Submitted:
06 November 2023
Posted:
08 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Implications for Viral Evolution (Target Enzymes and Receptors)
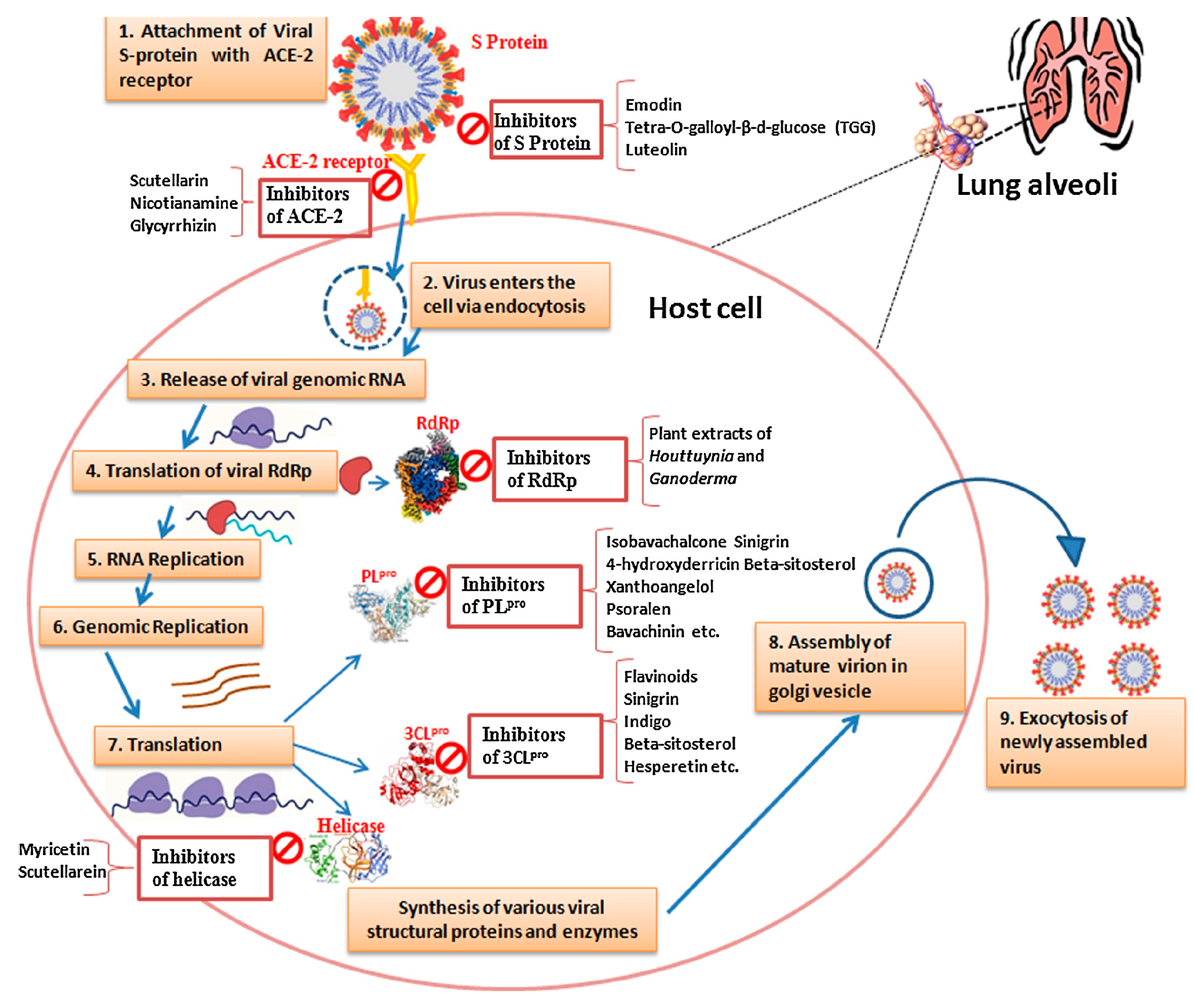
| Enzyme/Receptor | Function/Role | Target for Therapeutic Development | Virus-Host Interaction | Implication in Viral Evolution and SARS-CoV-2 Management | References |
|---|---|---|---|---|---|
| ACE2 | Viral Entry, Lung Protection | Therapeutic Target | Facilitates viral entry and cross-talk with host cells | Dual role in COVID-19 infections; potential protection from acute lung injury and ARDS; increased susceptibility due to high ACE2 expression | [50,51] |
| TMPRSS2 | Viral Entry, Priming | Potential Inhibitor | Mediates viral entry and spike protein cleavage | Important for viral entry; inhibitors may prevent infection and reduce viral spread | [52] |
| DPP4 | Possible Receptor | Role Uncertain | Possible binding to SARS-CoV-2 | Role in virus-host interaction needs further investigation; may be involved in viral entry | [53] |
| PLpro (Protease) | Viral Replication, Polyprotein Cleavage | Drug Target for Inhibition | Essential for viral replication | Critical for cleaving polyproteins; potential drug target for inhibiting viral replication | [54] |
| 3CLpro (Mpro) | Polyprotein Cleavage, Replication | Drug Target for Inhibition | Essential for viral replication | Key for cleaving polyproteins; promising target for antiviral strategies | [55,56] |
| RdRp (RNA Polymerase) | RNA Synthesis, Replication | Potential Drug Target | Crucial for viral genome replication | Required for replication; promising drug target for antiviral strategies | [44] |
| Helicase (NSP13) | RNA Unwinding, Replication | Potential Target | Facilitates RNA unwinding and replication | Important for viral genome replication; potential therapeutic target for anti-COVID-19 strategies | [57] |
| Cathepsin B/L | Viral Entry | Target for Inhibiting Entry | Involved in viral entry | Blocking cathepsin activity can prevent viral entry | [58,59,60] |
| Furin | S Protein Cleavage | Potential Drug Target | Cleavage of S protein and virus entry | Promising target for inhibiting viral entry and spread | [60,61] |
3. Structure and Function of SARS-CoV-2 PLpro
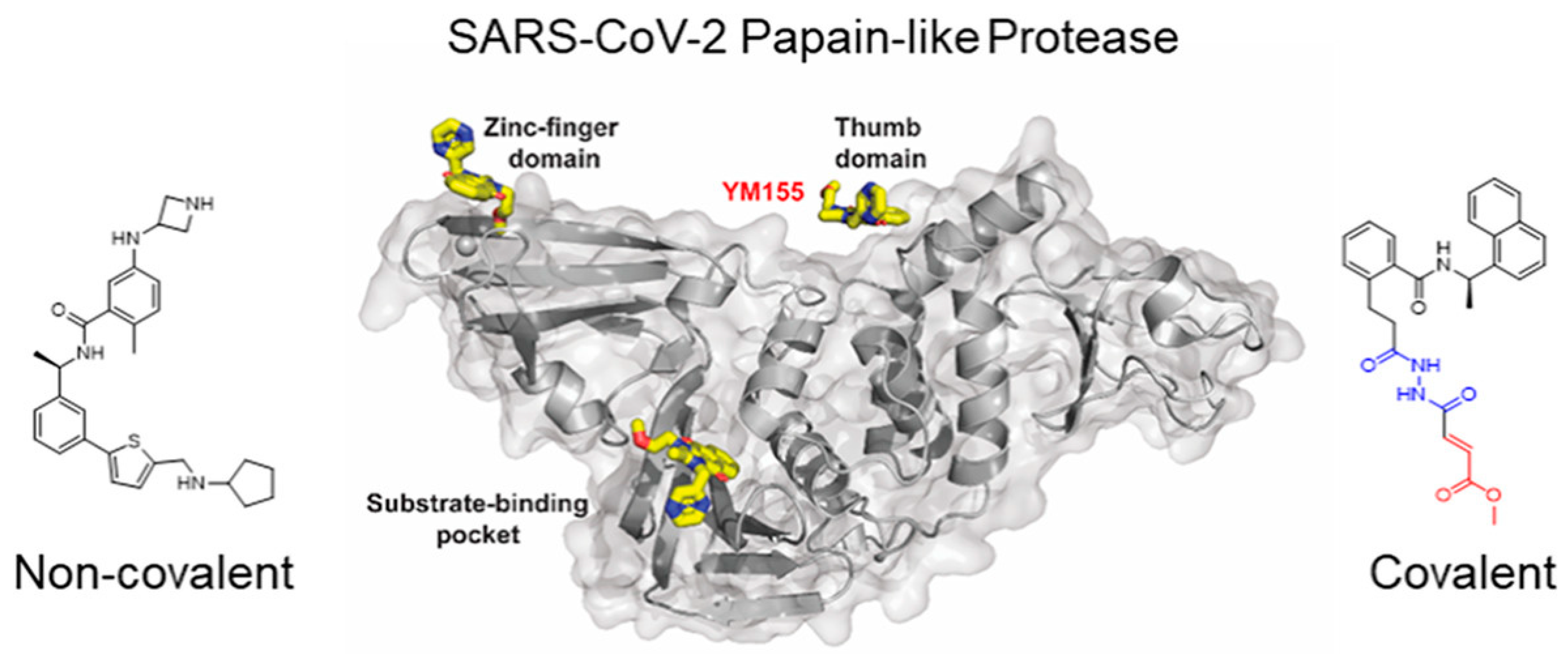
4. Multifaceted Approach with MD Simulations Targeting SARS-CoV-2 Papain-Like Protease (PLpro)
| Inhibitor/Drug Candidate | Simulation Method | Key/Type of Interaction | Simulation Length | Force Field Used | Binding Free Energy | Mechanism of Action | Ref. |
|---|---|---|---|---|---|---|---|
| GRL-0617 | Molecular Dynamics Simulation | Non-covalent binding | 100 ns | AMBER | -21.5 kcal/mol | Non-covalent inhibition of PLpro. | [99] |
| VIR250 and VIR251 | Molecular Dynamics Simulation | Irreversible binding | 50 ns | OPLS-AA | Not available | Irreversible inhibitors of PLpro. | [100,101] |
| Neobavaisoflavone | Molecular Dynamics Simulation | Low energy binding | 75 ns | CHARMM36 | Not available | Binding to the catalytic triad of PLpro. | [102] |
| Ritonavir | Molecular Dynamics Simulation | Binding analysis | 50 ns | GROMOS | -8.2 kcal/mol | Investigated for potential PLpro inhibition. | [20] |
| Dasabuvir (A17) | Molecular Dynamics Simulation | Stable binding | 100 ns | CHARMM27 | -11.7 kcal/mol | Stable binding with PLpro. | [103] |
| Methisazone (A34) | Molecular Dynamics Simulation | Stable binding | 75 ns | AMBER | -12.3 kcal/mol | Exhibits stable dynamic behavior in complex. | [103] |
| Vaniprevir (A53) | Molecular Dynamics Simulation | High binding affinity | 100 ns | OPLS-AA | Not available | Shows high binding affinity to PLpro. | [103] |
| Baicalein | Molecular Dynamics Simulation | Binding to active site | 50 ns | CHARMM36 | -12.8 kcal/mol | Binds to the active site of PLpro. | [104] |
| Disulfiram | Molecular Dynamics Simulation | Inhibition analysis | 75 ns | GROMOS | Not available | Repurposed for potential PLpro inhibition. | [105] |
| Carmofur | Molecular Dynamics Simulation | Binding to PLpro | 100 ns | AMBER | -10.5 kcal/mol | Demonstrates binding to PLpro. | [106] |
| Ebselen | Molecular Dynamics Simulation | Antiviral activity | 75 ns | CHARMM27 | Not available | Investigated for its antiviral activity. | [107] |
| Tideglusib | Molecular Dynamics Simulation | Potential inhibitor | 50 ns | CHARMM36 | Not available | Explored for its potential as an inhibitor. | [106] |
| Shikonin | Molecular Dynamics Simulation | Active site binding | 100 ns | AMBER | -15.6 kcal/mol | Binds to the active site of PLpro. | [108] |
| PX-12 (Belinostat) | Molecular Dynamics Simulation | Inhibition potential | 75 ns | GROMOS | Not available | Investigated for its inhibition potential. | [49] |
