Submitted:
22 November 2023
Posted:
23 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Oxidative Stress Produces DNA Lesions, Turning OGG1 into an Epigenetic Eeader
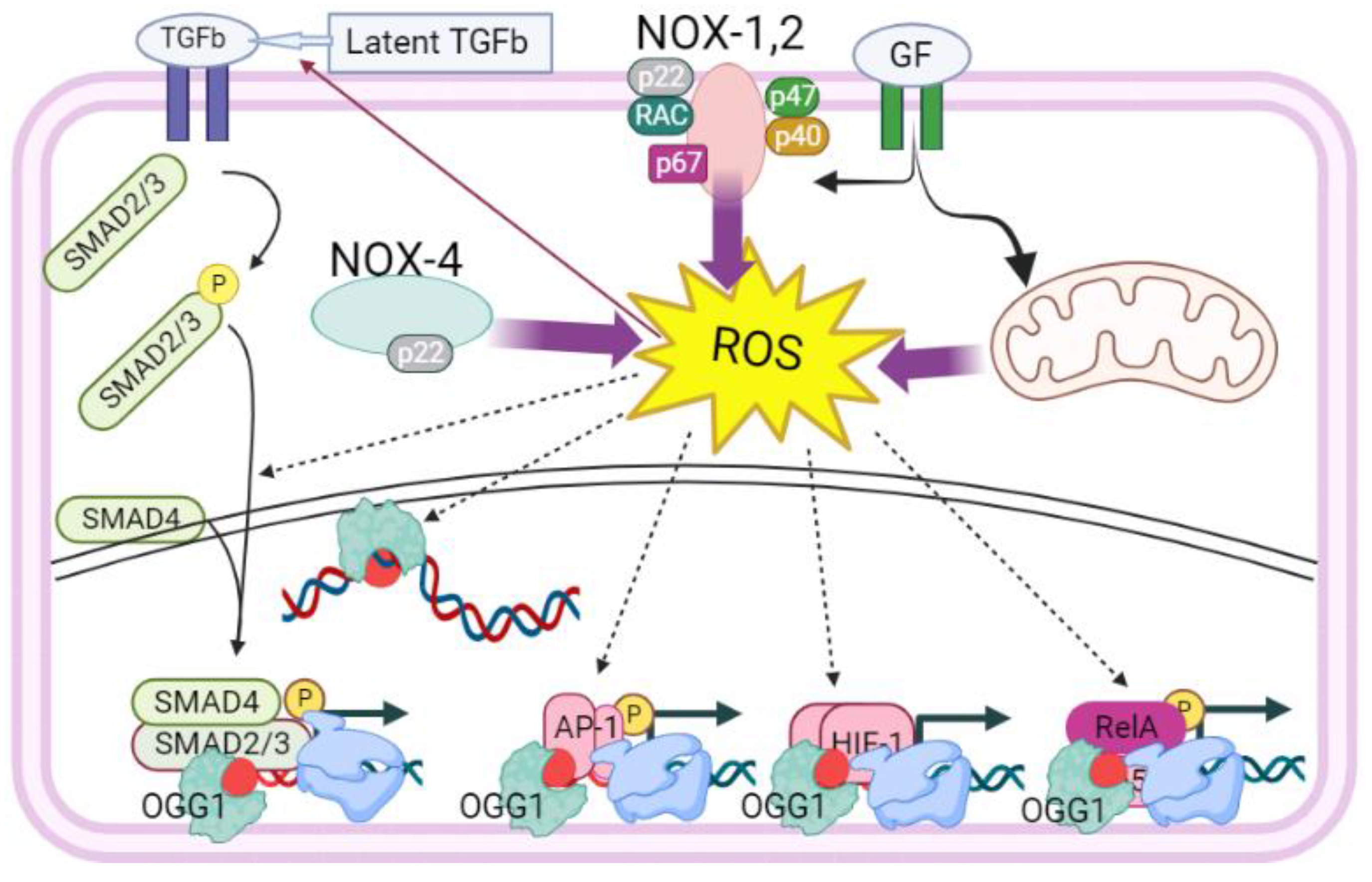
3. OGG1-Induced DNA Remodeling in Proximity of Transacting Factor’s cis Elements
4. Direct Impact of the OGG1 Interaction with NFκB
5. A Convoluted Mode of Regulation for NFκB Leads to a Complex Function in Cancer
6. Oxidative Stress as a Driver for Cancer Plasticity
7. A Model for Malignant Progression for Cancers Driven by Changes in Oxidative Stress Response of OGG1
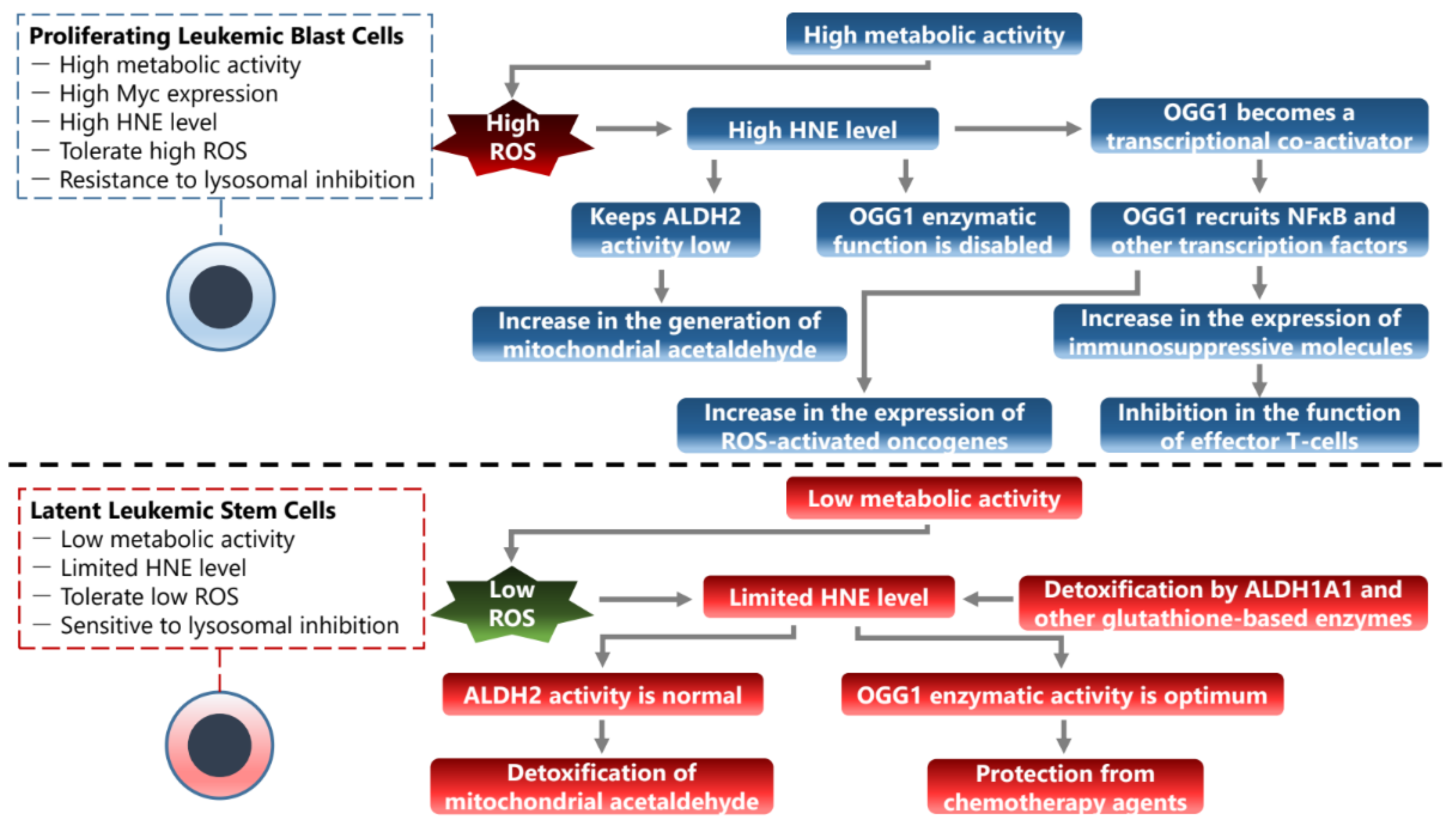
8. Phenotypic Transitions in the Microenvironment
9. Conclusions
Funding
Conflicts of Interest
References
- Gregory, C.D. Hijacking Homeostasis: Regulation of the Tumor Microenvironment by Apoptosis. Immunol. Rev. 2023. [Google Scholar] [CrossRef] [PubMed]
- Tabu, K.; Taga, T. Cancer Ego-System in Glioma: An Iron-Replenishing Niche Network Systemically Self-Organized by Cancer Stem Cells. Inflamm. Regen. 2022, 42, 54. [Google Scholar] [CrossRef] [PubMed]
- Vlahopoulos, S.A.; Cen, O.; Hengen, N.; Agan, J.; Moschovi, M.; Critselis, E.; Adamaki, M.; Bacopoulou, F.; Copland, J.A.; Boldogh, I.; et al. Dynamic Aberrant NF-κB Spurs Tumorigenesis: A New Model Encompassing the Microenvironment. Cytokine Growth Factor Rev. 2015, 26, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Lambrou, G.I.; Hatziagapiou, K.; Vlahopoulos, S. Inflammation and Tissue Homeostasis: The NF-κB System in Physiology and Malignant Progression. Mol. Biol. Rep. 2020. [Google Scholar] [CrossRef] [PubMed]
- Vlahopoulos, S.A. Aberrant Control of NF-κB in Cancer Permits Transcriptional and Phenotypic Plasticity, to Curtail Dependence on Host Tissue: Molecular Mode. Cancer Biol. Med. 2017, 14, 254–270. [Google Scholar] [CrossRef] [PubMed]
- Neganova, M.; Liu, J.; Aleksandrova, Y.; Klochkov, S.; Fan, R. Therapeutic Influence on Important Targets Associated with Chronic Inflammation and Oxidative Stress in Cancer Treatment. Cancers 2021, 13, 6062. [Google Scholar] [CrossRef] [PubMed]
- Beury, D.W.; Carter, K.A.; Nelson, C.; Sinha, P.; Hanson, E.; Nyandjo, M.; Fitzgerald, P.J.; Majeed, A.; Wali, N.; Ostrand-Rosenberg, S. Myeloid-Derived Suppressor Cell Survival and Function Are Regulated by the Transcription Factor Nrf2. J. Immunol. Baltim. Md 1950 2016, 196, 3470–3478. [Google Scholar] [CrossRef]
- Zhao, L.; He, R.; Long, H.; Guo, B.; Jia, Q.; Qin, D.; Liu, S.-Q.; Wang, Z.; Xiang, T.; Zhang, J.; et al. Late-Stage Tumors Induce Anemia and Immunosuppressive Extramedullary Erythroid Progenitor Cells. Nat. Med. 2018, 24, 1536–1544. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Chen, H.; Liang, J.; Li, Y.; Yang, J.; Luo, C.; Tang, Y.; Ding, Y.; Liu, X.; Yuan, Q.; et al. Dual Role of Reactive Oxygen Species and Their Application in Cancer Therapy. J. Cancer 2021, 12, 5543–5561. [Google Scholar] [CrossRef]
- Burrows, C.J.; Muller, J.G. Oxidative Nucleobase Modifications Leading to Strand Scission. Chem. Rev. 1998, 98, 1109–1152. [Google Scholar] [CrossRef]
- Cheng, K.C.; Cahill, D.S.; Kasai, H.; Nishimura, S.; Loeb, L.A. 8-Hydroxyguanine, an Abundant Form of Oxidative DNA Damage, Causes G----T and A----C Substitutions. J. Biol. Chem. 1992, 267, 166–172. [Google Scholar] [CrossRef]
- Hazra, T.K.; Das, A.; Das, S.; Choudhury, S.; Kow, Y.W.; Roy, R. Oxidative DNA Damage Repair in Mammalian Cells: A New Perspective. DNA Repair 2007, 6, 470–480. [Google Scholar] [CrossRef]
- McCullough, A.K.; Dodson, M.L.; Lloyd, R.S. Initiation of Base Excision Repair: Glycosylase Mechanisms and Structures. Annu. Rev. Biochem. 1999, 68, 255–285. [Google Scholar] [CrossRef]
- Mitra, S.; Izumi, T.; Boldogh, I.; Bhakat, K.K.; Hill, J.W.; Hazra, T.K. Choreography of Oxidative Damage Repair in Mammalian Genomes. Free Radic. Biol. Med. 2002, 33, 15–28. [Google Scholar] [CrossRef]
- Bessho, T.; Roy, R.; Yamamoto, K.; Kasai, H.; Nishimura, S.; Tano, K.; Mitra, S. Repair of 8-Hydroxyguanine in DNA by Mammalian N-Methylpurine-DNA Glycosylase. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 8901–8904. [Google Scholar] [CrossRef]
- Izumi, T.; Hazra, T.K.; Boldogh, I.; Tomkinson, A.E.; Park, M.S.; Ikeda, S.; Mitra, S. Requirement for Human AP Endonuclease 1 for Repair of 3’-Blocking Damage at DNA Single-Strand Breaks Induced by Reactive Oxygen Species. Carcinogenesis 2000, 21, 1329–1334. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.J.; Wilson, S.H. DNA Scanning by Base Excision Repair Enzymes and Implications for Pathway Coordination. DNA Repair 2018, 71, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Sampath, H.; Lloyd, R.S. Roles of OGG1 in Transcriptional Regulation and Maintenance of Metabolic Homeostasis. DNA Repair 2019, 81, 102667. [Google Scholar] [CrossRef] [PubMed]
