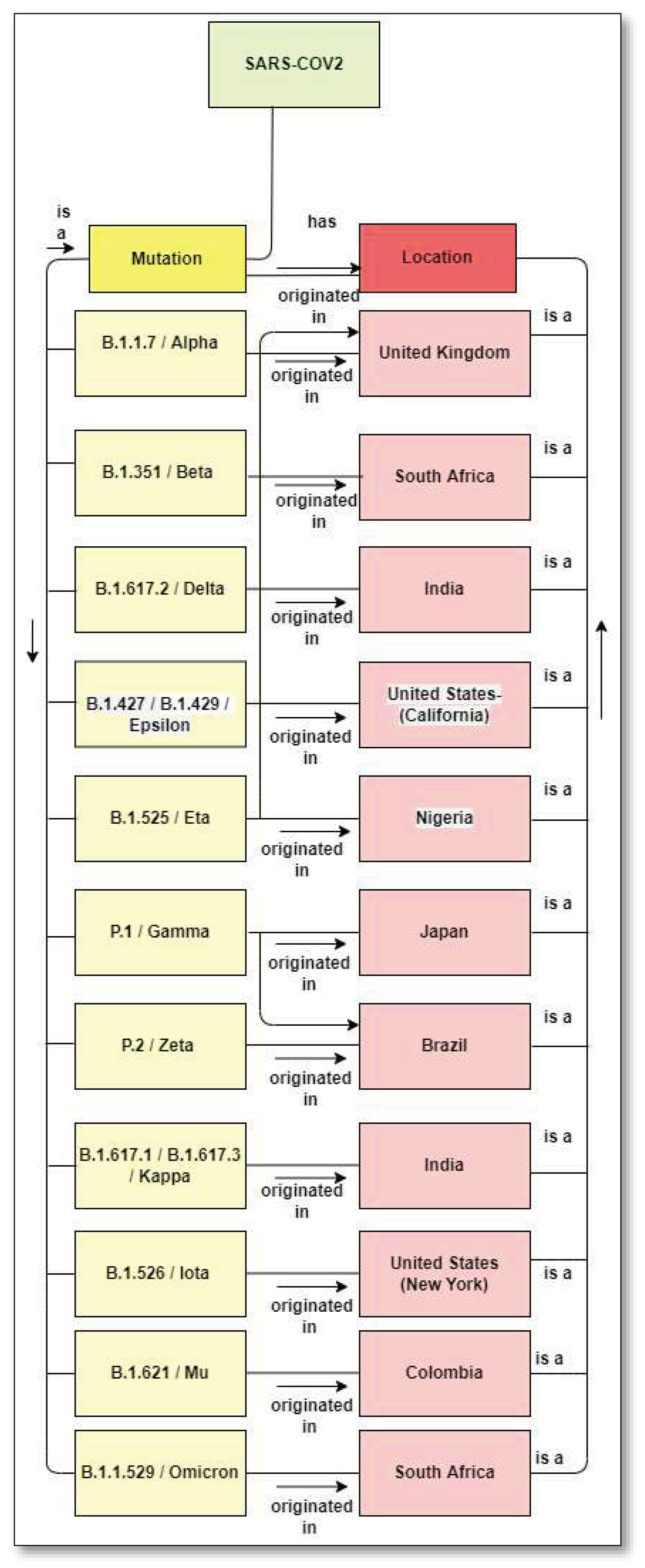
1.3. Mutation
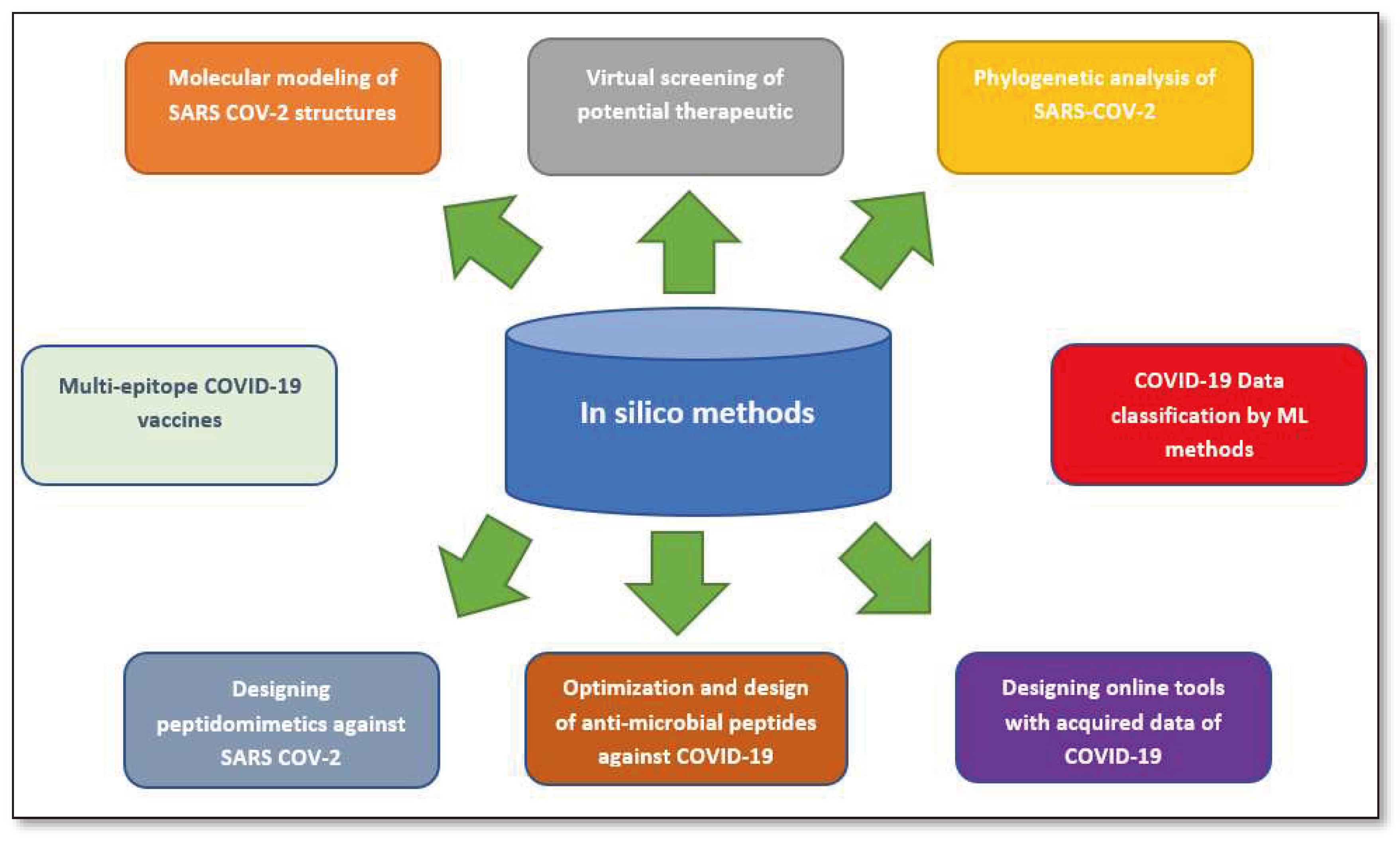
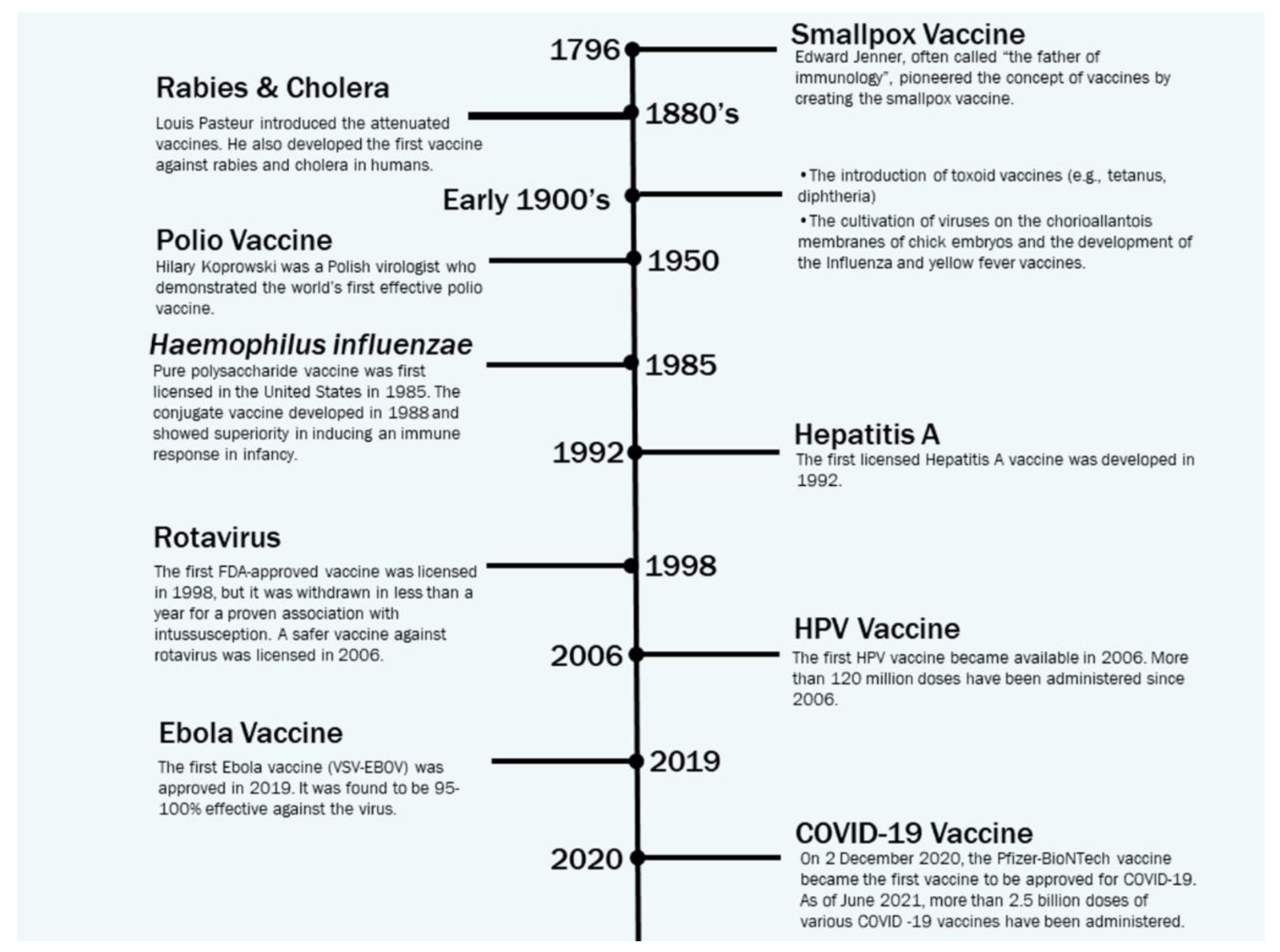
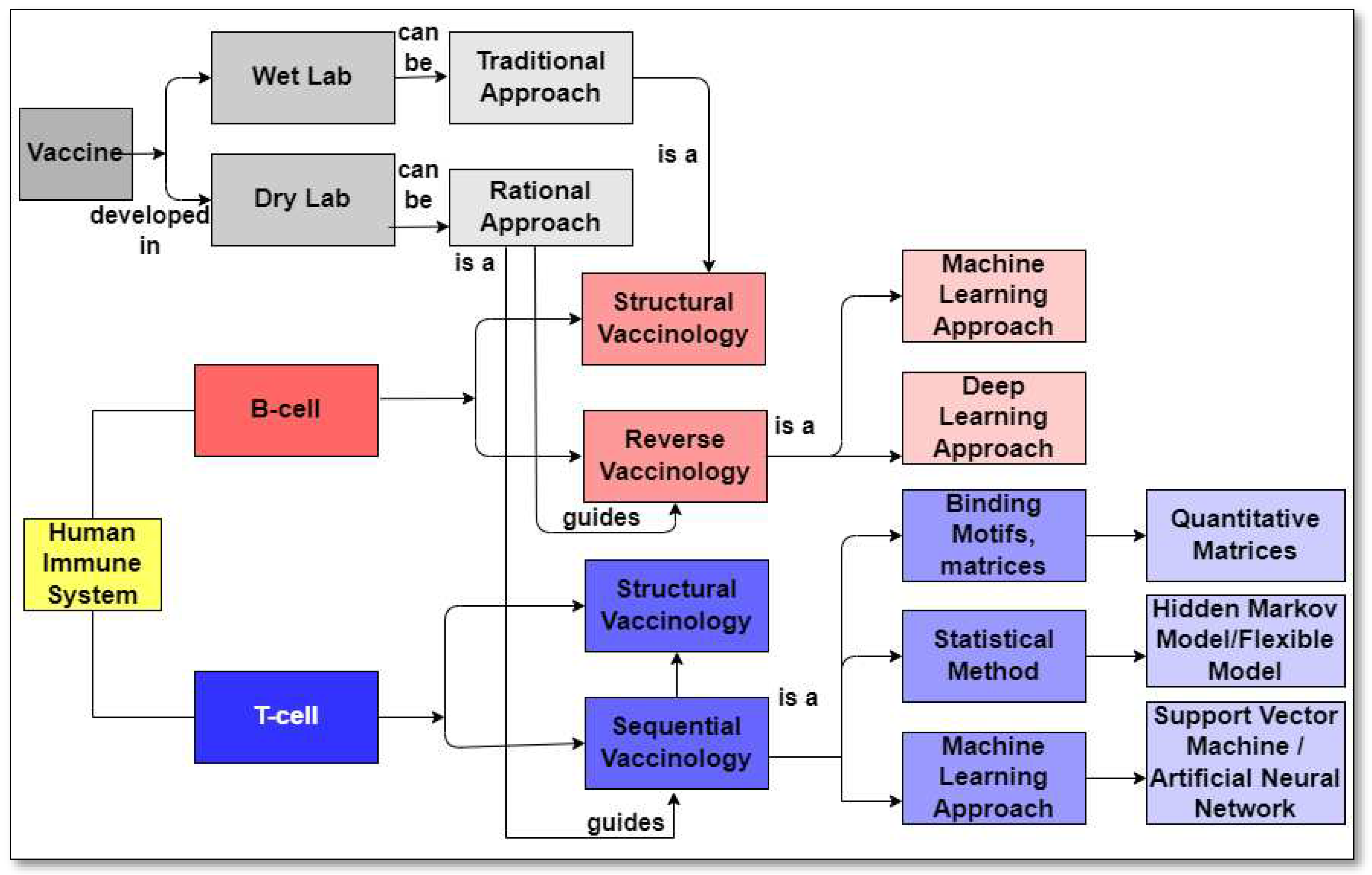
2. This publication provides a concise review of the existing literature on in silico strategies for combating COVID-19, focusing on SARS-CoV-2 structure predictions, phylogenetic analysis, drug virtual screening, natural compound production, vaccine development, and machine learning and AI. It also discusses potential challenges in using machine learning for future pandemics.Vaccine Development Strategies: Historical Outline
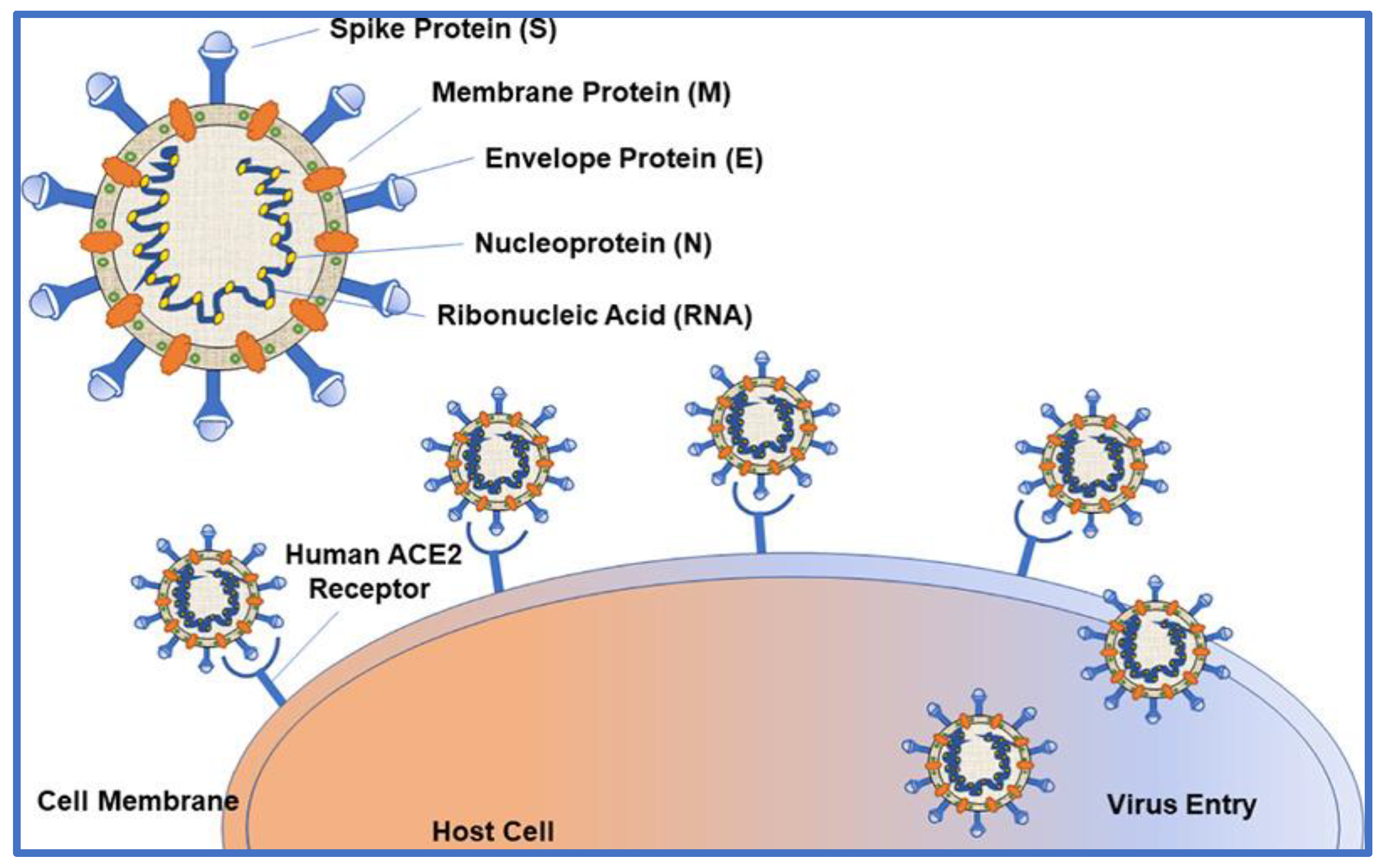
3. Molecular Architecture of SARS CoV-2
4. The COVID-19 Vaccine Landscape
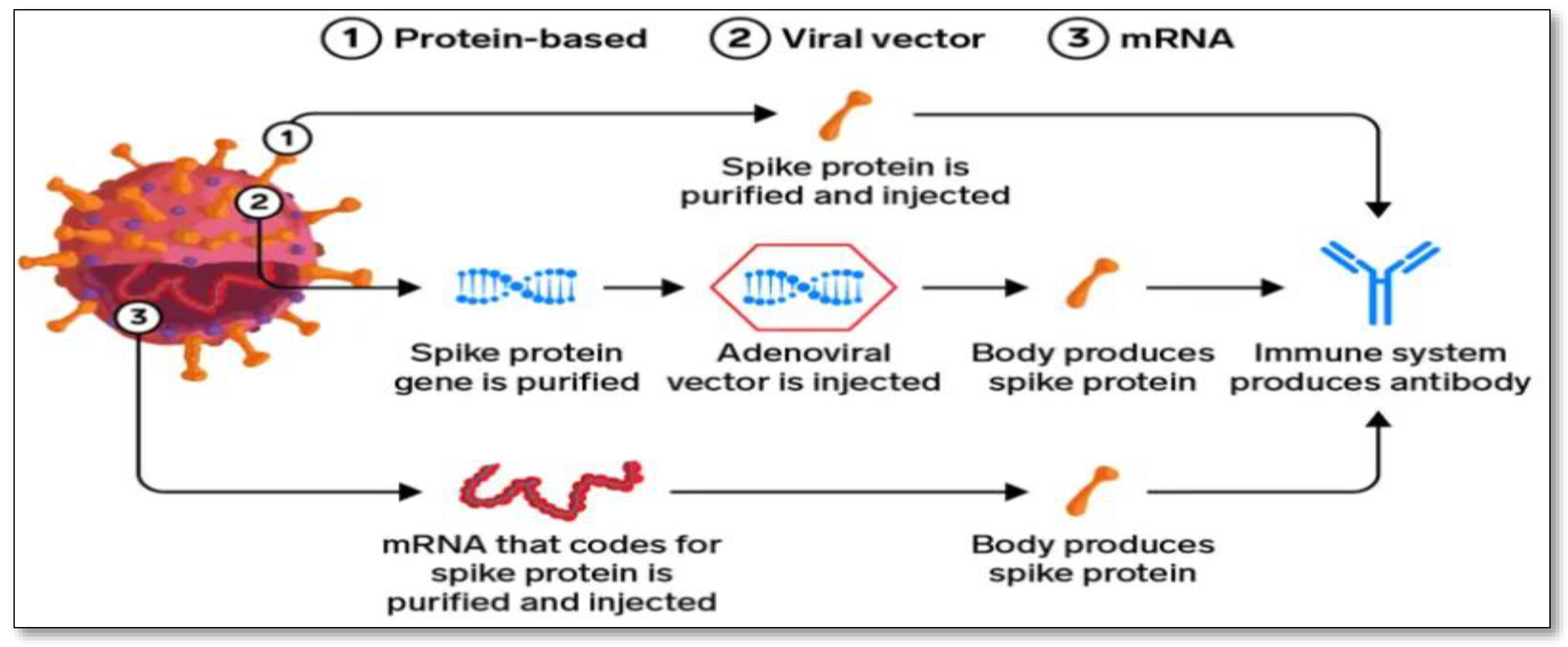
5. Different Coronavirus Vaccines in Development
5.1. mRNA Vaccine: A New Possibility
5.2. Protein-based Vaccine: The Slower, Traditional Method
5.3. Viral Vector Vaccine: The Powerful Immune Response
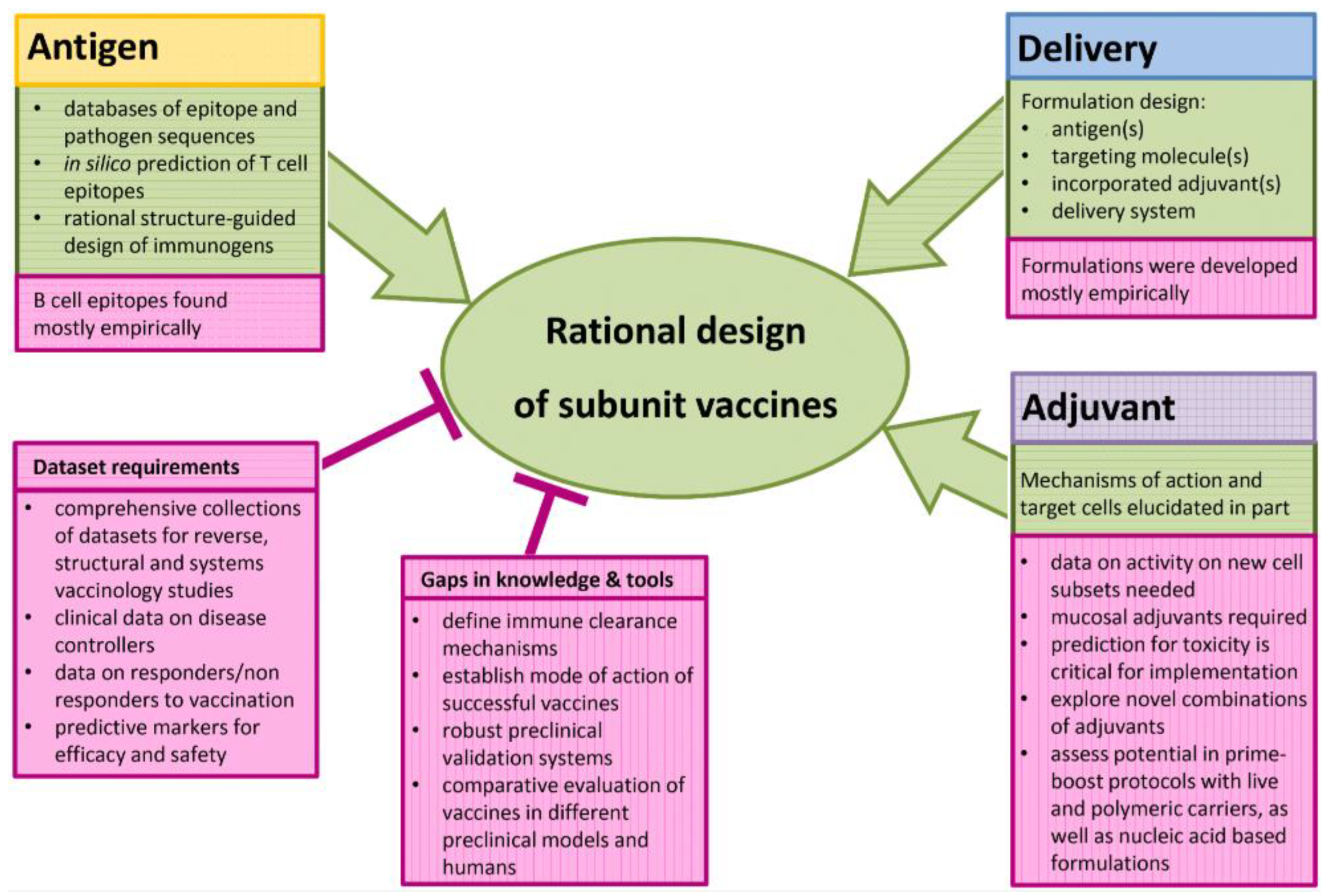
6. The Rational Approach to develop a SARS-CoV-2 Vaccine
7. A Rational Approach to Pan-coronavirus Vaccines
8. Machine Learning Approaches
8.1. Introduction
8.2. Machine Learning Algorithms to Combat COVID-19
8.3. Machine Learning Approaches for COVID-19 Forecast
| Organization | Principals | Advantages | Disadvantages |
|---|---|---|---|