| Sub-structurally Similar Compounds with Ritonavir | Molecular Dynamics Simulation | Antiviral drug potential | 50 ns | OPLS-AA | Not available | Explored for their antiviral potential. | [109] |
5. Clinical and Preclinical Studies: Integrating MD Simulation Insights
6. Challenges and Future Directions: Guided by MD Simulations
7. Conclusions and Author Insights on Targeting SARS-CoV-2 Papain-Like Protease
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pal, M.; Berhanu, G.; Desalegn, C.; Kandi, V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Lian, X.; Su, X.; Wu, W.; Marraro, G.A.; Zeng, Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respiratory research 2020, 21, 1–14. [Google Scholar] [CrossRef]
- Acter, T.; Uddin, N.; Das, J.; Akhter, A.; Choudhury, T.R.; Kim, S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: A global health emergency. Science of the Total Environment 2020, 730, 138996. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, G.; Cai, X.-p.; Deng, J.-w.; Zheng, L.; Zhu, H.-h.; Zheng, M.; Yang, B.; Chen, Z. An overview of COVID-19. Journal of Zhejiang University. Science. B 2020, 21, 343. [Google Scholar] [CrossRef]
- Satarker, S.; Nampoothiri, M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Archives of medical research 2020, 51, 482–491. [Google Scholar] [CrossRef]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: implications for SARS-CoV-2. Nature Reviews Microbiology 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Astuti, I. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes & Metabolic Syndrome: Clinical Research & Reviews 2020, 14, 407–412. [Google Scholar]
- Ni, W.; Yang, X.; Yang, D.; Bao, J.; Li, R.; Xiao, Y.; Hou, C.; Wang, H.; Liu, J.; Yang, D. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Critical Care 2020, 24, 1–10. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.-f.; Xu, W.; Liu, S.-w. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Applied microbiology and biotechnology 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Enjuanes, L.; Ziebuhr, J.; Snijder, E.J. Nidovirales: evolving the largest RNA virus genome. Virus research 2006, 117, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, Z.; Lai, M.; Shu, S.; Du, Y.; Zhou, Z.H.; Sun, R. In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature 2017, 541, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Alturki, S.O.; Alturki, S.O.; Connors, J.; Cusimano, G.; Kutzler, M.A.; Izmirly, A.M.; Haddad, E.K. The 2020 pandemic: current SARS-CoV-2 vaccine development. Frontiers in immunology 2020, 11, 1880. [Google Scholar] [CrossRef]
- Rodrigues, C.M.; Plotkin, S.A. Impact of vaccines; health, economic and social perspectives. Frontiers in microbiology 2020, 11, 1526. [Google Scholar] [CrossRef] [PubMed]
- Gusev, E.; Sarapultsev, A.; Solomatina, L.; Chereshnev, V. SARS-CoV-2-specific immune response and the pathogenesis of COVID-19. International journal of molecular sciences 2022, 23, 1716. [Google Scholar] [CrossRef] [PubMed]
- Onomoto, K.; Onoguchi, K.; Yoneyama, M. Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors. Cellular & molecular immunology 2021, 18, 539–555. [Google Scholar]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, S.; Selvaraj, C.; Singh, S.K.; Dubey, V.K.; Kumar, S.; Fialho, A.M.; Saudagar, P. Bacterial protein azurin and derived peptides as potential anti-SARS-CoV-2 agents: insights from molecular docking and molecular dynamics simulations. Journal of Biomolecular Structure and Dynamics 2021, 39, 5706–5721. [Google Scholar] [CrossRef]
- Muralidharan, N.; Sakthivel, R.; Velmurugan, D.; Gromiha, M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics 2021, 39, 2673–2678. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Brindisi, M.; Shahabi, D.; Chapman, M.E.; Mesecar, A.D. Drug development and medicinal chemistry efforts toward SARS-coronavirus and Covid-19 therapeutics. ChemMedChem 2020, 15, 907–932. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020, 370, eabe9403. [Google Scholar] [CrossRef] [PubMed]
- Beyerstedt, S.; Casaro, E.B.; Rangel, É.B. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. European journal of clinical microbiology & infectious diseases 2021, 40, 905–919. [Google Scholar]
- Bost, P.; Giladi, A.; Liu, Y.; Bendjelal, Y.; Xu, G.; David, E.; Blecher-Gonen, R.; Cohen, M.; Medaglia, C.; Li, H. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell 2020, 181, 1475–1488.e1412. [Google Scholar] [CrossRef] [PubMed]
- Klemm, T.; Ebert, G.; Calleja, D.J.; Allison, C.C.; Richardson, L.W.; Bernardini, J.P.; Lu, B.G.; Kuchel, N.W.; Grohmann, C.; Shibata, Y. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. The EMBO journal 2020, 39, e106275. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Chaudhary, J.K.; Jain, N.; Chaudhary, P.K.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. [Google Scholar] [CrossRef]