- Lebraud, E.; Pinna, G.; Siberchicot, C.; Depagne, J.; Busso, D.; Fantini, D.; Irbah, L.; Robeska, E.; Kratassiouk, G.; Ravanat, J.-L.; et al. Chromatin Recruitment of OGG1 Requires Cohesin and Mediator and Is Essential for Efficient 8-oxoG Removal. Nucleic Acids Res. 2020, 48, 9082–9097. [Google Scholar] [CrossRef]
- D’Augustin, O.; Huet, S.; Campalans, A.; Radicella, J.P. Lost in the Crowd: How Does Human 8-Oxoguanine DNA Glycosylase 1 (OGG1) Find 8-Oxoguanine in the Genome? Int. J. Mol. Sci. 2020, 21, 8360. [Google Scholar] [CrossRef]
- Dizdaroglu, M.; Coskun, E.; Jaruga, P. Repair of Oxidatively Induced DNA Damage by DNA Glycosylases: Mechanisms of Action, Substrate Specificities and Excision Kinetics. Mutat. Res. Rev. Mutat. Res. 2017, 771, 99–127. [Google Scholar] [CrossRef] [PubMed]
- Grishko, V.; Solomon, M.; Breit, J.F.; Killilea, D.W.; Ledoux, S.P.; Wilson, G.L.; Gillespie, M.N. Hypoxia Promotes Oxidative Base Modifications in the Pulmonary Artery Endothelial Cell VEGF Gene. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2001, 15, 1267–1269. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Ding, Y.; Burrows, C.J. Oxidative DNA Damage Is Epigenetic by Regulating Gene Transcription via Base Excision Repair. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 2604–2609. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Burrows, C.J. 8-Oxo-7,8-Dihydroguanine, Friend and Foe: Epigenetic-like Regulator versus Initiator of Mutagenesis. DNA Repair 2017, 56, 75–83. [Google Scholar] [CrossRef]
- Fleming, A.M.; Zhu, J.; Ding, Y.; Burrows, C.J. 8-Oxo-7,8-Dihydroguanine in the Context of a Gene Promoter G-Quadruplex Is an On-Off Switch for Transcription. ACS Chem. Biol. 2017, 12, 2417–2426. [Google Scholar] [CrossRef]
- Ba, X.; Boldogh, I. 8-Oxoguanine DNA Glycosylase 1: Beyond Repair of the Oxidatively Modified Base Lesions. Redox Biol. 2018, 14, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Forneris, F.; Binda, C.; Vanoni, M.A.; Mattevi, A.; Battaglioli, E. Histone Demethylation Catalysed by LSD1 Is a Flavin-Dependent Oxidative Process. FEBS Lett. 2005, 579, 2203–2207. [Google Scholar] [CrossRef]
- Perillo, B.; Ombra, M.N.; Bertoni, A.; Cuozzo, C.; Sacchetti, S.; Sasso, A.; Chiariotti, L.; Malorni, A.; Abbondanza, C.; Avvedimento, E.V. DNA Oxidation as Triggered by H3K9me2 Demethylation Drives Estrogen-Induced Gene Expression. Science 2008, 319, 202–206. [Google Scholar] [CrossRef]
- Amente, S.; Bertoni, A.; Morano, A.; Lania, L.; Avvedimento, E.V.; Majello, B. LSD1-Mediated Demethylation of Histone H3 Lysine 4 Triggers Myc-Induced Transcription. Oncogene 2010, 29, 3691–3702. [Google Scholar] [CrossRef]
- Bazopoulou, D.; Knoefler, D.; Zheng, Y.; Ulrich, K.; Oleson, B.J.; Xie, L.; Kim, M.; Kaufmann, A.; Lee, Y.-T.; Dou, Y.; et al. Developmental ROS Individualizes Organismal Stress Resistance and Lifespan. Nature 2019, 576, 301–305. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. DNA Modifications Walk a Fine Line between Epigenetics and Mutagenesis. Nat. Rev. Mol. Cell Biol. 2023, 24, 449–450. [Google Scholar] [CrossRef]
- Pan, L.; Zhu, B.; Hao, W.; Zeng, X.; Vlahopoulos, S.A.; Hazra, T.K.; Hegde, M.L.; Radak, Z.; Bacsi, A.; Brasier, A.R.; et al. Oxidized Guanine Base Lesions Function in 8-Oxoguanine DNA Glycosylase-1-Mediated Epigenetic Regulation of Nuclear Factor κB-Driven Gene Expression. J. Biol. Chem. 2016, 291, 25553–25566. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Fleming, A.M.; Burrows, C.J. Sequencing the Mouse Genome for the Oxidatively Modified Base 8-Oxo-7,8-Dihydroguanine by OG-Seq. J. Am. Chem. Soc. 2017, 139, 2569–2572. [Google Scholar] [CrossRef] [PubMed]
- Amente, S.; Di Palo, G.; Scala, G.; Castrignanò, T.; Gorini, F.; Cocozza, S.; Moresano, A.; Pucci, P.; Ma, B.; Stepanov, I.; et al. Genome-Wide Mapping of 8-Oxo-7,8-Dihydro-2’-Deoxyguanosine Reveals Accumulation of Oxidatively-Generated Damage at DNA Replication Origins within Transcribed Long Genes of Mammalian Cells. Nucleic Acids Res. 2019, 47, 221–236. [Google Scholar] [CrossRef]
- Gorini, F.; Scala, G.; Ambrosio, S.; Majello, B.; Amente, S. OxiDIP-Seq for Genome-Wide Mapping of Damaged DNA Containing 8-Oxo-2’-Deoxyguanosine. Bio-Protoc. 2022, 12, e4540. [Google Scholar] [CrossRef] [PubMed]