| MIT | OptiVax works by looking for all possible peptide fragments from a set of viral or tumor proteins that could be used as vaccine candidates. | This method improves the presentation likelihood of a diverse group of vaccine peptides based on the HLA haplotype distribution of a target human population and predicted epitope drift. | Getting excellent data on how people differ in their genetic composition, particularly in important genes that affect the response to a vaccine or viral infection, was one of the hurdles. |
| EvalVax is a complimentary technology they created that predicts vaccine coverage and allows others to evaluate different vaccine formulations. | |||
| TCS | For the de novo design of small compounds capable of blocking the 3CL protease, they used deep neural network-based generative and predictive models. The small compounds that were created were filtered and screened against the binding site of SARS-3CL CoV-2's protease structure. | They identified 31 possible compounds as good candidates for further production and testing against SARS-CoV-2 based on the screening results and additional research. | This was done to make the challenge resemble the class of natural language processing (NLP) problems for which sophisticated AI models and architectures have been built throughout time. |
| Benevolent AI | Rather than focusing primarily on medications that could directly affect the virus, they investigated strategies to stop the virus from infecting human cells through biological processes. | Find approved medications that could potentially stop COVID-19 from progressing, suppress the "cytokine storm," and minimize the inflammatory damage caused by the disease. | Takes 1.5 hours to process. |
| UK-based company Exscientia and Diamond Light Source and Calibr, a division of Scripps Research, US | Applied improved biosensor platforms to screen the collection of 15,000 clinically ready molecules. | The first objective is to find any current medications that can be repurposed to protect humans. | Discovering potential prospects among the currently available medications is difficult and time-consuming. |
| Insilco Medicine, a Hong Kong-based pharmaceutical research company | The seven promising compounds against COVID-19 were discovered, and two of them have already been synthesized for testing. | Used Virtual Reality to Fine-Tune New AI-Generated COVID-19 Drugs | Synthesis and validation could take a long time and cost a lot of money. |
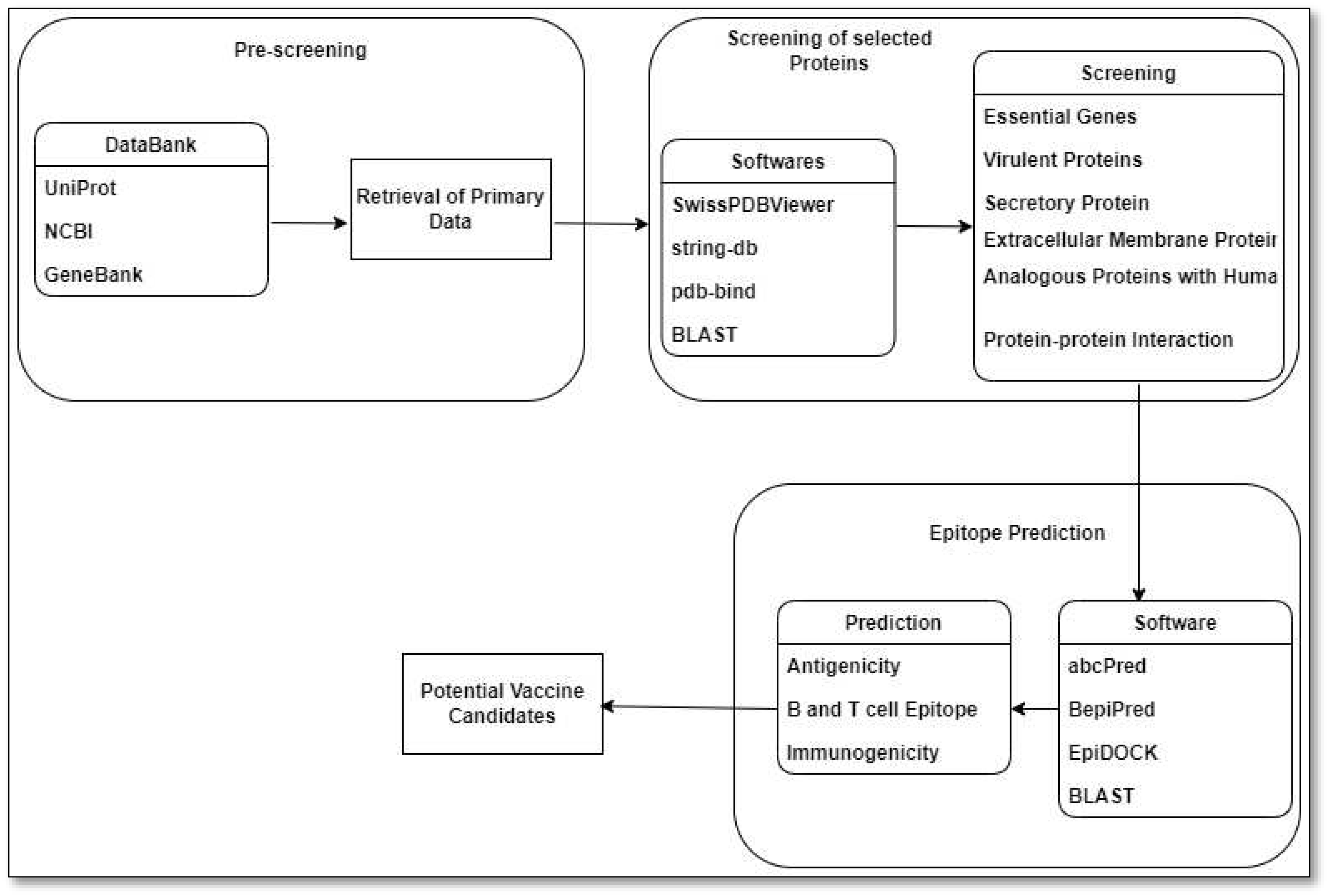
9. Reverse Vaccinology Approach
| Research Group | Database | Target Proteins | B-cell and T-cell Epitope Prediction |
Antigenicity and Allergenicity test | Molecular Docking | Immunogenicity Evaluation |
Energy Minimization and Binding Affinity Prediction | In Silico Cloning and Codon Optimization | ML Approach |