- Cannalire, R.; Cerchia, C.; Beccari, A.R.; Di Leva, F.S.; Summa, V. Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities. Journal of medicinal chemistry 2020, 65, 2716–2746. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.K.; Singh, A.; Prakash, S.; Kumar, M.; Singh, A.K. Race to arsenal COVID-19 therapeutics: Current alarming status and future directions. Chemico-Biological Interactions 2020, 332, 109298. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nature reviews Molecular cell biology 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Jiang, H.; Yang, P.; Zhang, J. Potential inhibitors targeting papain-like protease of SARS-CoV-2: two birds with one stone. Frontiers in chemistry 2022, 10, 822785. [Google Scholar] [CrossRef]
- Huang, J.; Song, W.; Huang, H.; Sun, Q. Pharmacological therapeutics targeting RNA-dependent RNA polymerase, proteinase and spike protein: from mechanistic studies to clinical trials for COVID-19. Journal of clinical medicine 2020, 9, 1131. [Google Scholar] [CrossRef] [PubMed]
- Drucker, D.J. Diabetes, obesity, metabolism, and SARS-CoV-2 infection: the end of the beginning. Cell metabolism 2021, 33, 479–498. [Google Scholar] [CrossRef] [PubMed]
- Kai, H.; Kai, M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors—lessons from available evidence and insights into COVID-19. Hypertension Research 2020, 43, 648–654. [Google Scholar] [CrossRef] [PubMed]
- South, A.M.; Diz, D.I.; Chappell, M.C. COVID-19, ACE2, and the cardiovascular consequences. American Journal of Physiology-Heart and Circulatory Physiology 2020. [Google Scholar]
- Verdecchia, P.; Cavallini, C.; Spanevello, A.; Angeli, F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. European journal of internal medicine 2020, 76, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-Y.; Zheng, B.; Zhang, Y.; Li, J.-P. Role and mechanism of angiotensin-converting enzyme 2 in acute lung injury in coronavirus disease 2019. Chronic Diseases and Translational Medicine 2020, 6, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Lukassen, S.; Chua, R.L.; Trefzer, T.; Kahn, N.C.; Schneider, M.A.; Muley, T.; Winter, H.; Meister, M.; Veith, C.; Boots, A.W. SARS-CoV-2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. The EMBO journal 2020, 39, e105114. [Google Scholar] [CrossRef] [PubMed]
- Mohamadian, M.; Chiti, H.; Shoghli, A.; Biglari, S.; Parsamanesh, N.; Esmaeilzadeh, A. COVID-19: Virology, biology and novel laboratory diagnosis. The journal of gene medicine 2021, 23, e3303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, J.; Liu, X.; Xu, D.; Shi, L.; Liu, J.; Zeng, Q.; Wang, X. 5-Aminolaevulinic acid photodynamic therapy amplifies intense inflammatory response in the treatment of acne vulgaris via CXCL8. Experimental Dermatology 2021, 30, 923–931. [Google Scholar] [CrossRef]
- Shapira, T.; Monreal, I.A.; Dion, S.P.; Buchholz, D.W.; Imbiakha, B.; Olmstead, A.D.; Jager, M.; Désilets, A.; Gao, G.; Martins, M. A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature 2022, 605, 340–348. [Google Scholar] [CrossRef]
- Bestle, D.; Heindl, M.R.; Limburg, H.; Pilgram, O.; Moulton, H.; Stein, D.A.; Hardes, K.; Eickmann, M.; Dolnik, O.; Rohde, C. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life science alliance 2020, 3. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 2020, 1866, 165878. [Google Scholar] [CrossRef] [PubMed]
- Rohaim, M.A.; El Naggar, R.F.; Clayton, E.; Munir, M. Structural and functional insights into non-structural proteins of coronaviruses. Microbial pathogenesis 2021, 150, 104641. [Google Scholar] [CrossRef] [PubMed]
- Romano, M.; Ruggiero, A.; Squeglia, F.; Maga, G.; Berisio, R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells 2020, 9, 1267. [Google Scholar] [CrossRef]
- Ma, Y.; Frutos-Beltrán, E.; Kang, D.; Pannecouque, C.; De Clercq, E.; Menéndez-Arias, L.; Liu, X.; Zhan, P. Medicinal chemistry strategies for discovering antivirals effective against drug-resistant viruses. Chemical Society Reviews 2021, 50, 4514–4540. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Khan, S.A.; Zia, K.; Ashraf, S.; Uddin, R.; Ul-Haq, Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics 2021, 39, 2607–2616. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B 2020, 10, 766–788. [Google Scholar] [CrossRef]
- Gil, C.; Ginex, T.; Maestro, I.; Nozal, V.; Barrado-Gil, L.; Cuesta-Geijo, M.Á.; Urquiza, J.; Ramírez, D.; Alonso, C.; Campillo, N.E. COVID-19: drug targets and potential treatments. Journal of medicinal chemistry 2020, 63, 12359–12386. [Google Scholar] [CrossRef]
