Submitted:
09 November 2023
Posted:
09 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
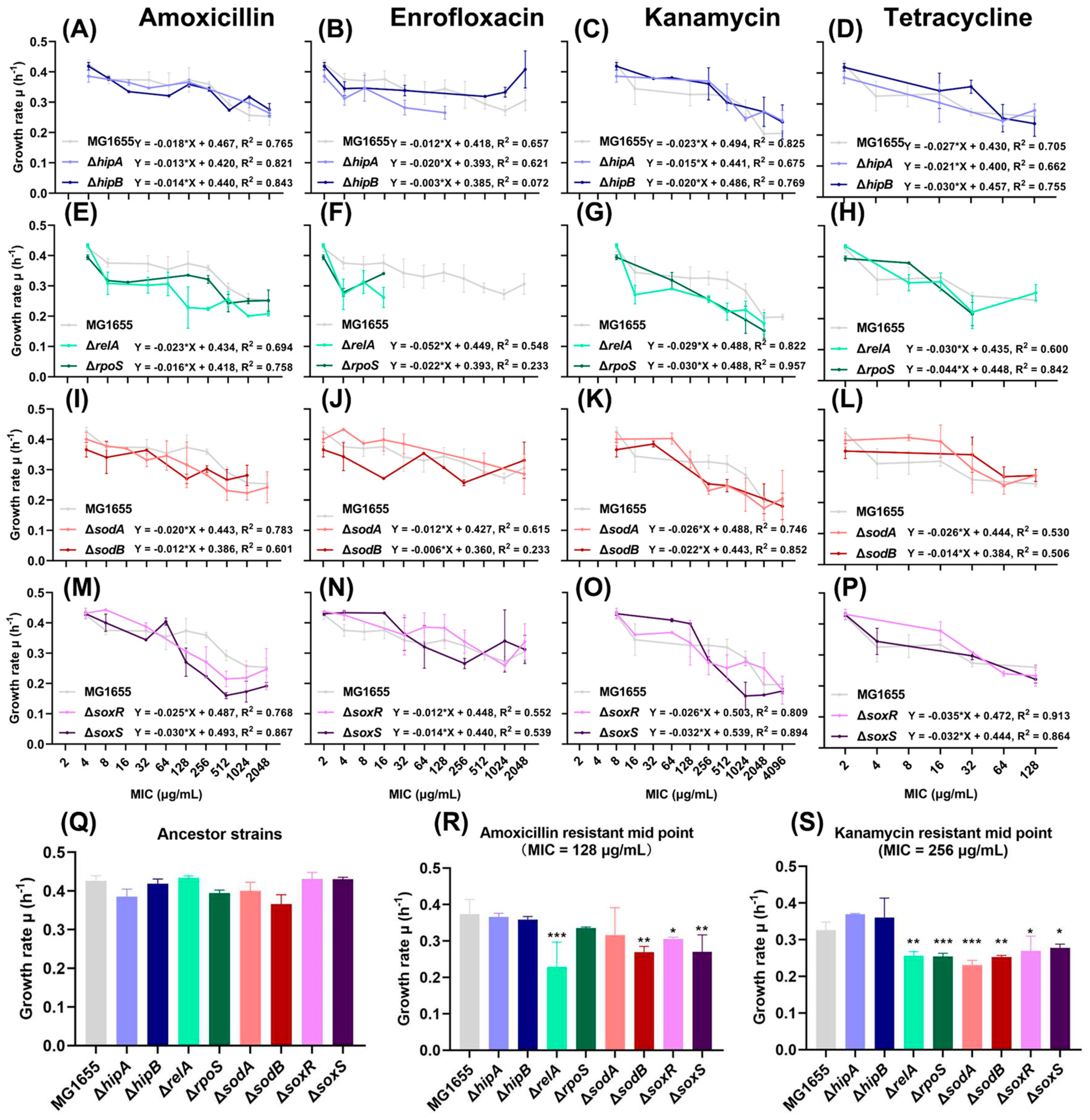
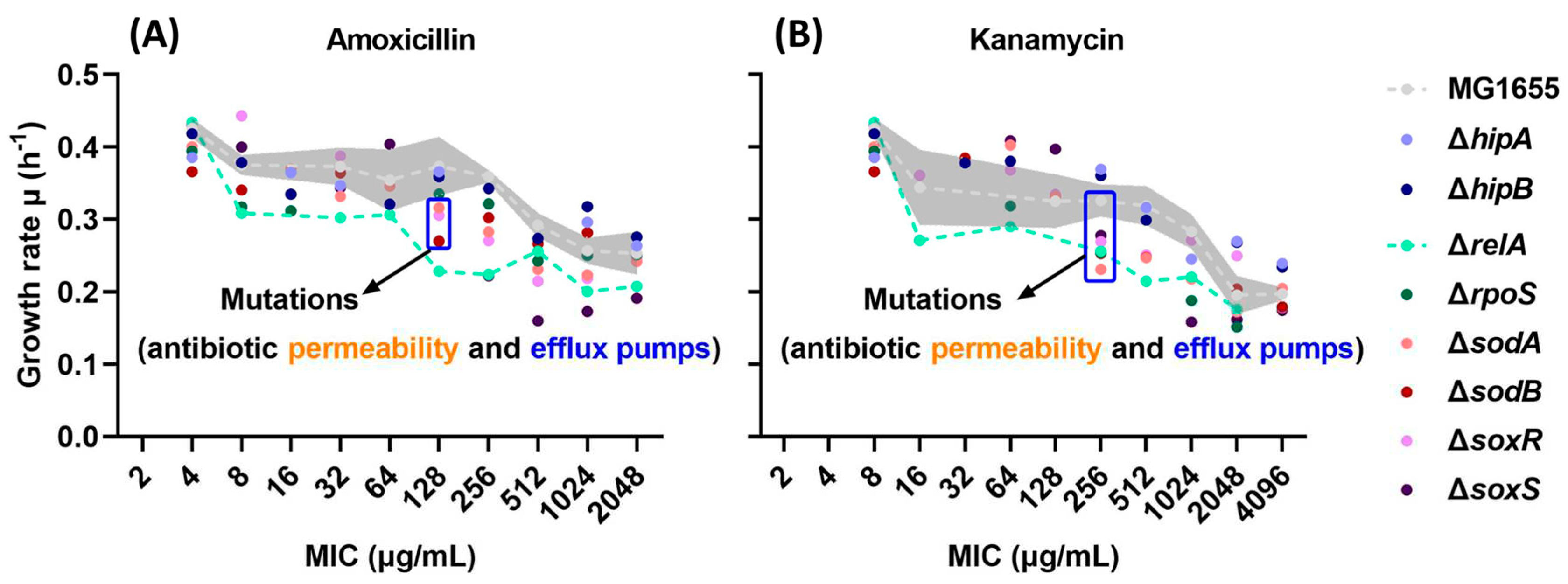
2.1. Growth Rates Gradually Decreased during the Evolution of Resistance.
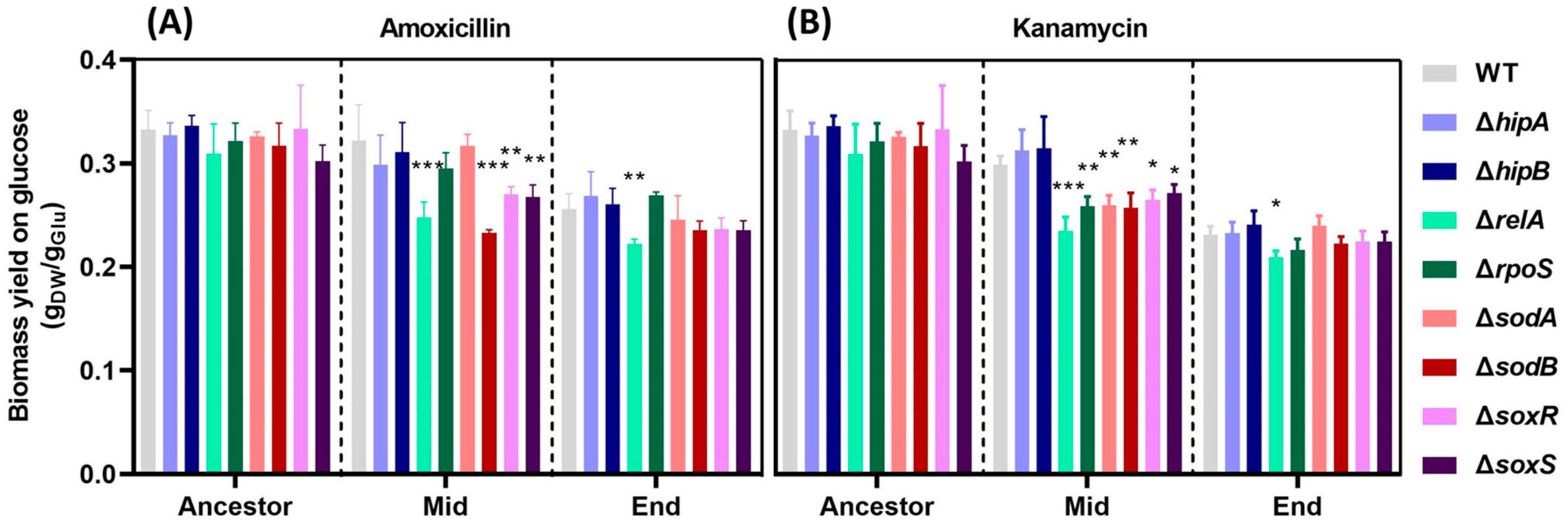
2.2. Decrease in Biomass Yield on Glucose with Increased Resistance.
2.3. Mutations in the Amoxicillin-Resistant Strains during the Resistance Evolution.
2.4. Mutations in the Kanamycin-Resistant Strains during the Resistance Evolution.
3. Discussion
4. Conclusion
5. Materials and Methods
5.1. Bacterial Strains, Growth Media, and Culture Conditions
5.2. MIC Determination
5.3. Growth Rate Measurements
5.4. Biomass Yield on Glucose Measurements
5.5. Whole Genome Sequencing
5.6. Quantification and Statistical Analysis
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Llor, C.; Bjerrum, L. Antimicrobial Resistance: Risk Associated with Antibiotic Overuse and Initiatives to Reduce the Problem. Ther. Adv. drug Saf. 2014, 5, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Mendelsohn, E.; Ross, N.; Zambrana-Torrelio, C.; Van Boeckel, T.P.; Laxminarayan, R.; Daszak, P. Global Patterns and Correlates in the Emergence of Antimicrobial Resistance in Humans. Proceedings. Biol. Sci. 2023, 290, 20231085. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Antibiotic Resistance and Its Cost: Is It Possible to Reverse Resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef]
- Pinheiro, F.; Warsi, O.; Andersson, D.I.; Lässig, M. Metabolic Fitness Landscapes Predict the Evolution of Antibiotic Resistance. Nat. Ecol. Evol. 2021, 5, 677–687. [Google Scholar] [CrossRef]