- Gorini, F.; Scala, G.; Cooke, M.S.; Majello, B.; Amente, S. Towards a Comprehensive View of 8-Oxo-7,8-Dihydro-2’-Deoxyguanosine: Highlighting the Intertwined Roles of DNA Damage and Epigenetics in Genomic Instability. DNA Repair 2021, 97, 103027. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Qi, T.; Pan, L.; Wang, R.; Zhu, B.; Aguilera-Aguirre, L.; Radak, Z.; Hazra, T.K.; Vlahopoulos, S.A.; Bacsi, A.; et al. Effects of the Stimuli-Dependent Enrichment of 8-Oxoguanine DNA Glycosylase1 on Chromatinized DNA. Redox Biol. 2018, 18, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Pezone, A.; Taddei, M.L.; Tramontano, A.; Dolcini, J.; Boffo, F.L.; De Rosa, M.; Parri, M.; Stinziani, S.; Comito, G.; Porcellini, A.; et al. Targeted DNA Oxidation by LSD1-SMAD2/3 Primes TGF-Β1/ EMT Genes for Activation or Repression. Nucleic Acids Res. 2020, 48, 8943–8958. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Hao, W.; Xue, Y.; Wang, K.; Zheng, X.; Luo, J.; Ba, X.; Xiang, Y.; Qin, X.; Bergwik, J.; et al. 8-Oxoguanine Targeted by 8-Oxoguanine DNA Glycosylase 1 (OGG1) Is Central to Fibrogenic Gene Activation upon Lung Injury. Nucleic Acids Res. 2023, 51, 1087–1102. [Google Scholar] [CrossRef]
- Bruner, S.D.; Norman, D.P.; Verdine, G.L. Structural Basis for Recognition and Repair of the Endogenous Mutagen 8-Oxoguanine in DNA. Nature 2000, 403, 859–866. [Google Scholar] [CrossRef]
- Boiteux, S.; Radicella, J.P. The Human OGG1 Gene: Structure, Functions, and Its Implication in the Process of Carcinogenesis. Arch. Biochem. Biophys. 2000, 377, 1–8. [Google Scholar] [CrossRef]
- Hitomi, K.; Iwai, S.; Tainer, J.A. The Intricate Structural Chemistry of Base Excision Repair Machinery: Implications for DNA Damage Recognition, Removal, and Repair. DNA Repair 2007, 6, 410–428. [Google Scholar] [CrossRef]
- Ba, X.; Bacsi, A.; Luo, J.; Aguilera-Aguirre, L.; Zeng, X.; Radak, Z.; Brasier, A.R.; Boldogh, I. 8-Oxoguanine DNA Glycosylase-1 Augments Proinflammatory Gene Expression by Facilitating the Recruitment of Site-Specific Transcription Factors. J. Immunol. Baltim. Md 1950 2014, 192, 2384–2394. [Google Scholar] [CrossRef]
- Xue, Y.; Li, C.; Deng, S.; Chen, X.; Han, J.; Zheng, X.; Tian, M.; Hao, W.; Pan, L.; Boldogh, I.; et al. 8-Oxoguanine DNA Glycosylase 1 Selectively Modulates ROS-Responsive NF-κB Targets through Recruitment of MSK1 and Phosphorylation of RelA/P65 at Ser276. J. Biol. Chem. 2023, 299, 105308. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Vlahopoulos, S.; Tanner, L.; Bergwik, J.; Bacsi, A.; Radak, Z.; Egesten, A.; Ba, X.; Brasier, A.R.; Boldogh, I. Substrate-Specific Binding of 8-Oxoguanine DNA Glycosylase 1 (OGG1) Reprograms Mucosal Adaptations to Chronic Airway Injury. Front. Immunol. 2023, 14, 1186369. [Google Scholar] [CrossRef]
- Bangalore, D.M.; Tessmer, I. Direct hOGG1-Myc Interactions Inhibit hOGG1 Catalytic Activity and Recruit Myc to Its Promoters under Oxidative Stress. Nucleic Acids Res. 2022, 50, 10385–10398. [Google Scholar] [CrossRef] [PubMed]
- Visnes, T.; Cázares-Körner, A.; Hao, W.; Wallner, O.; Masuyer, G.; Loseva, O.; Mortusewicz, O.; Wiita, E.; Sarno, A.; Manoilov, A.; et al. Small-Molecule Inhibitor of OGG1 Suppresses Proinflammatory Gene Expression and Inflammation. Science 2018, 362, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wu, S.; Wu, G.; Yang, J. Inhibition of 8-Oxoguanine DNA Glycosylase (OGG1) Expression Suppresses Polycystic Ovarian Syndrome via the NF-κB Signaling Pathway. Reprod. Biol. 2022, 22, 100679. [Google Scholar] [CrossRef]
- Wibisana, J.N.; Inaba, T.; Shinohara, H.; Yumoto, N.; Hayashi, T.; Umeda, M.; Ebisawa, M.; Nikaido, I.; Sako, Y.; Okada, M. Enhanced Transcriptional Heterogeneity Mediated by NF-κB Super-Enhancers. PLoS Genet. 2022, 18, e1010235. [Google Scholar] [CrossRef]
- Xu, J.-W.; Ling, S.; Liu, J. Higher-Order Chromatin Regulation of Inflammatory Gene Expression. Mediators Inflamm. 2017, 2017, 7848591. [Google Scholar] [CrossRef]
- Wong, R.W.J.; Tan, T.K.; Amanda, S.; Ngoc, P.C.T.; Leong, W.Z.; Tan, S.H.; Asamitsu, K.; Hibi, Y.; Ueda, R.; Okamoto, T.; et al. Feed-Forward Regulatory Loop Driven by IRF4 and NF-κB in Adult T-Cell Leukemia/Lymphoma. Blood 2020, 135, 934–947. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Zhao, M.; Long, B.; Li, H. Super-Enhancer-Associated Gene CAPG Promotes AML Progression. Commun. Biol. 2023, 6, 622. [Google Scholar] [CrossRef] [PubMed]
- Daenthanasanmak, A.; Bamford, R.N.; Yoshioka, M.; Yang, S.-M.; Homan, P.; Karim, B.; Bryant, B.R.; Petrus, M.N.; Thomas, C.J.; Green, P.L.; et al. Triple Combination of BET plus PI3K and NF-κB Inhibitors Exhibit Synergistic Activity in Adult T-Cell Leukemia/Lymphoma. Blood Adv. 2022, 6, 2346–2360. [Google Scholar] [CrossRef] [PubMed]
- Vlahopoulos, S.; Adamaki, M.; Khoury, N.; Zoumpourlis, V.; Boldogh, I. Roles of DNA Repair Enzyme OGG1 in Innate Immunity and Its Significance for Lung Cancer. Pharmacol. Ther. 2018. [Google Scholar] [CrossRef] [PubMed]