- Bourgonje, A.R.; Abdulle, A.E.; Timens, W.; Hillebrands, J.L.; Navis, G.J.; Gordijn, S.J.; Bolling, M.C.; Dijkstra, G.; Voors, A.A.; Osterhaus, A.D. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19). The Journal of pathology 2020, 251, 228–248. [Google Scholar] [CrossRef]
- Davidson, A.M.; Wysocki, J.; Batlle, D. Interaction of SARS-CoV-2 and other coronavirus with ACE (angiotensin-converting enzyme)-2 as their main receptor: therapeutic implications. Hypertension 2020, 76, 1339–1349. [Google Scholar] [CrossRef]
- Sarker, J.; Das, P.; Sarker, S.; Roy, A.K.; Momen, A.R. A review on expression, pathological roles, and inhibition of TMPRSS2, the serine protease responsible for SARS-CoV-2 spike protein activation. Scientifica 2021, 2021, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cameron, K.; Rozano, L.; Falasca, M.; Mancera, R.L. Does the SARS-CoV-2 spike protein receptor binding domain interact effectively with the DPP4 (CD26) receptor? A molecular docking study. International Journal of Molecular Sciences 2021, 22, 7001. [Google Scholar] [CrossRef] [PubMed]
- Mouffouk, C.; Mouffouk, S.; Mouffouk, S.; Hambaba, L.; Haba, H. Flavonols as potential antiviral drugs targeting SARS-CoV-2 proteases (3CLpro and PLpro), spike protein, RNA-dependent RNA polymerase (RdRp) and angiotensin-converting enzyme II receptor (ACE2). European journal of pharmacology 2021, 891, 173759. [Google Scholar] [CrossRef] [PubMed]
- Chitranshi, N.; Gupta, V.K.; Rajput, R.; Godinez, A.; Pushpitha, K.; Shen, T.; Mirzaei, M.; You, Y.; Basavarajappa, D.; Gupta, V. Evolving geographic diversity in SARS-CoV2 and in silico analysis of replicating enzyme 3CL pro targeting repurposed drug candidates. Journal of Translational Medicine 2020, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Mitra, D.; Paul, M.; Chaudhary, P.; Kamboj, A.; Thatoi, H.; Janmeda, P.; Jain, D.; Panneerselvam, P.; Shrivastav, R. Potential inhibitors of SARS-CoV-2 (COVID 19) proteases PLpro and Mpro/3CLpro: molecular docking and simulation studies of three pertinent medicinal plant natural components. Current Research in Pharmacology and Drug Discovery 2021, 2, 100038. [Google Scholar] [CrossRef] [PubMed]
- Ariumi, Y. Host cellular RNA helicases regulate SARS-CoV-2 infection. Journal of Virology 2022, 96, e00002–00022. [Google Scholar] [CrossRef]
- Padmanabhan, P.; Desikan, R.; Dixit, N.M. Targeting TMPRSS2 and Cathepsin B/L together may be synergistic against SARS-CoV-2 infection. PLoS computational biology 2020, 16, e1008461. [Google Scholar] [CrossRef]
- Pišlar, A.; Mitrović, A.; Sabotič, J.; Pečar Fonović, U.; Perišić Nanut, M.; Jakoš, T.; Senjor, E.; Kos, J. The role of cysteine peptidases in coronavirus cell entry and replication: The therapeutic potential of cathepsin inhibitors. PLoS Pathogens 2020, 16, e1009013. [Google Scholar] [CrossRef]
- Essalmani, R.; Jain, J.; Susan-Resiga, D.; Andréo, U.; Evagelidis, A.; Derbali, R.M.; Huynh, D.N.; Dallaire, F.; Laporte, M.; Delpal, A. Distinctive roles of furin and TMPRSS2 in SARS-CoV-2 infectivity. Journal of virology 2022, 96, e00128–00122. [Google Scholar] [CrossRef]
- Cheng, Y.-W.; Chao, T.-L.; Li, C.-L.; Chiu, M.-F.; Kao, H.-C.; Wang, S.-H.; Pang, Y.-H.; Lin, C.-H.; Tsai, Y.-M.; Lee, W.-H. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell reports 2020, 33. [Google Scholar] [CrossRef]
- Gao, X.; Qin, B.; Chen, P.; Zhu, K.; Hou, P.; Wojdyla, J.A.; Wang, M.; Cui, S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharmaceutica Sinica B 2021, 11, 237–245. [Google Scholar] [CrossRef]
- Gupta, A.; Pradhan, A.; Maurya, V.K.; Kumar, S.; Theengh, A.; Puri, B.; Saxena, S.K. Therapeutic approaches for SARS-CoV-2 infection. Methods 2021, 195, 29–43. [Google Scholar] [CrossRef]
- Dubey, R.; Dubey, K. SARS-CoV-2: Potential drug targets and its virtual screening. Modeling, control and drug development for COVID-19 outbreak prevention 2022, 203–244. [Google Scholar]
- Liu, J.; Cheng, Y.; Zheng, M.; Yuan, B.; Wang, Z.; Li, X.; Yin, J.; Ye, M.; Song, Y. Targeting the ubiquitination/deubiquitination process to regulate immune checkpoint pathways. Signal Transduction and Targeted Therapy 2021, 6, 28. [Google Scholar] [CrossRef]
- Hill, D.J.; Mio, M.J.; Prince, R.B.; Hughes, T.S.; Moore, J.S. A field guide to foldamers. Chemical Reviews 2001, 101, 3893–4012. [Google Scholar] [CrossRef]
- Sanders, B.C.; Pokhrel, S.; Labbe, A.D.; Mathews, I.I.; Cooper, C.J.; Davidson, R.B.; Phillips, G.; Weiss, K.L.; Zhang, Q.; O’Neill, H. Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2. Nature Communications 2023, 14, 1733. [Google Scholar] [CrossRef]
- de Paiva, R.E.; Neto, A.M.; Santos, I.A.; Jardim, A.C.; Corbi, P.P.; Bergamini, F.R. What is holding back the development of antiviral metallodrugs? A literature overview and implications for SARS-CoV-2 therapeutics and future viral outbreaks. Dalton Transactions 2020, 49, 16004–16033. [Google Scholar] [CrossRef]