- Nishimoto, A.T.; Dao, T.H.; Jia, Q.; Ortiz-Marquez, J.C.; Echlin, H.; Vogel, P.; van Opijnen, T.; Rosch, J.W. Interspecies Recombination, Not de Novo Mutation, Maintains Virulence after β-Lactam Resistance Acquisition in Streptococcus Pneumoniae. Cell Rep. 2022, 41, 111835. [Google Scholar] [CrossRef]
- Ogunlana, L.; Kaur, D.; Shaw, L.P.; Jangir, P.; Walsh, T.; Uphoff, S.; MacLean, R.C. Regulatory Fine-Tuning of Mcr-1 Increases Bacterial Fitness and Stabilises Antibiotic Resistance in Agricultural Settings. ISME J. 2023, 17, 2058–2069. [Google Scholar] [CrossRef]
- Vanacker, M.; Lenuzza, N.; Rasigade, J.P. The Fitness Cost of Horizontally Transferred and Mutational Antimicrobial Resistance in Escherichia Coli. Front. Microbiol. 2023, 14, 1186920. [Google Scholar] [CrossRef] [PubMed]
- Händel, N.; Schuurmans, J.M.; Brul, S.; Ter Kuilea, B.H. Compensation of the Metabolic Costs of Antibiotic Resistance by Physiological Adaptation in Escherichia Coli. Antimicrob. Agents Chemother. 2013, 57, 3752–3762. [Google Scholar] [CrossRef]
- Andersson, D.I. The Biological Cost of Mutational Antibiotic Resistance: Any Practical Conclusions? Curr. Opin. Microbiol. 2006, 9, 461–465. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J. V. Co-Selection of Antibiotic and Metal Resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Darphorn, T.S.; Koenders-Van Sintanneland, B.B.; Grootemaat, A.E.; van der Wel, N.N.; Brul, S.; ter Kuile, B.H. Transfer Dynamics of Multi-Resistance Plasmids in Escherichia Coli Isolated from Meat. PLoS One 2022, 17, e0270205. [Google Scholar] [CrossRef] [PubMed]
- Dawan, J.; Ahn, J. Bacterial Stress Responses as Potential Targets in Overcoming Antibiotic Resistance. Microorganisms 2022, 10, 1385. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Jonker, M.J.; de Leeuw, W.; Brul, S.; ter Kuile, B.H. Reactive Oxygen Species Accelerate de Novo Acquisition of Antibiotic Resistance in E. Coli. iScience 2023, 108373. [Google Scholar] [CrossRef]