- Aguilera-Aguirre, L.; Bacsi, A.; Radak, Z.; Hazra, T.K.; Mitra, S.; Sur, S.; Brasier, A.R.; Ba, X.; Boldogh, I. Innate Inflammation Induced by the 8-Oxoguanine DNA Glycosylase-1-KRAS-NF-κB Pathway. J. Immunol. Baltim. Md 1950 2014, 193, 4643–4653. [Google Scholar] [CrossRef] [PubMed]
- Gerke, M.B.; Christodoulou, I.; Karantanos, T. Definitions, Biology, and Current Therapeutic Landscape of Myelodysplastic/Myeloproliferative Neoplasms. Cancers 2023, 15, 3815. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lenardo, M.J.; Baltimore, D. 30 Years of NF-κB: A Blossoming of Relevance to Human Pathobiology. Cell 2017, 168, 37–57. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-H.; Lee, J.; Woo, J.; Lee, C.-H.; Yoo, C.-G. Proteasome Inhibitor-Induced IκB/NF-κB Activation Is Mediated by Nrf2-Dependent Light Chain 3B Induction in Lung Cancer Cells. Mol. Cells 2018, 41, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Li, L.; Zhang, M.; Song, N.; Zhao, Q.; Liu, Z.; Diao, A. TMEPAI Promotes Degradation of the NF-κB Signaling Pathway Inhibitory Protein IκBα and Contributes to Tumorigenesis. Int. J. Biol. Macromol. 2023, 235, 123859. [Google Scholar] [CrossRef]
- Han, Y.; Weinman, S.; Boldogh, I.; Walker, R.K.; Brasier, A.R. Tumor Necrosis Factor-Alpha-Inducible IkappaBalpha Proteolysis Mediated by Cytosolic m-Calpain. A Mechanism Parallel to the Ubiquitin-Proteasome Pathway for Nuclear Factor-Kappab Activation. J. Biol. Chem. 1999, 274, 787–794. [Google Scholar] [CrossRef]
- Varisli, L.; Cen, O.; Vlahopoulos, S. Dissecting Pharmacological Effects of Chloroquine in Cancer Treatment: Interference with Inflammatory Signaling Pathways. Immunology 2019. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-L.; Wang, X.; Mann, M.; Adamus, T.P.; Wang, D.; Moreira, D.F.; Zhang, Z.; Ouyang, C.; He, X.; Zhang, B.; et al. Myeloid Cell-Targeted miR-146a Mimic Inhibits NF-κB-Driven Inflammation and Leukemia Progression in Vivo. Blood 2020, 135, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Mehta, A.; de Boer, C.G.; Kowalczyk, M.S.; Lee, K.; Haldeman, P.; Rogel, N.; Knecht, A.R.; Farouq, D.; Regev, A.; et al. Heterogeneous Responses of Hematopoietic Stem Cells to Inflammatory Stimuli Are Altered with Age. Cell Rep. 2018, 25, 2992–3005.e5. [Google Scholar] [CrossRef] [PubMed]
- Jost, P.J.; Höckendorf, U. Necroinflammation Emerges as a Key Regulator of Hematopoiesis in Health and Disease. Cell Death Differ. 2019, 26, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Ghatak, D.; Datta, A.; Roychowdhury, T.; Chattopadhyay, S.; Roychoudhury, S. MicroRNA-324-5p-CUEDC2 Axis Mediates Gain-of-Function Mutant P53-Driven Cancer Stemness. Mol. Cancer Res. MCR 2021, 19, 1635–1650. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Li, P.; Wang, J.; Shu, Y.; Zhong, X.; Gao, Z.; Yang, J.; Jiang, Y.; Zhou, X.; et al. Long Noncoding RNA HOTAIR Regulates the Stemness of Breast Cancer Cells via Activation of the NF-κB Signaling Pathway. J. Biol. Chem. 2022, 298, 102630. [Google Scholar] [CrossRef]
- Dancik, G.M.; Varisli, L.; Vlahopoulos, S.A. The Molecular Context of Oxidant Stress Response in Cancer Establishes ALDH1A1 as a Critical Target: What This Means for Acute Myeloid Leukemia. Int. J. Mol. Sci. 2023, 24, 9372. [Google Scholar] [CrossRef]
- Moschovi, M.; Critselis, E.; Cen, O.; Adamaki, M.; Lambrou, G.I.; Chrousos, G.P.; Vlahopoulos, S. Drugs Acting on Homeostasis: Challenging Cancer Cell Adaptation. Expert Rev. Anticancer Ther. 2015, 15, 1405–1417. [Google Scholar] [CrossRef]
- Vlahopoulos, S.; Boldogh, I.; Casola, A.; Brasier, A.R. Nuclear Factor-kappaB-Dependent Induction of Interleukin-8 Gene Expression by Tumor Necrosis Factor Alpha: Evidence for an Antioxidant Sensitive Activating Pathway Distinct from Nuclear Translocation. Blood 1999, 94, 1878–1889. [Google Scholar] [CrossRef]
- Xue, Y.; Pan, L.; Vlahopoulos, S.; Wang, K.; Zheng, X.; Radak, Z.; Bacsi, A.; Tanner, L.; Brasier, A.R.; Ba, X.; et al. Epigenetic Control of Type III Interferon Expression by 8-Oxoguanine and Its Reader 8-Oxoguanine DNA Glycosylase1. Front. Immunol. 2023, 14, 1161160. [Google Scholar] [CrossRef]
- Wang, R.; Li, C.; Qiao, P.; Xue, Y.; Zheng, X.; Chen, H.; Zeng, X.; Liu, W.; Boldogh, I.; Ba, X. OGG1-Initiated Base Excision Repair Exacerbates Oxidative Stress-Induced Parthanatos. Cell Death Dis. 2018, 9, 628. [Google Scholar] [CrossRef] [PubMed]
- Hung, R.J.; Hall, J.; Brennan, P.; Boffetta, P. Genetic Polymorphisms in the Base Excision Repair Pathway and Cancer Risk: A HuGE Review. Am. J. Epidemiol. 2005, 162, 925–942. [Google Scholar] [CrossRef]
- Bravard, A.; Vacher, M.; Moritz, E.; Vaslin, L.; Hall, J.; Epe, B.; Radicella, J.P. Oxidation Status of Human OGG1-S326C Polymorphic Variant Determines Cellular DNA Repair Capacity. Cancer Res. 2009, 69, 3642–3649. [Google Scholar] [CrossRef]