|---|---|---|---|---|---|---|---|---|---|
| Ong et al.[116] | NCBI | S, E, M, N proteins | ✔ | ✔ | ● | ✔ | ● | ● | Vaxign-ML protegenicity (protective antigenicity) score calculation |
10. RNA Vaccine Design
11.1. Effectiveness of mRNA vaccines in preventing COVID-19
11.2. Reported Side Effects of mRNA Vaccines
11. Conclusions and Future Directions
12.1. Potential Drawbacks of Rational Vaccines for SARS-CoV-2
12.2. The ways to address challenges and drawbacks in the development and distribution of a rational vaccine for SARS-CoV-2
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- J. Boonyaratanakornkit and J. J. Taylor, “Techniques to Study Antigen-Specific B Cell Responses,” Frontiers in immunology. 2019. [CrossRef]
- J. L. Sanchez-Trincado, M. Gomez-Perosanz, and P. A. Reche, “Fundamentals and Methods for T- and B-Cell Epitope Prediction,” Journal of Immunology Research. 2017. [CrossRef]
- P. Forterre, “Defining Life: The Virus Viewpoint,” Orig. Life Evol. Biosph., 2010. [CrossRef]
- A. W. H. Chin et al., “Stability of SARS-CoV-2 in different environmental conditions,” The Lancet Microbe, 2020. [CrossRef]
- C. Arnold, “How computational immunology changed the face of COVID-19 vaccine development,” Nat. Med., Jul. 2020. [CrossRef]
- R. R. María, C. J. Arturo, J. A. Alicia, M. G. Paulina, and A. O. Gerardo, “The Impact of Bioinformatics on Vaccine Design and Development,” in Vaccines, 2017.