- Qi, W.; Jonker, M.J.; Leeuw, W. de; Brul, S.; Kuile, B.H. ter Role of RelA-Synthesized (p)PpGpp in de Novo Acquisition of Antibiotic Resistance in E. Coli. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A Common Mechanism of Cellular Death Induced by Bactericidal Antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef]
- Pribis, J.P.; García-Villada, L.; Zhai, Y.; Lewin-Epstein, O.; Wang, A.Z.; Liu, J.; Xia, J.; Mei, Q.; Fitzgerald, D.M.; Bos, J.; et al. Gamblers: An Antibiotic-Induced Evolvable Cell Subpopulation Differentiated by Reactive-Oxygen-Induced General Stress Response. Mol. Cell 2019, 74, 785–800. [Google Scholar] [CrossRef] [PubMed]
- Kaspy, I.; Rotem, E.; Weiss, N.; Ronin, I.; Balaban, N.Q.; Glaser, G. HipA-Mediated Antibiotic Persistence via Phosphorylation of the Glutamyl-TRNA-Synthetase. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef]
- Germain, E.; Castro-Roa, D.; Zenkin, N.; Gerdes, K. Molecular Mechanism of Bacterial Persistence by HipA. Mol. Cell 2013, 52, 248–254. [Google Scholar] [CrossRef]
- Magnusson, L.U.; Farewell, A.; Nyström, T. PpGpp: A Global Regulator in Escherichia Coli. Trends Microbiol. 2005, 13, 236–242. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Piro, K.M.; Xu, W.; Hansen, S.; Lewis, K.; Brennan, R.G. Molecular Mechanisms of HipA-Mediated Multidrug Tolerance and Its Neutralization by HipB. Science (80-. ). 2009, 323, 396–401. [Google Scholar] [CrossRef]
- Arenz, S.; Abdelshahid, M.; Sohmen, D.; Payoe, R.; Starosta, A.L.; Berninghausen, O.; Hauryliuk, V.; Beckmann, R.; Wilson, D.N. The Stringent Factor RelA Adopts an Open Conformation on the Ribosome to Stimulate PpGpp Synthesis. Nucleic Acids Res. 2016, 44, 6471–6481. [Google Scholar] [CrossRef] [PubMed]