- Hill, J.W.; Evans, M.K. Dimerization and Opposite Base-Dependent Catalytic Impairment of Polymorphic S326C OGG1 Glycosylase. Nucleic Acids Res. 2006, 34, 1620–1632. [Google Scholar] [CrossRef]
- Morreall, J.; Limpose, K.; Sheppard, C.; Kow, Y.W.; Werner, E.; Doetsch, P.W. Inactivation of a Common OGG1 Variant by TNF-Alpha in Mammalian Cells. DNA Repair 2015, 26, 15–22. [Google Scholar] [CrossRef]
- Peng, Y.; Li, Z.; Zhang, S.; Xiong, Y.; Cun, Y.; Qian, C.; Li, M.; Ren, T.; Xia, L.; Cheng, Y.; et al. Association of DNA Base Excision Repair Genes (OGG1, APE1 and XRCC1) Polymorphisms with Outcome to Platinum-Based Chemotherapy in Advanced Nonsmall-Cell Lung Cancer Patients. Int. J. Cancer 2014, 135, 2687–2696. [Google Scholar] [CrossRef] [PubMed]
- Le Marchand, L.; Donlon, T.; Lum-Jones, A.; Seifried, A.; Wilkens, L.R. Association of the hOGG1 Ser326Cys Polymorphism with Lung Cancer Risk. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2002, 11, 409–412. [Google Scholar]
- Smolarz, B.; Michalska, M.M.; Samulak, D.; Wójcik, L.; Romanowicz, H. Studies of Correlations Between Single Nucleotide Polymorphisms of DNA Repair Genes and Endometrial Cancer in Polish Women. Anticancer Res. 2018, 38, 5223–5229. [Google Scholar] [CrossRef]
- Kabzinski, J.; Walczak, A.; Dziki, A.; Mik, M.; Majsterek, I. Impact of the Ser326Cys Polymorphism of the OGG1 Gene on the Level of Oxidative DNA Damage in Patients with Colorectal Cancer. Pol. Przegl. Chir. 2018, 90, 13–15. [Google Scholar] [CrossRef]
- Alanazi, M.; Pathan, A.A.K.; Shaik, J.P.; Alhadheq, A.; Khan, Z.; Khan, W.; Al Naeem, A.; Parine, N.R. The hOGG1 Ser326Cys Gene Polymorphism and Breast Cancer Risk in Saudi Population. Pathol. Oncol. Res. POR 2017, 23, 525–535. [Google Scholar] [CrossRef]
- Costa, E.F.D.; Santos, E.S.; Liutti, V.T.; Leal, F.; Santos, V.C.A.; Rinck-Junior, J.A.; Mariano, F.V.; Coutinho-Camillo, C.M.; Altemani, A.; Lima, C.S.P.; et al. Association between Polymorphisms in Genes Related to DNA Base-Excision Repair with Risk and Prognosis of Oropharyngeal Squamous Cell Carcinoma. J. Cancer Res. Clin. Oncol. 2016, 142, 1917–1926. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Elahi, A.; Pow-Sang, J.; Lazarus, P.; Park, J. Association between Polymorphism of Human Oxoguanine Glycosylase 1 and Risk of Prostate Cancer. J. Urol. 2003, 170, 2471–2474. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zheng, S.L.; Turner, A.; Isaacs, S.D.; Wiley, K.E.; Hawkins, G.A.; Chang, B.; Bleecker, E.R.; Walsh, P.C.; Meyers, D.A.; et al. Associations between hOGG1 Sequence Variants and Prostate Cancer Susceptibility. Cancer Res. 2002, 62, 2253–2257. [Google Scholar] [PubMed]
- Gotoh, N.; Saitoh, T.; Takahashi, N.; Kasamatsu, T.; Minato, Y.; Lobna, A.; Oda, T.; Hoshino, T.; Sakura, T.; Shimizu, H.; et al. Association between OGG1 S326C CC Genotype and Elevated Relapse Risk in Acute Myeloid Leukemia. Int. J. Hematol. 2018, 108, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid Peroxidation: Production, Metabolism, and Signaling Mechanisms of Malondialdehyde and 4-Hydroxy-2-Nonenal. Oxid. Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef] [PubMed]
- Fazio, V.M.; Barrera, G.; Martinotti, S.; Farace, M.G.; Giglioni, B.; Frati, L.; Manzari, V.; Dianzani, M.U. 4-Hydroxynonenal, a Product of Cellular Lipid Peroxidation, Which Modulates c-Myc and Globin Gene Expression in K562 Erythroleukemic Cells. Cancer Res. 1992, 52, 4866–4871. [Google Scholar] [PubMed]
- Wang, W.; Ma, Y.; Huang, M.; Liang, W.; Zhao, X.; Li, Q.; Wang, S.; Hu, Z.; He, L.; Gao, T.; et al. Asymmetrical Arginine Dimethylation of Histone H4 by 8-Oxog/OGG1/PRMT1 Is Essential for Oxidative Stress-Induced Transcription Activation. Free Radic. Biol. Med. 2021, 164, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.-Y.; Wang, Y.; Cui, S.-Y.; Wu, X.-L.; Guo, Y.; Xu, R.-R. MicroRNA-125a Regulates Proliferation and Apoptosis of Acute Myeloid Leukemia through Targeting NF-κB Pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3594–3601. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.-H.; Zhang, W.-J.; Zhu, J.-F.; Cui, D.; Song, K.-D.; Qiang, P.; Mei, C.-Z.; Nie, Z.-C.; Ding, B.-S.; Han, Z.; et al. CaMKIIγ Regulates the Viability and Self-Renewal of Acute Myeloid Leukaemia Stem-like Cells by the Alox5/NF-κB Pathway. Int. J. Lab. Hematol. 2021, 43, 699–706. [Google Scholar] [CrossRef]
- Chen, X.-J.; Zhang, W.-N.; Chen, B.; Xi, W.-D.; Lu, Y.; Huang, J.-Y.; Wang, Y.-Y.; Long, J.; Wu, S.-F.; Zhang, Y.-X.; et al. Homoharringtonine Deregulates MYC Transcriptional Expression by Directly Binding NF-κB Repressing Factor. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 2220–2225. [Google Scholar] [CrossRef]