- Gupta, Y.; Maciorowski, D.; Zak, S.E.; Jones, K.A.; Kathayat, R.S.; Azizi, S.-A.; Mathur, R.; Pearce, C.M.; Ilc, D.J.; Husein, H. Bisindolylmaleimide IX: A novel anti-SARS-CoV2 agent targeting viral main protease 3CLpro demonstrated by virtual screening pipeline and in-vitro validation assays. Methods 2021, 195, 57–71. [Google Scholar] [CrossRef]
- Li, G.; Hilgenfeld, R.; Whitley, R.; De Clercq, E. Therapeutic strategies for COVID-19: progress and lessons learned. Nature Reviews Drug Discovery 2023, 1–27. [Google Scholar] [CrossRef]
- Osipiuk, J.; Azizi, S.-A.; Dvorkin, S.; Endres, M.; Jedrzejczak, R.; Jones, K.A.; Kang, S.; Kathayat, R.S.; Kim, Y.; Lisnyak, V.G. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nature communications 2021, 12, 743. [Google Scholar] [CrossRef]
- Arya, R.; Kumari, S.; Pandey, B.; Mistry, H.; Bihani, S.C.; Das, A.; Prashar, V.; Gupta, G.D.; Panicker, L.; Kumar, M. Structural insights into SARS-CoV-2 proteins. Journal of molecular biology 2021, 433, 166725. [Google Scholar] [CrossRef]
- Murgolo, N.; Therien, A.G.; Howell, B.; Klein, D.; Koeplinger, K.; Lieberman, L.A.; Adam, G.C.; Flynn, J.; McKenna, P.; Swaminathan, G. SARS-CoV-2 tropism, entry, replication, and propagation: Considerations for drug discovery and development. PLoS pathogens 2021, 17, e1009225. [Google Scholar] [CrossRef]
- Rut, W.; Lv, Z.; Zmudzinski, M.; Patchett, S.; Nayak, D.; Snipas, S.J.; El Oualid, F.; Huang, T.T.; Bekes, M.; Drag, M. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti–COVID-19 drug design. Science advances 2020, 6, eabd4596. [Google Scholar] [CrossRef]
- Nikolich-Zugich, J.; Knox, K.S.; Rios, C.T.; Natt, B.; Bhattacharya, D.; Fain, M.J. SARS-CoV-2 and COVID-19 in older adults: what we may expect regarding pathogenesis, immune responses, and outcomes. Geroscience 2020, 42, 505–514. [Google Scholar] [CrossRef]
- Prajapati, J.; Patel, R.; Rao, P.; Saraf, M.; Rawal, R.; Goswami, D. Perceiving SARS-CoV-2 Mpro and PLpro dual inhibitors from pool of recognized antiviral compounds of endophytic microbes: an in silico simulation study. Structural Chemistry 2022, 33, 1619–1643. [Google Scholar] [CrossRef]
- Malik, Y.A. Properties of coronavirus and SARS-CoV-2. The Malaysian journal of pathology 2020, 42, 3–11. [Google Scholar]
- Cao, W.; Cho, C.-C.D.; Geng, Z.Z.; Shaabani, N.; Ma, X.R.; Vatansever, E.C.; Alugubelli, Y.R.; Ma, Y.; Chaki, S.P.; Ellenburg, W.H. Evaluation of SARS-CoV-2 main protease inhibitors using a novel cell-based assay. ACS central science 2022, 8, 192–204. [Google Scholar] [CrossRef]
- Freitas, B.T.; Durie, I.A.; Murray, J.; Longo, J.E.; Miller, H.C.; Crich, D.; Hogan, R.J.; Tripp, R.A.; Pegan, S.D. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS infectious diseases 2020, 6, 2099–2109. [Google Scholar] [CrossRef]
- Yang, X.; Liu, Y.; Liu, Y.; Yang, Q.; Wu, X.; Huang, X.; Liu, H.; Cai, W.; Ma, G. Medication therapy strategies for the coronavirus disease 2019 (COVID-19): recent progress and challenges. Expert Review of Clinical Pharmacology 2020, 13, 957–975. [Google Scholar] [CrossRef]
- Narayanan, A.; Narwal, M.; Majowicz, S.A.; Varricchio, C.; Toner, S.A.; Ballatore, C.; Brancale, A.; Murakami, K.S.; Jose, J. Identification of SARS-CoV-2 inhibitors targeting Mpro and PLpro using in-cell-protease assay. Communications Biology 2022, 5, 169. [Google Scholar] [CrossRef]
- Steuten, K.; Kim, H.; Widen, J.C.; Babin, B.M.; Onguka, O.; Lovell, S.; Bolgi, O.; Cerikan, B.; Neufeldt, C.J.; Cortese, M. Challenges for targeting SARS-CoV-2 proteases as a therapeutic strategy for COVID-19. ACS infectious diseases 2021, 7, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Weiss, C.; Carriere, M.; Fusco, L.; Capua, I.; Regla-Nava, J.A.; Pasquali, M.; Scott, J.A.; Vitale, F.; Unal, M.A.; Mattevi, C. Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS nano 2020, 14, 6383–6406. [Google Scholar] [CrossRef]
- Selvaraj, C.; Dinesh, D.C.; Panwar, U.; Abhirami, R.; Boura, E.; Singh, S.K. Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19. Journal of Biomolecular Structure and Dynamics 2021, 39, 4582–4593. [Google Scholar] [CrossRef] [PubMed]
- Frances-Monerris, A.; Hognon, C.; Miclot, T.; Garcia-Iriepa, C.; Iriepa, I.; Terenzi, A.; Grandemange, S.; Barone, G.; Marazzi, M.; Monari, A. Molecular basis of SARS-CoV-2 infection and rational design of potential antiviral agents: modeling and simulation approaches. Journal of Proteome Research 2020, 19, 4291–4315. [Google Scholar] [CrossRef] [PubMed]
- Báez-Santos, Y.M.; John, S.E.S.; Mesecar, A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral research 2015, 115, 21–38. [Google Scholar] [CrossRef]