- Dalebroux, Z.D.; Swanson, M.S. PpGpp: Magic beyond RNA Polymerase. Nat. Rev. Microbiol. 2012, 10, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.J.; Kohanski, M.A.; Collins, J.J. Role of Reactive Oxygen Species in Antibiotic Action and Resistance. Curr. Opin. Microbiol. 2009, 12, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. The Molecular Mechanisms and Physiological Consequences of Oxidative Stress: Lessons from a Model Bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef]
- Darphorn, T.S.; Hu, Y.; Sintanneland, B.B.K.; Brul, S.; Kuile, B.H. ter Multiplication of AmpC upon Exposure to a Beta-Lactam Antibiotic Results in a Transferable Transposon in Escherichia Coli. Int. J. Mol. Sci. 2021, 22, 9230. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Xu, Y.; Jiang, B.; Qi, Q.; Tang, Y.-J.; Xian, M.; Wang, J.; Zhao, G. Systematic Identification of CpxRA-Regulated Genes and Their Roles in Escherichia Coli Stress Response. mSystems 2022, 7, e0041922. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.J.; Inouye, M. EnvZ-OmpR Interaction and Osmoregulation in Escherichia Coli. J. Biol. Chem. 2002, 277, 24155–24161. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.; Lee, C.R. Distinct Roles of Outer Membrane Porins in Antibiotic Resistance and Membrane Integrity in Escherichia Coli. Front. Microbiol. 2019, 10, 953. [Google Scholar] [CrossRef]
- Hobbs, J.K.; Boraston, A.B. (P)PpGpp and the Stringent Response: An Emerging Threat to Antibiotic Therapy. ACS Infect. Dis. 2019, 5, 1505–1517. [Google Scholar] [CrossRef]