- Reikvam, H. Inhibition of NF-κB Signaling Alters Acute Myelogenous Leukemia Cell Transcriptomics. Cells 2020, 9, 1677. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, B.; Verzella, D.; Capece, D.; Vecchiotti, D.; Di Vito Nolfi, M.; Flati, I.; Cornice, J.; Di Padova, M.; Angelucci, A.; Alesse, E.; et al. NF-κB: A Druggable Target in Acute Myeloid Leukemia. Cancers 2022, 14, 3557. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Q.; Zou, Z.; Huang, Z.; Chen, Y. TRIM10 Is Downregulated in Acute Myeloid Leukemia and Plays a Tumor Suppressive Role via Regulating NF-κB Pathway. Cancers 2023, 15, 417. [Google Scholar] [CrossRef] [PubMed]
- Dancik, G.M.; Varisli, L.; Tolan, V.; Vlahopoulos, S. Aldehyde Dehydrogenase Genes as Prospective Actionable Targets in Acute Myeloid Leukemia. Genes 2023, 14, 1807. [Google Scholar] [CrossRef] [PubMed]
- Gasparetto, M.; Pei, S.; Minhajuddin, M.; Khan, N.; Pollyea, D.A.; Myers, J.R.; Ashton, J.M.; Becker, M.W.; Vasiliou, V.; Humphries, K.R.; et al. Targeted Therapy for a Subset of Acute Myeloid Leukemias That Lack Expression of Aldehyde Dehydrogenase 1A1. Haematologica 2017, 102, 1054–1065. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yao, R.; Wang, H. Update of ALDH as a Potential Biomarker and Therapeutic Target for AML. BioMed Res. Int. 2018, 2018, 9192104. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.; Gasparetto, M.; Humphries, K.; Pollyea, D.A.; Vasiliou, V.; Jordan, C.T. Aldehyde Dehydrogenases in Acute Myeloid Leukemia. Ann. N. Y. Acad. Sci. 2014, 1310, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Gasparetto, M.; Smith, C.A. ALDHs in Normal and Malignant Hematopoietic Cells: Potential New Avenues for Treatment of AML and Other Blood Cancers. Chem. Biol. Interact. 2017, 276, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Dancik, G.M.; Voutsas, I.F.; Vlahopoulos, S. Aldehyde Dehydrogenase Enzyme Functions in Acute Leukemia Stem Cells. Front. Biosci. Sch. Ed. 2022, 14, 8. [Google Scholar] [CrossRef]
- Dancik, G.M.; Voutsas, I.F.; Vlahopoulos, S. Lower RNA Expression of ALDH1A1 Distinguishes the Favorable Risk Group in Acute Myeloid Leukemia. Mol. Biol. Rep. 2022, 49, 3321–3331. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Y.; Toubai, T.; Oravecz-Wilson, K.; Liu, C.; Mathewson, N.; Wu, J.; Rossi, C.; Cummings, E.; Wu, D.; et al. BET Bromodomain Inhibition Suppresses Graft-versus-Host Disease after Allogeneic Bone Marrow Transplantation in Mice. Blood 2015, 125, 2724–2728. [Google Scholar] [CrossRef] [PubMed]
- Latif, A.-L.; Newcombe, A.; Li, S.; Gilroy, K.; Robertson, N.A.; Lei, X.; Stewart, H.J.S.; Cole, J.; Terradas, M.T.; Rishi, L.; et al. BRD4-Mediated Repression of P53 Is a Target for Combination Therapy in AML. Nat. Commun. 2021, 12, 241. [Google Scholar] [CrossRef] [PubMed]
- Snyder, K.J.; Choe, H.K.; Gao, Y.; Sell, N.E.; Braunreiter, K.M.; Zitzer, N.C.; Neidemire-Colley, L.; Kalyan, S.; Dorrance, A.M.; Keller, A.; et al. Inhibition of Bromodomain and Extra Terminal (BET) Domain Activity Modulates the IL-23R/IL-17 Axis and Suppresses Acute Graft-Versus-Host Disease. Front. Oncol. 2021, 11, 760789. [Google Scholar] [CrossRef] [PubMed]
- Fong, C.Y.; Gilan, O.; Lam, E.Y.N.; Rubin, A.F.; Ftouni, S.; Tyler, D.; Stanley, K.; Sinha, D.; Yeh, P.; Morison, J.; et al. BET Inhibitor Resistance Emerges from Leukaemia Stem Cells. Nature 2015, 525, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Rathert, P.; Roth, M.; Neumann, T.; Muerdter, F.; Roe, J.-S.; Muhar, M.; Deswal, S.; Cerny-Reiterer, S.; Peter, B.; Jude, J.; et al. Transcriptional Plasticity Promotes Primary and Acquired Resistance to BET Inhibition. Nature 2015, 525, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Moreb, J.; Schweder, M.; Suresh, A.; Zucali, J.R. Overexpression of the Human Aldehyde Dehydrogenase Class I Results in Increased Resistance to 4-Hydroperoxycyclophosphamide. Cancer Gene Ther. 1996, 3, 24–30. [Google Scholar] [PubMed]
- Pan, G.; Deshpande, M.; Pang, H.; Stemmer, P.M.; Carruthers, N.J.; Shearn, C.T.; Backos, D.S.; Palaniyandi, S.S. 4-Hydroxy-2-Nonenal Attenuates 8-Oxoguanine DNA Glycosylase 1 Activity. J. Cell. Biochem. 2020. [Google Scholar] [CrossRef]
- Costa, R.G.A.; Silva, S.L.R.; Dias, I.R.S.B.; Oliveira, M. de S.; Rodrigues, A.C.B. da C.; Dias, R.B.; Bezerra, D.P. Emerging Drugs Targeting Cellular Redox Homeostasis to Eliminate Acute Myeloid Leukemia Stem Cells. Redox Biol. 2023, 62, 102692. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, N.; Gu, D.; Wickliffe, J.; Salazar, J.; Boldogh, I.; Xie, J. Genetic Evidence for XPC-KRAS Interactions During Lung Cancer Development. J. Genet. Genomics Yi Chuan Xue Bao 2015, 42, 589–596. [Google Scholar] [CrossRef]