- Maiti, B.K. Can Papain-like Protease Inhibitors Halt SARS-CoV-2 Replication? ACS Pharmacol Transl Sci 2020, 3, 1017–1019. [Google Scholar] [CrossRef] [PubMed]
- Lanz, J.; Biniaz-Harris, N.; Kuvaldina, M.; Jain, S.; Lewis, K.; Fallon, B.A. Disulfiram: Mechanisms, Applications, and Challenges. Antibiotics (Basel) 2023, 12, 524. [Google Scholar] [CrossRef]
- Maiti, B.K. Can papain-like protease inhibitors halt SARS-CoV-2 replication? ACS pharmacology & translational science 2020, 3, 1017–1019. [Google Scholar]
- Akram, M.W.; Hasannuzaman, M.; Cuce, E.; Cuce, P.M. Global technological advancement and challenges of glazed window, facade system and vertical greenery-based energy savings in buildings: A comprehensive review. Energy and Built Environment 2023, 4, 206–226. [Google Scholar] [CrossRef]
- Andreini, C.; Arnesano, F.; Rosato, A. The zinc proteome of SARS-CoV-2. Metallomics 2022, 14. [Google Scholar] [CrossRef]
- Debnath, S.K.; Debnath, M.; Srivastava, R.; Omri, A. Drugs repurposing for SARS-CoV-2: new insight of COVID-19 druggability. Expert Review of Anti-infective Therapy 2022, 20, 1187–1204. [Google Scholar] [CrossRef]
- Owji, H.; Negahdaripour, M.; Hajighahramani, N. Immunotherapeutic approaches to curtail COVID-19. International immunopharmacology 2020, 88, 106924. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Zheng, Y.; Zeng, X.; He, B.; Cheng, W. Structural biology of SARS-CoV-2: open the door for novel therapies. Signal transduction and targeted therapy 2022, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.U.; Parida, S.; Lingaraju, M.C.; Kesavan, M.; Kumar, D.; Singh, R.K. Drug repurposing approach to fight COVID-19. Pharmacological Reports 2020, 72, 1479–1508. [Google Scholar] [CrossRef] [PubMed]
- Elfiky, A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life sciences 2020, 253, 117592. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Cao, Z.; Han, M.; Wang, Z.; Chen, J.; Sun, W.; Wu, Y.; Xiao, W.; Liu, S.; Chen, E. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. bmj 2020, 369. [Google Scholar] [CrossRef] [PubMed]
- Martinotti, C.; Ruiz-Perez, L.; Deplazes, E.; Mancera, R.L. Molecular dynamics simulation of small molecules interacting with biological membranes. ChemPhysChem 2020, 21, 1486–1514. [Google Scholar] [CrossRef] [PubMed]
- Parmar, P.; Rao, P.; Sharma, A.; Shukla, A.; Rawal, R.M.; Saraf, M.; Patel, B.V.; Goswami, D. Meticulous assessment of natural compounds from NPASS database for identifying analogue of GRL0617, the only known inhibitor for SARS-CoV2 papain-like protease (PLpro) using rigorous computational workflow. Molecular diversity 2022, 26, 389–407. [Google Scholar] [CrossRef]
- Sanachai, K.; Mahalapbutr, P.; Sanghiran Lee, V.; Rungrotmongkol, T.; Hannongbua, S. In silico elucidation of potent inhibitors and rational drug design against SARS-CoV-2 papain-like protease. The Journal of Physical Chemistry B 2021, 125, 13644–13656. [Google Scholar] [CrossRef]
- Naidoo, D.; Kar, P.; Roy, A.; Mutanda, T.; Bwapwa, J.; Sen, A.; Anandraj, A. Structural insight into the binding of cyanovirin-n with the spike glycoprotein, mpro and PLpro of SARS-CoV-2: Protein–protein interactions, dynamics simulations and free energy calculations. Molecules 2021, 26, 5114. [Google Scholar] [CrossRef]
- Selvaraj, V.; Rathinavel, T.; Ammashi, S.; Nasir Iqbal, M. Polyphenolic phytochemicals exhibit promising SARS-COV-2 papain like protease (PLpro) inhibition validated through a computational approach. Polycyclic Aromatic Compounds 2023, 43, 5545–5566. [Google Scholar] [CrossRef]
- Bera, K.; Reeda, V.J.; Babila, P.; Dinesh, D.C.; Hritz, J.; Karthick, T. An in silico molecular dynamics simulation study on the inhibitors of SARS-CoV-2 proteases (3CLpro and PLpro) to combat COVID-19. Molecular Simulation 2021, 47, 1168–1184. [Google Scholar] [CrossRef]
- Yan, F.; Gao, F. An overview of potential inhibitors targeting non-structural proteins 3 (PLpro and Mac1) and 5 (3CLpro/Mpro) of SARS-CoV-2. Computational and structural biotechnology journal 2021, 19, 4868–4883. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Tong, J.; Wu, Y.; Zhao, S.; Lin, B.-L. A computational evaluation of targeted oxidation strategy (TOS) for potential inhibition of SARS-CoV-2 by disulfiram and analogues. Biophysical Chemistry 2021, 276, 106610. [Google Scholar] [CrossRef]
- Ma, C.; Hu, Y.; Townsend, J.A.; Lagarias, P.I.; Marty, M.T.; Kolocouris, A.; Wang, J. Ebselen, disulfiram, carmofur, PX-12, tideglusib, and shikonin are nonspecific promiscuous SARS-CoV-2 main protease inhibitors. ACS pharmacology & translational science 2020, 3, 1265–1277. [Google Scholar]