- Wu, L.; Wang, Z.; Guan, Y.; Huang, X.; Shi, H.; Liu, Y.; Zhang, X. The (p)PpGpp-Mediated Stringent Response Regulatory System Globally Inhibits Primary Metabolism and Activates Secondary Metabolism in Pseudomonas Protegens H78. Appl. Microbiol. Biotechnol. 2020, 104, 3061–3079. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.B.; Terry, D.S.; Altman, R.B.; Blanchard, S.C. Aminoglycoside Activity Observed on Single Pre-Translocation Ribosome Complexes. Nat. Chem. Biol. 2010, 6, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Mogre, A.; Veetil, R.T.; Seshasayee, A.S.N. Modulation of Global Transcriptional Regulatory Networks as a Strategy for Increasing Kanamycin Resistance of the Translational Elongation Factor-G Mutants in Escherichia Coli. G3 Genes, Genomes, Genet. 2017, 7, 3955–3966. [Google Scholar] [CrossRef]
- Oz, T.; Guvenek, A.; Yildiz, S.; Karaboga, E.; Tamer, Y.T.; Mumcuyan, N.; Ozan, V.B.; Senturk, G.H.; Cokol, M.; Yeh, P.; et al. Strength of Selection Pressure Is an Important Parameter Contributing to the Complexity of Antibiotic Resistance Evolution. Mol. Biol. Evol. 2014, 31, 2387. [Google Scholar] [CrossRef] [PubMed]
- Freeman, Z.N.; Dorus, S.; Waterfield, N.R. The KdpD/KdpE Two-Component System: Integrating K+ Homeostasis and Virulence. PLoS Pathog. 2013, 9, e1003201. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.J.; Kohanski, M.A.; Hayete, B.; Collins, J.J. Gyrase Inhibitors Induce an Oxidative Damage Cellular Death Pathway in Escherichia Coli. Mol. Syst. Biol. 2007, 3, 91. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.J.; Belenky, P.A.; Yang, J.H.; Cody MacDonald, I.; Martell, J.D.; Takahashi, N.; Chan, C.T.Y.; Lobritz, M.A.; Braff, D.; Schwarz, E.G.; et al. Antibiotics Induce Redox-Related Physiological Alterations as Part of Their Lethality. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, E2100–E2109. [Google Scholar] [CrossRef] [PubMed]