- Cogoi, S.; Ferino, A.; Miglietta, G.; Pedersen, E.B.; Xodo, L.E. The Regulatory G4 Motif of the Kirsten Ras (KRAS) Gene Is Sensitive to Guanine Oxidation: Implications on Transcription. Nucleic Acids Res. 2018, 46, 661–676. [Google Scholar] [CrossRef]
- Mali, V.R.; Ning, R.; Chen, J.; Yang, X.-P.; Xu, J.; Palaniyandi, S.S. Impairment of Aldehyde Dehydrogenase-2 by 4-Hydroxy-2-Nonenal Adduct Formation and Cardiomyocyte Hypertrophy in Mice Fed a High-Fat Diet and Injected with Low-Dose Streptozotocin. Exp. Biol. Med. Maywood NJ 2014, 239, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Kreuz, S.; Fischle, W. Oxidative Stress Signaling to Chromatin in Health and Disease. Epigenomics 2016, 8, 843–862. [Google Scholar] [CrossRef] [PubMed]
- Singhal, S.S.; Singh, S.P.; Singhal, P.; Horne, D.; Singhal, J.; Awasthi, S. Antioxidant Role of Glutathione S-Transferases: 4-Hydroxynonenal, a Key Molecule in Stress-Mediated Signaling. Toxicol. Appl. Pharmacol. 2015, 289, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Milkovic, L.; Zarkovic, N.; Marusic, Z.; Zarkovic, K.; Jaganjac, M. The 4-Hydroxynonenal-Protein Adducts and Their Biological Relevance: Are Some Proteins Preferred Targets? Antioxid. Basel Switz. 2023, 12, 856. [Google Scholar] [CrossRef] [PubMed]
- Folkerts, H.; Hilgendorf, S.; Wierenga, A.T.J.; Jaques, J.; Mulder, A.B.; Coffer, P.J.; Schuringa, J.J.; Vellenga, E. Inhibition of Autophagy as a Treatment Strategy for P53 Wild-Type Acute Myeloid Leukemia. Cell Death Dis. 2017, 8, e2927. [Google Scholar] [CrossRef] [PubMed]
- Liddiard, K.; Hills, R.; Burnett, A.K.; Darley, R.L.; Tonks, A. OGG1 Is a Novel Prognostic Indicator in Acute Myeloid Leukaemia. Oncogene 2010, 29, 2005–2012. [Google Scholar] [CrossRef] [PubMed]
- Owen, N.; Minko, I.G.; Moellmer, S.A.; Cammann, S.K.; Lloyd, R.S.; McCullough, A.K. Enhanced Cytarabine-Induced Killing in OGG1-Deficient Acute Myeloid Leukemia Cells. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, e2016833118. [Google Scholar] [CrossRef] [PubMed]
- Radpour, R.; Stucki, M.; Riether, C.; Ochsenbein, A.F. Epigenetic Silencing of Immune-Checkpoint Receptors in Bone Marrow- Infiltrating T Cells in Acute Myeloid Leukemia. Front. Oncol. 2021, 11, 663406. [Google Scholar] [CrossRef] [PubMed]
- Paczulla, A.M.; Rothfelder, K.; Raffel, S.; Konantz, M.; Steinbacher, J.; Wang, H.; Tandler, C.; Mbarga, M.; Schaefer, T.; Falcone, M.; et al. Absence of NKG2D Ligands Defines Leukaemia Stem Cells and Mediates Their Immune Evasion. Nature 2019, 572, 254–259. [Google Scholar] [CrossRef]
- Hassane, D.C.; Guzman, M.L.; Corbett, C.; Li, X.; Abboud, R.; Young, F.; Liesveld, J.L.; Carroll, M.; Jordan, C.T. Discovery of Agents That Eradicate Leukemia Stem Cells Using an in Silico Screen of Public Gene Expression Data. Blood 2008, 111, 5654–5662. [Google Scholar] [CrossRef]
- Karantanos, T.; Jones, R.J. Acute Myeloid Leukemia Stem Cell Heterogeneity and Its Clinical Relevance. Adv. Exp. Med. Biol. 2019, 1139, 153–169. [Google Scholar] [CrossRef]
- Vlahopoulos, S.; Wang, K.; Xue, Y.; Zheng, X.; Boldogh, I.; Pan, L. Endothelial Dysfunction through Oxidatively Generated Epigenetic Mark in Respiratory Viral Infections. Cells 2021, 10, 3067. [Google Scholar] [CrossRef] [PubMed]
- Terwoord, J.D.; Beyer, A.M.; Gutterman, D.D. Endothelial Dysfunction as a Complication of Anti-Cancer Therapy. Pharmacol. Ther. 2022, 237, 108116. [Google Scholar] [CrossRef]
- Feng, Y.; Luo, S.; Fan, D.; Guo, X.; Ma, S. The Role of Vascular Endothelial Cells in Tumor Metastasis. Acta Histochem. 2023, 125, 152070. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, X.; Tang, K.; Xin, Y.; Hu, G.; Zheng, Y.; Li, K.; Zhang, C.; Tan, Y. Adhesion to the Brain Endothelium Selects Breast Cancer Cells with Brain Metastasis Potential. Int. J. Mol. Sci. 2023, 24, 7087. [Google Scholar] [CrossRef] [PubMed]
- Podyacheva, E.; Danilchuk, M.; Toropova, Y. Molecular Mechanisms of Endothelial Remodeling under Doxorubicin Treatment. Biomed. Pharmacother. Biomedecine Pharmacother. 2023, 162, 114576. [Google Scholar] [CrossRef]
- Wang, S.; Kotamraju, S.; Konorev, E.; Kalivendi, S.; Joseph, J.; Kalyanaraman, B. Activation of Nuclear Factor-kappaB during Doxorubicin-Induced Apoptosis in Endothelial Cells and Myocytes Is pro-Apoptotic: The Role of Hydrogen Peroxide. Biochem. J. 2002, 367, 729–740. [Google Scholar] [CrossRef]
- Xu, A.; Deng, F.; Chen, Y.; Kong, Y.; Pan, L.; Liao, Q.; Rao, Z.; Xie, L.; Yao, C.; Li, S.; et al. NF-κB Pathway Activation during Endothelial-to-Mesenchymal Transition in a Rat Model of Doxorubicin-Induced Cardiotoxicity. Biomed. Pharmacother. Biomedecine Pharmacother. 2020, 130, 110525. [Google Scholar] [CrossRef] [PubMed]