- Zmudzinski, M.; Rut, W.; Olech, K.; Granda, J.; Giurg, M.; Burda-Grabowska, M.; Zhang, L.; Sun, X.; Lv, Z.; Nayak, D. Ebselen derivatives are very potent dual inhibitors of SARS-CoV-2 proteases-PLpro and Mpro in in vitro studies. BioRxiv 2020. 2020.2008. 2030.273979. [Google Scholar]
- Wang, Z.; Yang, L.; Zhao, X.-E. Co-crystallization and structure determination: An effective direction for anti-SARS-CoV-2 drug discovery. Computational and Structural Biotechnology Journal 2021, 19, 4684–4701. [Google Scholar] [CrossRef]
- Arwansyah, A.; Arif, A.; Ramli, I.; Kurniawan, I.; Sukarti, S.; Nur Alam, M.; Illing, I.; Farid Lewa, A.; Manguntungi, B. Molecular modelling on SARS-CoV-2 papain-like protease: an integrated study with homology modelling, molecular docking, and molecular dynamics simulations. SAR and QSAR in Environmental Research 2021, 32, 699–718. [Google Scholar] [CrossRef] [PubMed]
- Talevi, A.; Bellera, C.L. Challenges and opportunities with drug repurposing: finding strategies to find alternative uses of therapeutics. Expert Opinion on Drug Discovery 2020, 15, 397–401. [Google Scholar] [CrossRef]
- Pang, J.; Gao, S.; Sun, Z.; Yang, G. Discovery of small molecule PLpro inhibitor against COVID-19 using structure-based virtual screening, molecular dynamics simulation, and molecular mechanics/Generalized Born surface area (MM/GBSA) calculation. Struct Chem 2021, 32, 879–886. [Google Scholar] [CrossRef]
- Hu, X.; Zeng, Z.; Zhang, J.; Wu, D.; Li, H.; Geng, F. Molecular dynamics simulation of the interaction of food proteins with small molecules. Food Chemistry 2023, 405, 134824. [Google Scholar] [CrossRef] [PubMed]
- Wouters, O.J.; McKee, M.; Luyten, J. Estimated research and development investment needed to bring a new medicine to market, 2009-2018. Jama 2020, 323, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Zrieq, R.; Ahmad, I.; Snoussi, M.; Noumi, E.; Iriti, M.; Algahtani, F.D.; Patel, H.; Saeed, M.; Tasleem, M.; Sulaiman, S. Tomatidine and patchouli alcohol as inhibitors of SARS-CoV-2 enzymes (3CLpro, PLpro and NSP15) by molecular docking and molecular dynamics simulations. International Journal of Molecular Sciences 2021, 22, 10693. [Google Scholar] [CrossRef] [PubMed]
- Thangavel, N.; Albratty, M. Benchmarked molecular docking integrated molecular dynamics stability analysis for prediction of SARS-CoV-2 papain-like protease inhibition by olive secoiridoids. J King Saud Univ Sci 2023, 35, 102402. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Athar, M.; Jha, P.C. Computational investigation of binding of chloroquinone and hydroxychloroquinone against PLPro of SARS-CoV-2. J Biomol Struct Dyn 2022, 40, 3071–3081. [Google Scholar] [CrossRef]
- Kandeel, M.; Abdelrahman, A.; Oh-Hashi, K.; Ibrahim, A.; Venugopala, K.; Morsy, M.; Ibrahim, M. Repurposing of FDA-approved antivirals, antibiotics, anthelmintics, antioxidants, and cell protectives against SARS-CoV-2 papain-like protease. Journal of Biomolecular Structure and Dynamics 2020, 39, 1–8. [Google Scholar] [CrossRef]
- Joshi, S.; Joshi, M.; Degani, M.S. Tackling SARS-CoV-2: proposed targets and repurposed drugs. Future medicinal chemistry 2020, 12, 1579–1601. [Google Scholar] [CrossRef]
- Zhou, Y.-W.; Xie, Y.; Tang, L.-S.; Pu, D.; Zhu, Y.-J.; Liu, J.-Y.; Ma, X.-L. Therapeutic targets and interventional strategies in COVID-19: mechanisms and clinical studies. Signal transduction and targeted therapy 2021, 6, 317. [Google Scholar] [CrossRef]
- Russo, M.; Moccia, S.; Spagnuolo, C.; Tedesco, I.; Russo, G.L. Roles of flavonoids against coronavirus infection. Chemico-biological interactions 2020, 328, 109211. [Google Scholar] [CrossRef]
- Kumar, B.K.; Faheem, n.; Sekhar, K.V.G.C.; Ojha, R.; Prajapati, V.K.; Pai, A.; Murugesan, S. Pharmacophore based virtual screening, molecular docking, molecular dynamics and MM-GBSA approach for identification of prospective SARS-CoV-2 inhibitor from natural product databases. Journal of Biomolecular Structure and Dynamics 2022, 40, 1363–1386. [Google Scholar] [CrossRef]
- Kumar, B.K.; Faheem, *!!! REPLACE !!!*; Sekhar, K.; Ojha, R.; Prajapati, V.K.; Pai, A.; Murugesan, S. Pharmacophore based virtual screening, molecular docking, molecular dynamics and MM-GBSA approach for identification of prospective SARS-CoV-2 inhibitor from natural product databases. J Biomol Struct Dyn 2022, 40, 1363–1386. [Google Scholar] [CrossRef] [PubMed]