- Foti, J.J.; Devadoss, B.; Winkler, J.A.; Collins, J.J.; Walker, G.C. Oxidation of the Guanine Nucleotide Pool Underlies Cell Death by Bactericidal Antibiotics. Science (80-. ). 2012, 336, 315–319. [Google Scholar] [CrossRef]
- Kohanski, M.A.; DePristo, M.A.; Collins, J.J. Sublethal Antibiotic Treatment Leads to Multidrug Resistance via Radical-Induced Mutagenesis. Mol. Cell 2010, 37, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Goodman, M.F.; Mcdonald, J.P.; Jaszczur, M.M.; Woodgate, R. Insights into the Complex Levels of Regulation Imposed on Escherichia Coli DNA Polymerase V. DNA Repair (Amst). 2016, 44, 42–50. [Google Scholar] [CrossRef]
- Crane, J.K.; Alvarado, C.L.; Sutton, M.D. Role of the SOS Response in the Generation of Antibiotic Resistance in Vivo. Antimicrob. Agents Chemother. 2021, 65, e0001321. [Google Scholar] [CrossRef]
- Sinha, A.K.; Winther, K.S. The RelA Hydrolase Domain Acts as a Molecular Switch for (p)PpGpp Synthesis. Commun. Biol. 2021, 4, 434. [Google Scholar] [CrossRef]
- Irving, S.E.; Choudhury, N.R.; Corrigan, R.M. The Stringent Response and Physiological Roles of (Pp)PGpp in Bacteria. Nat. Rev. Microbiol. 2020, 19, 256–271. [Google Scholar] [CrossRef] [PubMed]
- Hauryliuk, V.; Atkinson, G.C.; Murakami, K.S.; Tenson, T.; Gerdes, K. Recent Functional Insights into the Role of (p)PpGpp in Bacterial Physiology. Nat. Rev. Microbiol. 2015, 13, 298–309. [Google Scholar] [CrossRef]
- Pérez-Gallego, M.; Torrens, G.; Castillo-Vera, J.; Moya, B.; Zamorano, L.; Cabot, G.; Hultenby, K.; Albertí, S.; Mellroth, P.; Henriques-Normark, B.; et al. Impact of AmpC Derepression on Fitness and Virulence: The Mechanism or the Pathway? MBio 2016, 7, e01783–16. [Google Scholar] [CrossRef] [PubMed]