- V. C. C. Cheng, S. K. P. Lau, P. C. Y. Woo, and Y. Y. Kwok, “Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection,” Clinical Microbiology Reviews. 2007. [CrossRef]
- J. Cui, F. Li, and Z. L. Shi, “Origin and evolution of pathogenic coronaviruses,” Nature Reviews Microbiology. 2019. [CrossRef]
- M. A. Shereen, S. Khan, A. Kazmi, N. Bashir, and R. Siddique, “COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses,” Journal of Advanced Research. 2020. [CrossRef]
- M. Cascella, M. Rajnik, A. Cuomo, S. C. Dulebohn, and R. Di Napoli, Features, Evaluation and Treatment Coronavirus (COVID-19). 2020.
- I. Alam et al., “CovMT: an interactive SARS-CoV-2 mutation tracker, with a focus on critical variants,” The Lancet Infectious Diseases. 2021. [CrossRef]
- M. Moradi, R. Golmohammadi, A. Najafi, M. Moosazadeh Moghaddam, M. Fasihi-Ramandi, and R. Mirnejad, “A contemporary review on the important role of in silico approaches for managing different aspects of COVID-19 crisis,” Informatics in Medicine Unlocked. 2022. [CrossRef]
- Mani, S. Viruses, Vaccines and the Race for a Covid-19 Vaccine: A Tutorial. (n.d.) Retrieved October 24, 2023, from www.preprints.org/manuscript/202009.0153.
- Koch, T., Fathi, A., Addo, M. The COVID-19 Vaccine Landscape. (n.d.) Retrieved October 24, 2023, from link.springer.com/chapter/10.1007/978-3-030-63761-3_31. [CrossRef]
- Smitha, T., Thomas, A. A brief outlook on the current emerging trends of COVID 19 vaccines. (n.d.) Retrieved October 24, 2023, from www.ncbi.nlm.nih.gov/pmc/articles/PMC7802844/.
- Few, M. Epidemics, indigenous communities, and public health in the COVID-19 era: views from smallpox inoculation campaigns in colonial Guatemala. (n.d.) Retrieved October 24, 2023, from www.cambridge.org.
- Saleh A, Qamar S, Tekin A, et al. (July 26, 2021) Vaccine Development Throughout History. Cureus 13(7): e16635. [CrossRef]
- C. Rueckert and C. A. Guzmán, “Vaccines: From Empirical Development to Rational Design,” PLoS Pathogens, vol. 8, no. 11, p. e1003001, Nov. 2012. [CrossRef]
- K. G. Andersen, A. Rambaut, W. I. Lipkin, E. C. Holmes, and R. F. Garry, “The proximal origin of SARS-CoV-2,” Nature Medicine. 2020. [CrossRef]
- S. Belouzard, J. K. Millet, B. N. Licitra, and G. R. Whittaker, “Mechanisms of coronavirus cell entry mediated by the viral spike protein.,” Viruses. 2012. [CrossRef]
- A. T. Ton, F. Gentile, M. Hsing, F. Ban, and A. Cherkasov, “Rapid Identification of Potential Inhibitors of SARS-CoV-2 Main Protease by Deep Docking of 1.3 Billion Compounds,” Mol. Inform., 2020. [CrossRef]
- L. Zhang et al., “Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors,” Science (80-. )., 2020. [CrossRef]
- A. Shamsi et al., “Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: Possible implication in COVID-19 therapy,” Biosci. Rep., 2020. [CrossRef]
- R. L. Graham, J. S. Sparks, L. D. Eckerle, A. C. Sims, and M. R. Denison, “SARS coronavirus replicase proteins in pathogenesis,” Virus Res., 2008. [CrossRef]
- A. A. T. Naqvi et al., “Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach,” Biochimica et Biophysica Acta - Molecular Basis of Disease. 2020. [CrossRef]
- J. Shang et al., “Structural basis of receptor recognition by SARS-CoV-2,” Nature, 2020. [CrossRef]
- D. Wrapp et al., “Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation,” Science (80-. )., 2020. [CrossRef]
- T. Suzuki, Y. Otake, S. Uchimoto, A. Hasebe, and Y. Goto, “Genomic characterization and phylogenetic classification of bovine coronaviruses through whole genome sequence analysis,” Viruses, 2020. [CrossRef]
- H. Higuchi, S. F. Bronk, A. Bateman, K. Harrington, R. G. Vile, and G. J. Gores, “Viral fusogenic membrane glycoprotein expression causes syncytia formation with bioenergetic cell death: Implications for gene therapy,” Cancer Res., 2000.
- B. Alberts, A. Johnson, J. Lewis, K. Raff, Martin Roberts, and P. Walter, “Transport from the ER through the Golgi Apparatus,” Mol. Biol. Cell, 2002.
- J. S. Morse, T. Lalonde, S. Xu, and W. R. Liu, “Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV,” ChemBioChem, 2020. [CrossRef]
- H. Iqbal, “The importance of cell-mediated immunity in COVID-19 – An opinion,” Medical Hypotheses. 2020. [CrossRef]
- N. C. Kyriakidis, A. López-Cortés, E. V. González, A. B. Grimaldos, and E. O. Prado, “SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates,” npj Vaccines. 2021. [CrossRef]
- D. R. Burton and L. M. Walker, “Rational Vaccine Design in the Time of COVID-19,” Cell Host Microbe, 2020. [CrossRef]
- F. X. Heinz and K. Stiasny, “Distinguishing features of current COVID-19 vaccines: knowns and unknowns of antigen presentation and modes of action,” npj Vaccines. 2021. [CrossRef]
- T. Thanh Le et al., “The COVID-19 vaccine development landscape,” Nature reviews. Drug discovery. 2020. [CrossRef]
- T. T. Le, J. P. Cramer, R. Chen, and S. Mayhew, “Evolution of the COVID-19 vaccine development landscape,” Nature reviews. Drug discovery. 2020. [CrossRef]
- World Health Organization (WHO)., “COVID-19 - Landscape of novel coronavirus candidate vaccine development worldwide,” Who, 2020.
- World Health Organization, “COVID-19 vaccine tracker and landscape,” Who. 2021.
- [41] Quinlan et al., 2020 B.D. Quinlan, H. Mou, L. Zhang, Y. Guo, W. He, A. Ojha, M.S. Parcells, G. Luo, W. Li, G. Zhong, et al. The SARS-CoV-2 receptor-binding domain elicits a potent neutralizing response without antibody-dependent enhancement bioRxiv (2020). [CrossRef]
- D.R. Burton What Are the Most Powerful Immunogen Design Vaccine Strategies? Reverse Vaccinology 2.0 Shows Great Promise Cold Spring Harb. Perspect. Biol., 9 (2017), p. 030262.