- Mi, J.; Zhang, X.; Rabbani, Z.N.; Liu, Y.; Reddy, S.K.; Su, Z.; Salahuddin, F.K.; Viles, K.; Giangrande, P.H.; Dewhirst, M.W.; et al. RNA Aptamer-Targeted Inhibition of NF-Kappa B Suppresses Non-Small Cell Lung Cancer Resistance to Doxorubicin. Mol. Ther. J. Am. Soc. Gene Ther. 2008, 16, 66–73. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Zuo, N.; Yang, H.; Fang, S.; Shi, J. Extracellular Traps Increase Burden of Bleeding by Damaging Endothelial Cell in Acute Promyelocytic Leukaemia. Front. Immunol. 2022, 13, 841445. [Google Scholar] [CrossRef]
- Martínez-Rey, D.; Carmona-Rodríguez, L.; Fernández-Aceñero, M.J.; Mira, E.; Mañes, S. Extracellular Superoxide Dismutase, the Endothelial Basement Membrane, and the WNT Pathway: New Players in Vascular Normalization and Tumor Infiltration by T-Cells. Front. Immunol. 2020, 11, 579552. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Huang, Y.; Li, Z.; Pan, G.; Zheng, L.; Xiao, X.; Wang, F.; Chen, J.; Chen, X.; Lin, X.; et al. Glioblastoma Vascular Plasticity Limits Effector T-Cell Infiltration and Is Blocked by cAMP Activation. Cancer Immunol. Res. 2023, 11, 1351–1366. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Song, C.; Fang, Y.; Yin, Y.; Wu, Z.; Wang, Y.; Xu, Z.; Gao, S.; Li, A.; Liu, G. TH5487, a Small Molecule Inhibitor of OGG1, Attenuates Pulmonary Fibrosis by NEDD4L-Mediated OGG1 Degradation. Chem. Biol. Interact. 2022, 362, 109999. [Google Scholar] [CrossRef] [PubMed]
- Tanner, L.; Single, A.B.; Bhongir, R.K.V.; Heusel, M.; Mohanty, T.; Karlsson, C. a. Q.; Pan, L.; Clausson, C.-M.; Bergwik, J.; Wang, K.; et al. Small-Molecule-Mediated OGG1 Inhibition Attenuates Pulmonary Inflammation and Lung Fibrosis in a Murine Lung Fibrosis Model. Nat. Commun. 2023, 14, 643. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-Y.; Jeon, S.; Yoo, Y.J.; Jin, H.; Won, H.Y.; Yoon, K.; Hwang, E.S.; Lee, Y.-J.; Na, Y.; Cho, J.; et al. The Hsp27-Mediated IkBα-NFκB Signaling Axis Promotes Radiation-Induced Lung Fibrosis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 5364–5375. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, P.; Dey, P.; Ghosh, S.; Sarma, A.; Ghosh, U. Reduction of Metastatic Potential by Inhibiting EGFR/Akt/P38/ERK Signaling Pathway and Epithelial-Mesenchymal Transition after Carbon Ion Exposure Is Potentiated by PARP-1 Inhibition in Non-Small-Cell Lung Cancer. BMC Cancer 2019, 19, 829. [Google Scholar] [CrossRef] [PubMed]
- Asgarova, A.; Asgarov, K.; Godet, Y.; Peixoto, P.; Nadaradjane, A.; Boyer-Guittaut, M.; Galaine, J.; Guenat, D.; Mougey, V.; Perrard, J.; et al. PD-L1 Expression Is Regulated by Both DNA Methylation and NF-kB during EMT Signaling in Non-Small Cell Lung Carcinoma. Oncoimmunology 2018, 7, e1423170. [Google Scholar] [CrossRef] [PubMed]
- Labelle, M.; Begum, S.; Hynes, R.O. Direct Signaling between Platelets and Cancer Cells Induces an Epithelial-Mesenchymal-like Transition and Promotes Metastasis. Cancer Cell 2011, 20, 576–590. [Google Scholar] [CrossRef]
- Gao, D.; Vahdat, L.T.; Wong, S.; Chang, J.C.; Mittal, V. Microenvironmental Regulation of Epithelial-Mesenchymal Transitions in Cancer. Cancer Res. 2012, 72, 4883–4889. [Google Scholar] [CrossRef]
- Gao, D.; Joshi, N.; Choi, H.; Ryu, S.; Hahn, M.; Catena, R.; Sadik, H.; Argani, P.; Wagner, P.; Vahdat, L.T.; et al. Myeloid Progenitor Cells in the Premetastatic Lung Promote Metastases by Inducing Mesenchymal to Epithelial Transition. Cancer Res. 2012, 72, 1384–1394. [Google Scholar] [CrossRef]
- Tanner, L.; Bergwik, J.; Bhongir, R.K.V.; Pan, L.; Dong, C.; Wallner, O.; Kalderén, C.; Helleday, T.; Boldogh, I.; Adner, M.; et al. Pharmacological OGG1 Inhibition Decreases Murine Allergic Airway Inflammation. Front. Pharmacol. 2022, 13, 999180. [Google Scholar] [CrossRef] [PubMed]
- Jetten, N.; Verbruggen, S.; Gijbels, M.J.; Post, M.J.; De Winther, M.P.J.; Donners, M.M.P.C. Anti-Inflammatory M2, but Not pro-Inflammatory M1 Macrophages Promote Angiogenesis in Vivo. Angiogenesis 2014, 17, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Mia, S.; Warnecke, A.; Zhang, X.-M.; Malmström, V.; Harris, R.A. An Optimized Protocol for Human M2 Macrophages Using M-CSF and IL-4/IL-10/TGF-β Yields a Dominant Immunosuppressive Phenotype. Scand. J. Immunol. 2014, 79, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Trombetti, S.; Cesaro, E.; Catapano, R.; Sessa, R.; Lo Bianco, A.; Izzo, P.; Grosso, M. Oxidative Stress and ROS-Mediated Signaling in Leukemia: Novel Promising Perspectives to Eradicate Chemoresistant Cells in Myeloid Leukemia. Int. J. Mol. Sci. 2021, 22, 2470. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