- Macvanin, M.; Björkman, J.; Eriksson, S.; Rhen, M.; Andersson, D.I.; Hughes, D. Fusidic Acid-Resistant Mutants of Salmonella Enterica Serovar Typhimurium with Low Fitness in Vivo Are Defective in RpoS Induction. Antimicrob. Agents Chemother. 2003, 47, 3743–3749. [Google Scholar] [CrossRef]
- Knopp, M.; Andersson, D.I. Amelioration of the Fitness Costs of Antibiotic Resistance Due To Reduced Outer Membrane Permeability by Upregulation of Alternative Porins. Mol. Biol. Evol. 2015, 32, 3252–3263. [Google Scholar] [CrossRef]
- Nicoloff, H.; Perreten, V.; McMurry, L.M.; Levy, S.B. Role for Tandem Duplication and Lon Protease in AcrAB-TolC- Dependent Multiple Antibiotic Resistance (Mar) in an Escherichia Coli Mutant without Mutations in MarRAB or AcrRAB. J. Bacteriol. 2006, 188, 4413–4423. [Google Scholar] [CrossRef] [PubMed]
- Keeney, D.; Ruzin, A.; Mcaleese, F.; Murphy, E.; Bradford, P.A. MarA-Mediated Overexpression of the AcrAB Efflux Pump Results in Decreased Susceptibility to Tigecycline in Escherichia Coli. J. Antimicrob. Chemother. 2008, 61, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; Sanz-García, F.; Blanco, P.; Martínez, J.L. Fitness Costs Associated with the Acquisition of Antibiotic Resistance. Essays Biochem. 2017, 61, 37–48. [Google Scholar] [CrossRef]
- Lind, P.A.; Tobin, C.; Berg, O.G.; Kurland, C.G.; Andersson, D.I. Compensatory Gene Amplification Restores Fitness after Inter-Species Gene Replacements. Mol. Microbiol. 2010, 75, 1078–1089. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Antibiotic Resistance and Its Cost: Is It Possible to Reverse Resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, M.; Enke, T.; Chubukov, V.; Ricci, V.; Piddock, L.; Sauer, U. Metabolic Constraints on the Evolution of Antibiotic Resistance. Mol. Syst. Biol. 2017, 13, 917. [Google Scholar] [CrossRef]