- L.M. Walker, D.R. Burton Passive immunotherapy of viral infections: ‘super-antibodies’ enter the fray Nat. Rev. Immunol., 18 (2018), pp. 297-308. [CrossRef]
- D. Corti, A. Lanzavecchia Broadly neutralizing antiviral antibodies Annu. Rev. Immunol., 31 (2013), pp. 705-742. [CrossRef]
- J. Pallesen, N. Wang, K.S. Corbett, D. Wrapp, R.N. Kirchdoerfer, H.L. Turner, C.A. Cottrell, M.M. Becker, L. Wang, W. Shi, et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen Proc. Natl. Acad. Sci. USA, 114 (2017), pp. E7348-E7357. [CrossRef]
- D. Wrapp, N. Wang, K.S. Corbett, J.A. Goldsmith, C.L. Hsieh, O. Abiona, B.S. Graham, J.S. McLellan Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation Science, 367 (2020), pp. 1260-1263. [CrossRef]
- J.L. Slon-Campos, W. Dejnirattisai, B.W. Jagger, C. López-Camacho, W. Wongwiwat, L.A. Durnell, E.S. Winkler, R.E. Chen, A. Reyes Sandoval, F.A. Rey, et al. A protective Zika virus E-dimer-based subunit vaccine engineered to abrogate antibody-dependent enhancement of dengue infection Nat. Immunol., 20 (2019), pp. 1291-1298. [CrossRef]
- P.G. Choe, R.A.P.M. Perera, W.B. Park, K.H. Song, J.H. Bang, E.S. Kim, H.B. Kim, L.W.R. Ko, S.W. Park, N.J. Kim, et al. MERS-CoV Antibody Responses 1 Year after Symptom Onset, South Korea, 2015 Emerg. Infect. Dis., 23 (2017), pp. 1079-1084. [CrossRef]
- L.P. Wu, N.C. Wang, Y.H. Chang, X.Y. Tian, D.Y. Na, L.Y. Zhang, L. Zheng, T. Lan, L.F. Wang, G.D. Liang Duration of antibody responses after severe acute respiratory syndrome Emerg. Infect. Dis., 13 (2007), pp. 1562-1564. [CrossRef]
- H.H. Tam, M.B. Melo, M. Kang, J.M. Pelet, V.M. Ruda, M.H. Foley, J.K. Hu, S. Kumari, J. Crampton, A.D. Baldeon, et al. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination Proc. Natl. Acad. Sci. U S A, 113 (2016), pp. E6639-E6648. [CrossRef]
- K.M. Cirelli, D.G. Carnathan, B. Nogal, J.T. Martin, O.L. Rodriguez, A.A. Upadhyay, C.A. Enemuo, E.H. Gebru, Y. Choe, F. Viviano, et al. Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance Cell, 177 (2019), pp. 1153-1171.e1128. [CrossRef]
- T.J. Moyer, Y. Kato, W. Abraham, J.Y.H. Chang, D.W. Kulp, N. Watson, H.L. Turner, S. Menis, R.K. Abbott, J.N. Bhiman, et al. Engineered immunogen binding to alum adjuvant enhances humoral immunity Nat. Med., 26 (2020), pp. 430-440. [CrossRef]
- L.M. Walker, D.R. Burton Passive immunotherapy of viral infections: ‘super-antibodies’ enter the fray Nat. Rev. Immunol., 18 (2018), pp. 297-308. [CrossRef]
- D. Pinto, Y.J. Park, M. Beltramello, A.C. Walls, M.A. Tortorici, S. Bianchi, S. Jaconi, K. Culap, F. Zatta, A. De Marco, et al. Structural and functional analysis of a potent sarbecovirus neutralizing antibody bioRxiv (2020). [CrossRef]
- A.C. Walls, M.A. Tortorici, B.J. Bosch, B. Frenz, P.J.M. Rottier, F. DiMaio, F.A. Rey, D. Veesler Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer Nature, 531 (2016), pp. 114-117. [CrossRef]
- G. Mahalakshmi, S. Sridevi, and S. Rajaram, “A survey on forecasting of time series data,” 2016. [CrossRef]
- M. Qin, Z. Li, and Z. Du, “Red tide time series forecasting by combining ARIMA and deep belief network,” Knowledge-Based Syst., 2017. [CrossRef]
- Z. Zhao, W. Chen, X. Wu, P. C. V. Chen, and J. Liu, “LSTM network: A deep learning approach for short-term traffic forecast,” IET Image Process., 2017. [CrossRef]
- E. M. de Oliveira and F. L. Cyrino Oliveira, “Forecasting mid-long term electric energy consumption through bagging ARIMA and exponential smoothing methods,” Energy, 2018. [CrossRef]
- M. Längkvist, L. Karlsson, and A. Loutfi, “A review of unsupervised feature learning and deep learning for time-series modeling,” Pattern Recognit. Lett., 2014. [CrossRef]
- F. A. Gers, J. Schmidhuber, and F. Cummins, “Learning to forget: Continual prediction with LSTM,” Neural Comput., 2000. [CrossRef]
- K. Fukushima, “Neocognitron: A self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position,” Biol. Cybern., 1980. [CrossRef]
- X. Yao, “Evolving artificial neural networks,” Proc. IEEE, 1999. [CrossRef]
- N. L. W. S. R. Ginantra, I. G. A. D. Indradewi, and E. Hartono, “Machine learning approach for Acute Respiratory Infections (ISPA) prediction: Case study Indonesia,” 2020. [CrossRef]
- R. Yin, V. H. Tran, X. Zhou, J. Zheng, and C. K. Kwoh, “Predicting antigenic variants of H1N1 influenza virus based on epidemics and pandemics using a stacking model,” PLoS One, 2018. [CrossRef]
- S. B. Cho, “Exploiting machine learning techniques for location recognition and prediction with smartphone logs,” Neurocomputing, 2016. [CrossRef]
- K. Mori, T. Wada, and K. Ohtsuki, “A New Disaster Recognition Algorithm Based on SVM for ERESS: Buffering and Bagging-SVM,” 2016. [CrossRef]
- D. N. Le, V. S. Parvathy, D. Gupta, A. Khanna, J. J. P. C. Rodrigues, and K. Shankar, “IoT enabled depthwise separable convolution neural network with deep support vector machine for COVID-19 diagnosis and classification,” Int. J. Mach. Learn. Cybern., 2021. [CrossRef]
- S. Sadhukhan, S. Banerjee, P. Das, and A. K. Sangaiah, “Producing better disaster management plan in post-disaster situation using social media mining,” in Computational Intelligence for Multimedia Big Data on the Cloud with Engineering Applications, 2018. [CrossRef]
- N. Assery, Y. Xiaohong, S. Almalki, R. Kaushik, and Q. Xiuli, “Comparing learning-based methods for identifying disaster-related tweets,” 2019. [CrossRef]
- F. Mohammadi et al., “Artificial neural network and logistic regression modelling to characterize COVID-19 infected patients in local areas of Iran,” Biomed. J., 2021. [CrossRef]
- Bhandari et al., “Logistic regression analysis to predict mortality risk in COVID-19 patients from routine hematologic parameters,” Ibnosina J. Med. Biomed. Sci., 2020. [CrossRef]
- A. M. Almeshal, A. I. Almazrouee, M. R. Alenizi, and S. N. Alhajeri, “Forecasting the spread of COVID-19 in kuwait using compartmental and logistic regression models,” Appl. Sci., 2020. [CrossRef]
- M. Elhoseny, “Multi-object Detection and Tracking (MODT) Machine Learning Model for Real-Time Video Surveillance Systems,” Circuits, Syst. Signal Process., 2020. [CrossRef]
- Y. Chen and Y. Liu, “Which risk factors matter more for psychological distress during the covid-19 pandemic? An application approach of gradient boosting decision trees,” Int. J. Environ. Res. Public Health, 2021. [CrossRef]
- M. Loey, G. Manogaran, M. H. N. Taha, and N. E. M. Khalifa, “A hybrid deep transfer learning model with machine learning methods for face mask detection in the era of the COVID-19 pandemic,” Meas. J. Int. Meas. Confed., 2021. [CrossRef]
- C. Iwendi et al., “COVID-19 patient health prediction using boosted random forest algorithm,” Front. Public Heal., 2020. [CrossRef]
- C. M. YEŞİLKANAT, “Spatio-temporal estimation of the daily cases of COVID-19 in worldwide using random forest machine learning algorithm,” Chaos, Solitons and Fractals, 2020. [CrossRef]
- L. Luoh, “ZigBee-based intelligent indoor positioning system soft computing,” Soft Comput., 2014. [CrossRef]
- M. Polese, R. Jana, V. Kounev, K. Zhang, S. Deb, and M. Zorzi, “Machine Learning at the Edge: A Data-Driven Architecture with Applications to 5G Cellular Networks,” IEEE Trans. Mob. Comput., 2021. [CrossRef]
- P. Chhikara, R. Tekchandani, N. Kumar, V. Chamola, and M. Guizani, “DCNN-GA: A Deep Neural Net Architecture for Navigation of UAV in Indoor Environment,” IEEE Internet Things J., 2021. [CrossRef]
- S. More et al., “Security Assured CNN-Based Model for Reconstruction of Medical Images on the Internet of Healthcare Things,” IEEE Access, 2020. [CrossRef]
- S. Rath, A. Tripathy, and A. R. Tripathy, “Prediction of new active cases of coronavirus disease (COVID-19) pandemic using multiple linear regression model,” Diabetes Metab. Syndr. Clin. Res. Rev., 2020. [CrossRef]
- R. O. Ogundokun, A. F. Lukman, G. B. M. Kibria, J. B. Awotunde, and B. B. Aladeitan, “Predictive modelling of COVID-19 confirmed cases in Nigeria,” Infect. Dis. Model., 2020. [CrossRef]
- R. Gupta, G. Pandey, P. Chaudhary, and S. K. Pal, “Machine Learning Models for Government to Predict COVID-19 Outbreak,” Digit. Gov. Res. Pract., 2020. [CrossRef]
- E. Matthew and O. Adeyinka, “Application of Hierarchical Polynomial Regression Models to Predict Transmission of COVID-19 at Global Level,” Int. J. Clin. Biostat. Biometrics, 2020. [CrossRef]
- M. H. D. M. Ribeiro, R. G. da Silva, V. C. Mariani, and L. dos S. Coelho, “Short-term forecasting COVID-19 cumulative confirmed cases: Perspectives for Brazil,” Chaos, Solitons and Fractals, 2020. [CrossRef]
- M. Saqib, “Forecasting COVID-19 outbreak progression using hybrid polynomial-Bayesian ridge regression model,” Appl. Intell., 2021. [CrossRef]
- S. R. Vadyala, S. N. Betgeri, E. A. Sherer, and A. Amritphale, “Prediction of the number of COVID-19 confirmed cases based on K-means-LSTM,” Array, 2021. [CrossRef]
- T. Zhang and G. Lin, “Generalized k-means in GLMs with applications to the outbreak of COVID-19 in the United States,” Comput. Stat. Data Anal., 2021. [CrossRef]
- Z. R. S. Elsi et al., “Utilization of Data Mining Techniques in National Food Security during the Covid-19 Pandemic in Indonesia,” 2020. [CrossRef]
- A. M. Anter, D. Oliva, A. Thakare, and Z. Zhang, “AFCM-LSMA: New intelligent model based on Lévy slime mould algorithm and adaptive fuzzy C-means for identification of COVID-19 infection from chest X-ray images,” Adv. Eng. Informatics, 2021. [CrossRef]
- K. Chadaga, S. Prabhu, B. K. Vivekananda, S. Niranjana, and S. Umakanth, “Battling COVID-19 using machine learning: A review,” Cogent Engineering. 2021. [CrossRef]
- B. Sahu, P. K. Das, M. R. Kabat, and R. Kumar, “Prevention of Covid-19 affected patient using multi robot cooperation and Q-learning approach: a solution,” Qual. Quant., 2022. [CrossRef]
- L. Miralles-Pechuán, F. Jiménez, H. Ponce, and L. Martínez-Villaseñor, “A Methodology Based on Deep Q-Learning/Genetic Algorithms for Optimizing COVID-19 Pandemic Government Actions,” 2020. [CrossRef]
- W. Zhang et al., “Analysis of COVID-19 epidemic and clinical risk factors of patients under epidemiological Markov model,” Results Phys., 2021. [CrossRef]
- Liu et al., “Computationally Optimized SARS-CoV-2 MHC Class I and II Vaccine Formulations Predicted to Target Human Haplotype Distributions,” Cell Syst., 2020. [CrossRef]
- A. Chandra Kaushik and U. Raj, “AI-driven drug discovery: A boon against COVID-19?,” AI Open, 2020. [CrossRef]
- “COVID-19: Role of Artificial Intelligent in Drug Discovery.” https://www.tcs.com/company-overview/tcs-artificial-intelligence-cure-covid-19 (accessed Nov. 02, 2020).
- “Potential new treatment for COVID-19 uncovered by BenevolentAI enters trials | TechCrunch.” https://techcrunch.com/2020/04/14/potential-new-treatment-for-covid-19-uncovered-by-benevolentai-enters-trials/?guccounter=1 (accessed Nov. 02, 2020).
- “AI technology to screen existing drugs for use against COVID-19.” https://www.drugtargetreview.com/news/59188/ai-technology-to-screen-existing-drugs-for-use-against-covid-19/ (accessed Nov. 02, 2020).
- “AI Speeds Drug Discovery to fight COVID-19 | by Arijit Roy | Towards Data Science.” https://towardsdatascience.com/ai-speeds-drug-discovery-to-fight-covid-19-b853a3f93e82 (accessed Nov. 02, 2020).
- A. Nandy and S. C. Basak, “Bioinformatics in design of antiviral vaccines,” in Encyclopedia of Biomedical Engineering, 2018. [CrossRef]
- R. Rappuoli, “Reverse vaccinology,” Current Opinion in Microbiology. 2000. [CrossRef]
- G. P. Monterrubio-López, J. A. González-Y-Merchand, and R. M. Ribas-Aparicio, “Identification of novel potential vaccine candidates against tuberculosis based on reverse vaccinology,” Biomed Res. Int., 2015. [CrossRef]
- S. Vivona, F. Bernante, and F. Filippini, “NERVE: New Enhanced Reverse Vaccinology Environment,” BMC Biotechnol., 2006. [CrossRef]
- Y. Hisham and Y. Ashhab, “Identification of cross-protective potential antigens against pathogenic brucella spp. through combining pan-genome analysis with reverse vaccinology,” J. Immunol. Res., 2018. [CrossRef]
- M. Tahir Ul Qamar et al., “Reverse vaccinology assisted designing of multiepitope-based subunit vaccine against SARS-CoV-2,” Infect. Dis. Poverty, 2020. [CrossRef]
- H. Z. Chen, L. L. Tang, X. L. Yu, J. Zhou, Y. F. Chang, and X. Wu, “Bioinformatics analysis of epitope-based vaccine design against the novel SARS-CoV-2,” Infect. Dis. Poverty, 2020. [CrossRef]
- M. Bhattacharya et al., “Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach,” J. Med. Virol., 2020. [CrossRef]
- A. Rakib et al., “Immunoinformatics-guided design of an epitope-based vaccine against severe acute respiratory syndrome coronavirus 2 spike glycoprotein,” Comput. Biol. Med., 2020. [CrossRef]
- A. Naz, F. Shahid, T. T. Butt, F. M. Awan, A. Ali, and A. Malik, “Designing Multi-Epitope Vaccines to Combat Emerging Coronavirus Disease 2019 (COVID-19) by Employing Immuno-Informatics Approach,” Front. Immunol., 2020. [CrossRef]
- T. Kar et al., “A candidate multi-epitope vaccine against SARS-CoV-2,” Sci. Rep., 2020. [CrossRef]
- B. Robson, “COVID-19 Coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed achilles’ heel conserved region to minimize probability of escape mutations and drug resistance,” Comput. Biol. Med., 2020. [CrossRef]
- M. Enayatkhani et al., “Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study,” J. Biomol. Struct. Dyn., 2020. [CrossRef]
- B. Sarkar, M. A. Ullah, F. T. Johora, M. A. Taniya, and Y. Araf, “Immunoinformatics-guided designing of epitope-based subunit vaccines against the SARS Coronavirus-2 (SARS-CoV-2),” Immunobiology, 2020. [CrossRef]
- Park, J., Lagniton, P., Liu, Y. mRNA vaccines for COVID-19: what, why and how. (n.d.) Retrieved October 26, 2023, from www.ncbi.nlm.nih.gov/pmc/articles/PMC8071766/.
- Fang, E., Liu, X., Li, M., Zhang, Z., Song, L., Zhu, B. Advances in COVID-19 mRNA vaccine development. (n.d.) Retrieved October 26, 2023, from www.nature.com/articles/s41392-022-00950-y.
- Anand, P., Stahel, V. The safety of Covid-19 mRNA vaccines: a review. (n.d.) Retrieved October 26, 2023, from pssjournal.biomedcentral.com.
- Vitiello, A., Ferrara, F. Brief review of the mRNA vaccines COVID-19. (n.d.) Retrieved October 26, 2023, from link.springer.com/article/10.1007/s10787-021-00811-0. [CrossRef]
- Tenforde, M., Self, W., Adams, K., Gaglani, M., Ginde, A. Association Between mRNA Vaccination and COVID-19 Hospitalization and Disease Severity. (n.d.) Retrieved October 26, 2023, from jamanetwork.com/journals/jama/article-abstract/2786039.
- Huang, Q., Zeng, J., Yan, J. Review COVID-19 mRNA vaccines. (n.d.) Retrieved October 26, 2023, from www.sciencedirect.com/science/article/pii/S167385272100045X.