- Durão, P.; Balbontín, R.; Gordo, I. Evolutionary Mechanisms Shaping the Maintenance of Antibiotic Resistance. Trends Microbiol. 2018, 26, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.G.T.; Herbert, D.; Tempest, D.W. Chapter XIII The Continuous Cultivation of Micro-Organisms: 2. Construction of a Chemostat. Methods Microbiol. 1970, 2, 277–327. [Google Scholar] [CrossRef]
- Chen, T.; Brul, S.; Hugenholtz, J. Exploring the Potential of Bacillus Subtilis as Cell Factory for Food Ingredients and Special Chemicals. Microb. Cell Fact. 2023, 22, 200. [Google Scholar] [CrossRef] [PubMed]
- Link, H.; Anselment, B.; Weuster-Botz, D. Leakage of Adenylates during Cold Methanol/Glycerol Quenching of Escherichia Coli. Metabolomics 2008, 4, 240–247. [Google Scholar] [CrossRef]
- Qi, W.; Jonker, M.J.; Teichmann, L.; Wortel, M.; ter Kuile, B.H. The Influence of Oxygen and Oxidative Stress on de Novo Acquisition of Antibiotic Resistance in E. Coli and Lactobacillus Lactis. BMC Microbiol. 2023, 23, 279. [Google Scholar] [CrossRef]
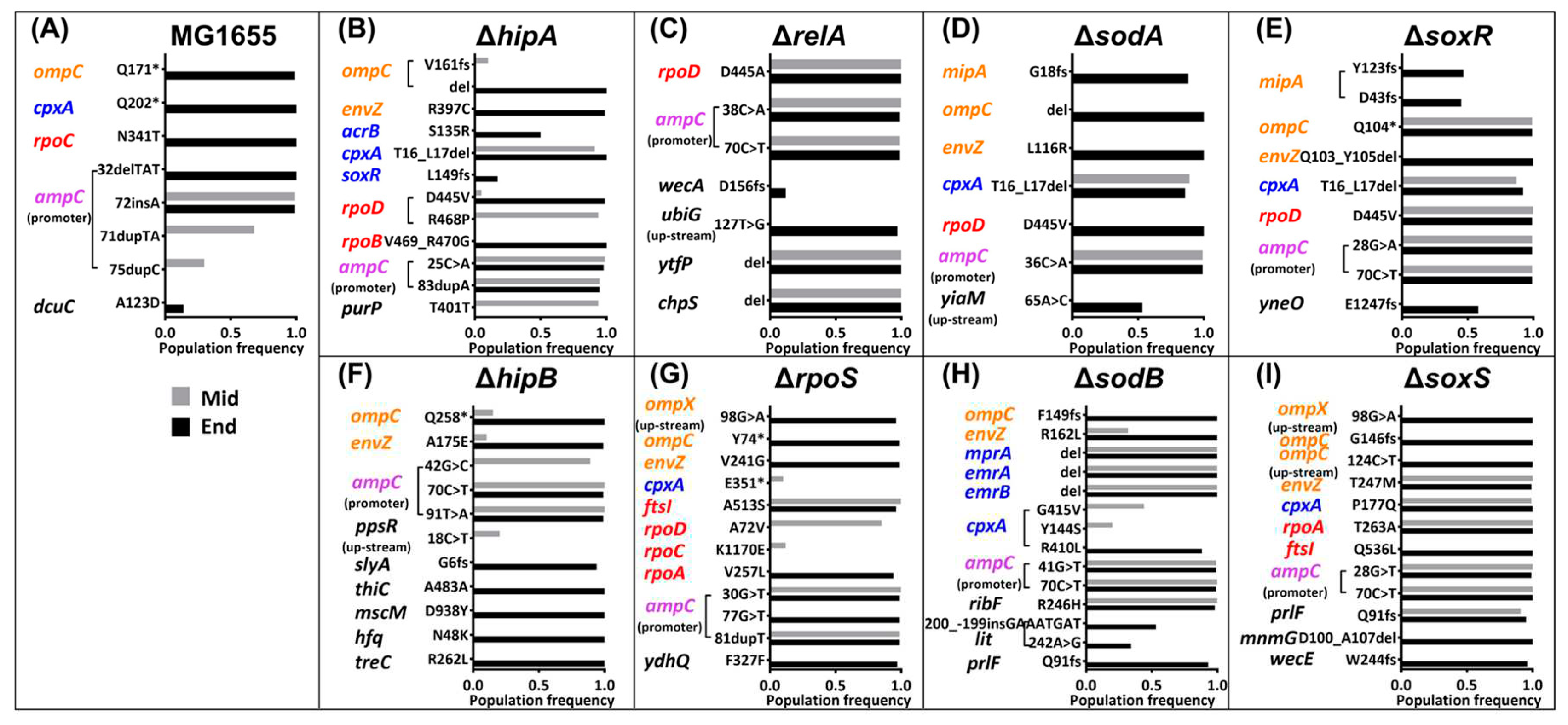
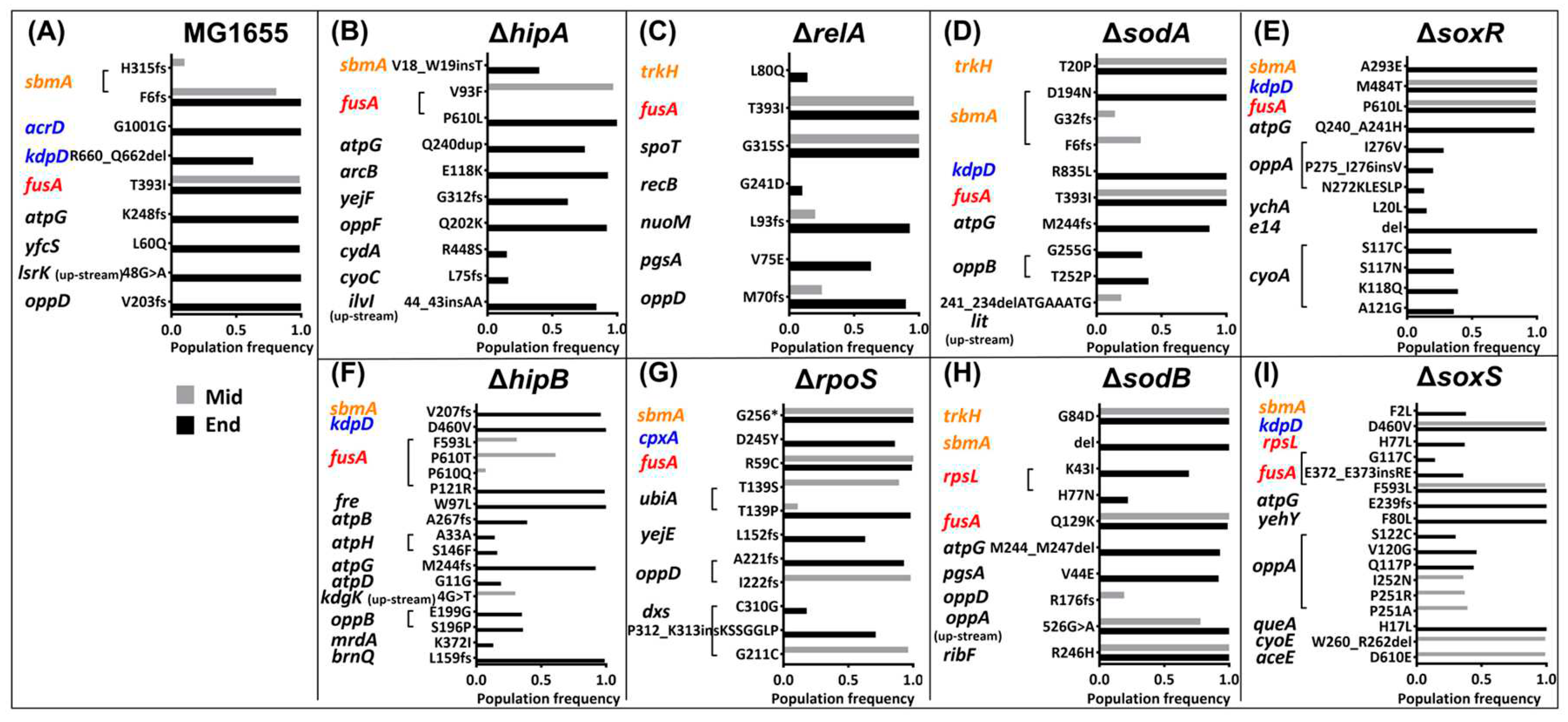
| Times | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|
| Mid |
ompC envZ |
rpoD | cpxA | ampC | ||||
| End |
ompX mipA rpoA ftsI prlF |
rpoD | cpxA | envZ | ompC | ampC |
| Times | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|
| Mid |
trkH kdpD oppA |
sbmA oppD |
fusA | |||||
| End |
rpsL oppB pgsA |
trkH oppA oppD |
kdpD | atpG | sbmA | fusA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