- Kashte, S., Gulbake, A., El-Amin III, S., Gupta, A. COVID-19 vaccines: rapid development, implications, challenges and future prospects. (n.d.) Retrieved October 26, 2023, from link.springer.com/article/10.1007/s13577-021-00512-4. [CrossRef]
- Huang, Y., Sun, H., Yu, H., Li, S., Zheng, Q. Neutralizing antibodies against SARS-CoV-2: current understanding, challenge and perspective | Antibody Therapeutics | Oxford Academic. (n.d.) Retrieved October 26, 2023, from academic.oup.com/abt/article-abstract/3/4/285/6053820.
| Country | Stabilizing Mutations | Carrier | Type | Mechanism |
|---|---|---|---|---|
| Germany,USA/BioNTech/Pfizer | Yes | Simian Monkey A Virus | mRNA | Following injection, the vaccine particles collide with cells and fuse with them, releasing mRNA. Molecules in the cell read the sequence and construct spike proteins. The cell eventually destroys the vaccine's mRNA, leaving no permanent trace. |
| USA/Moderna | Yes | Human AA Virus | mRNA | The vaccine's nucleoside-modified mRNA is formulated in lipid particles, allowing delivery of the nucleoside-modified mRNA into host cells and expression of the SARS CoV 2 Spike antigen. |
| UK, Sweden/Oxford/AstraZeneca | No | Monkey AA Virus | Adenovector | The adenoviruses collide with cells and latch onto proteins on their surfaces after being injected into a person's arm. The virus is engulfed in a bubble by the cell, which pulls it inside. Once inside, the adenovirus escapes the bubble and travels to the nucleus, which houses the cell's DNA. |
| China/Cansino | No | Adenovector | The new coronavirus is carried into the human body using a modified common cold virus. | |
| Russia/Sputnik V | No | Human AA Virus | Adenovector | A weakened virus is used to deliver small amounts of a pathogen and to stimulate an immune response. |
| China/Sinopharm | Not applicable | Inactivated | It works by exposing the body's immune system to the virus via killing viral particles without risking a serious disease response. | |
| India/BharatCovaxin | Not applicable | Inactivated | It is made from a weakened version of a common cold virus (called an adenovirus) derived from chimps. It has been altered to resemble a coronavirus. | |
| China/Sinovac CoronaVac | Not applicable | Inactivated | It works by exposing the body's immune system to the virus via killing viral particles without risking a serious disease response. | |
| Russia/Vector Institute EpiVacCorona | Yes | Subunit | The vaccine is based on chemically synthesized SARS-CoV-2 protein-peptide antigens conjugated to a carrier protein and adsorbed on an aluminum-containing adjuvant (aluminum hydroxide). | |
| Germany/CureVac | Yes | mRNA | Following vaccination, the body recognizes the protein as potentially harmful and activates the immune system, producing antibodies and T cells to combat it. In this way, we mimic natural viral infection and activate the immune system. | |
| USA/Johnson & Johnson | Yes | Adenovector | An adenovirus serves as a vehicle for the transmission of coronavirus genetic material (DNA). The adenovirus delivers the small piece of DNA to the cell, which then produces the spike protein. |
| Algorithms | Subcategory | Methods | Recent Publications |
|---|---|---|---|
| Supervised Learning | Classification | K-nearest Neighbours (KNN) | KNN was used to identify respiratory diseases by Ginantra et al. [62] KNN was utilized by Yin et al. [63] to identify severe influenza. In (Cho, 2016) [64], KNN was used to track the infected users' locations. |
| Support Vector Machine (SVM) | SVM was employed by Mori et al. [65] to forecast the onset of a disaster in a particular area. For the diagnosis of COVID-19, SVM was used with IoT (Internet of Things) and CNN (Convolutional Neural Networks) by Le et al. [66]. | ||
| Naïve Bayes | Naive Bayes classifier has been used by Sadhukhan et al. [67] and Assery et al. [68] to group tweets. During the pandemic, it assisted in managing social networking issues. | ||
| Logistic Regression | To categorize COVID-19 patients in Iran, Mohammadi et al. [69] employed ANN and LR. Various blood and clinical indicators were employed by Bhandari et al. [70] to use Logistic Regression to predict the death rate. COVID-19 in Kuwait was predicted using LR by Almeshal et al. [71] | ||
| Decision Trees | During pandemics, decision trees have been used to determine the location of the users by Elhoseny [72]. Decision trees were used to discuss the prediction of several aspects that contributed to psychological suffering during the pandemic by Chen & Liu [73]. Using decision trees, a hybrid face mask detection application was created by Loey et al.[74]. | ||
| Random Forest | RF method was used to predict COVID-19 health by Iwendi et al. [75] The spatial-temporal distribution of COVID-19 daily cases worldwide was estimated using the random forest machine learning approach by Yeşilkanat [76]. | ||
| Artificial Neural Network (ANN) | IoT and ANN were combined to determine the user's position by Luoh [77]. Although there was little data available, the accuracy was great. ANN was employed by Polese et al. [78] to identify the precise group of persons who were present in a location. | ||
| Deep Neural Network (DNN) | DNN was employed by Chhikara et al. [79] to evacuate a crowd in a crisis. Transferring healthy individuals to areas free of infectious diseases is crucial. | ||
| Convolution Neural Network (CNN) | People with COVID-19 were diagnosed using an IoHT (Internet of Health Things) and CNN-based model by More et al. [80] | ||
| Regression | Linear | To anticipate new COVID-19 coronavirus illness cases that are currently active, a multivariate linear regression model was applied by Rath et al. [81] A linear regression model was used to predict COVID-19 positive samples in Nigeria by Ogundokun et al.[82] | |
| Polynomial | Based on the polynomial regression model, estimates of the COVID-19 epidemic in India were examined by Pandey et al.[83]. To predict the global spread of COVID-19, hierarchical polynomial regression models were used by Ekum & Ogunsanya [84]. | ||
| Support Vector | It was applied in Brazil to anticipate COVID-19 confirmed cases in the short term by Ribeiro et al.[85] | ||
| Ridge | It was explored in utilizing a hybrid polynomial-Bayesian ridge regression model to predict the progression of the COVID-19 pandemic by Saqib [86]. | ||
| Unsupervised Learning | K-Means | The use of k-means to project the number of COVID-19 cases reported was covered by Vadyala et al. [87] In the United States of America, the K-Means method was used to forecast COVID-19 instances by Zhang & Lin [88]. | |
| K-Medoids | Indonesia's National Food Security during the COVID-19 epidemic utilized data mining techniques, including the k-medoids algorithm, as was discussed in by Elsi et al.[89] | ||
| Fuzzy C-Means | For detecting COVID-19 infection in chest X-rays, an intelligent model based on the Lévy slime mould algorithm and adaptive fuzzy C-means has been constructed by Anter et al. [90] Using Fast Fuzzy C means clustering, ROI extraction in COVID-19 CT lung images was mentioned by Chadaga et al. [91] The radiologists and other medical specialists benefited from this. | ||
| Reinforcement Learning | Q-Learning | Using multi-robot cooperation and the Q-learning approach, a strategy for the prevention of COVID-19 affected patients was provided by Sahu et al. [92] Miralles-Pechuán et al. [93] presented a novel methodology based on Deep Q-learning/Genetic Algorithms for optimizing Covid-19 pandemic government activities. | |
| Markov’s Decision Process | Clinical risk factors of patients and the COVID-19 pandemic were analyzed using the epidemiological Markov model by W. Zhang et al. [94] |
| Research Group | Database | Target Proteins | B-cell and T-cell Epitope Prediction |
Antigenicity and Allergenicity Test | Molecular Docking | Immunogenicity Evaluation |
Energy Minimization and Binding Affinity Prediction | In Silico Cloning and Codon Optimization |
|---|---|---|---|---|---|---|---|---|
| Qamar et al.[106] | GenBank | S, E, M proteins | ✔ | ✔ | ✔ | ✔ | ● | ✔ |
| Chen et al.[107] | NCBI | S, N proteins | ✔ | ✔ | ● | ● | ● | ● |
| Bhattacharya et al.[108] | NCBI | S protein | ✔ | ✔ | ✔ | ● | ● | ● |
| Ahmed et al.[109] | UniProt | S protein | ✔ | ✔ | ✔ | ● | ● | ● |
| Naz et al.[110] | NCBI | S protein | ✔ | ✔ | ✔ | ● | ✔ | ● |
| Kar et al.[111] | PDB (6VSB) | S protein | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Robson[112] | GenBank and the Protein Data Bank | S protein | ✔ | ● | ● | ● | ● | ● |
| Enayatkhani et al. [113] | NCBI | First S, E, M, N, ORF10, ORF8, ORF3a; then after the antigenicity test N, ORF3a, and M | ✔ | ✔ | ✔ | ● | ✔ | ● |
| Sarkar et al.[114] | NCBI | N, M, S, ORF3a | ✔ | ✔ | ✔ | ● | ● | ✔ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