- Jupudi, S.; Rajagopal, K.; Murugesan, S.; Kumar, B.K.; Raman, K.; Byran, G.; Chennaiah, J.; pillai Muthiah, V.; Sankaran, S. Identification of Papain-Like Protease inhibitors of SARS CoV-2 through HTVS, Molecular docking, MMGBSA and Molecular dynamics approach. South African Journal of Botany 2022, 151, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Vardhan, S.; Sahoo, S.K. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Computers in biology and medicine 2020, 124, 103936. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Luan, J.; Zhang, L. Molecular docking of potential SARS-CoV-2 papain-like protease inhibitors. Biochemical and Biophysical Research Communications 2021, 538, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Kandeel, M.; Abdelrahman, A.H.; Oh-Hashi, K.; Ibrahim, A.; Venugopala, K.N.; Morsy, M.A.; Ibrahim, M.A. Repurposing of FDA-approved antivirals, antibiotics, anthelmintics, antioxidants, and cell protectives against SARS-CoV-2 papain-like protease. Journal of Biomolecular Structure and Dynamics 2021, 39, 5129–5136. [Google Scholar] [CrossRef] [PubMed]
- Çalışkaner, Z.O. Determination of Binding Potential of HCV Protease Inhibitors Against to SARS-CoV-2 Papain-like Protease wtih Computational Docking. Letters in Drug Design & Discovery 2021, 18, 949–960. [Google Scholar]
- Fu, Z.; Huang, B.; Tang, J.; Liu, S.; Liu, M.; Ye, Y.; Liu, Z.; Xiong, Y.; Zhu, W.; Cao, D. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nature communications 2021, 12, 488. [Google Scholar] [CrossRef] [PubMed]
- Tregoning, J.S.; Flight, K.E.; Higham, S.L.; Wang, Z.; Pierce, B.F. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nature reviews immunology 2021, 21, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Sharun, K.; Tiwari, R.; Dadar, M.; Malik, Y.S.; Singh, K.P.; Chaicumpa, W. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Human vaccines & immunotherapeutics 2020, 16, 1232–1238. [Google Scholar]
- Gurung, A.B.; Ali, M.A.; Lee, J.; Farah, M.A.; Al-Anazi, K.M. An updated review of computer-aided drug design and its application to COVID-19. BioMed research international 2021, 2021. [Google Scholar] [CrossRef]
- Asif, M.; Saleem, M.; Saadullah, M.; Yaseen, H.S.; Al Zarzour, R. COVID-19 and therapy with essential oils having antiviral, anti-inflammatory, and immunomodulatory properties. Inflammopharmacology 2020, 28, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Süntar, I. Importance of ethnopharmacological studies in drug discovery: role of medicinal plants. Phytochemistry Reviews 2020, 19, 1199–1209. [Google Scholar] [CrossRef]
- Kaliamurthi, S.; Selvaraj, G.; Selvaraj, C.; Singh, S.K.; Wei, D.-Q.; Peslherbe, G.H. Structure-based virtual screening reveals ibrutinib and zanubrutinib as potential repurposed drugs against COVID-19. International Journal of Molecular Sciences 2021, 22, 7071. [Google Scholar] [CrossRef]
- Freire, M.C.; Noske, G.D.; Bitencourt, N.V.; Sanches, P.R.; Santos-Filho, N.A.; Gawriljuk, V.O.; de Souza, E.P.; Nogueira, V.H.; de Godoy, M.O.; Nakamura, A.M. Non-toxic dimeric peptides derived from the bothropstoxin-I are potent SARS-CoV-2 and papain-like protease inhibitors. Molecules 2021, 26, 4896. [Google Scholar] [CrossRef]
- Sohraby, F.; Aryapour, H. Unraveling the unbinding pathways of SARS-CoV-2 Papain-like proteinase known inhibitors by Supervised Molecular Dynamics simulation. PLoS ONE 2021, 16, e0251910. [Google Scholar] [CrossRef] [PubMed]
- Rudrapal, M.; Khairnar, S.J.; Jadhav, A.G. Drug repurposing (DR): an emerging approach in drug discovery. Drug repurposing-hypothesis, molecular aspects and therapeutic applications 2020, 10. [Google Scholar]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal transduction and targeted therapy 2021, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Fu, W.; Jiang, D.; Sun, H.; Wang, J.; Zhang, X.; Weng, G.; Liu, H.; Tao, P.; Hou, T. VAD-MM/GBSA: A Variable Atomic Dielectric MM/GBSA Model for Improved Accuracy in Protein–Ligand Binding Free Energy Calculations. Journal of Chemical Information and Modeling 2021, 61, 2844–2856. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering precision nanoparticles for drug delivery. Nature reviews drug discovery 2021, 20, 101–124. [Google Scholar] [CrossRef]
- Consortium, W.S.T. Repurposed antiviral drugs for Covid-19—interim WHO solidarity trial results. New England journal of medicine 2021, 384, 497–511. [Google Scholar] [CrossRef]
- Abd El-Aziz, T.M.; Stockand, J.D. Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2)-an update on the status. Infection, Genetics and Evolution 2020, 83, 104327. [Google Scholar] [CrossRef] [PubMed]
- Citarella, A.; Scala, A.; Piperno, A.; Micale, N. SARS-CoV-2 Mpro: A potential target for peptidomimetics and small-molecule inhibitors. Biomolecules 2021, 11, 607. [Google Scholar] [CrossRef] [PubMed]
- Gentile, D.; Patamia, V.; Scala, A.; Sciortino, M.T.; Piperno, A.; Rescifina, A. Putative inhibitors of SARS-CoV-2 main protease from a library of marine natural products: a virtual screening and molecular modeling study. Marine drugs 2020, 18, 225. [Google Scholar] [CrossRef] [PubMed]
- Minetti, C.A.; Remeta, D.P. Forces driving a magic bullet to its target: revisiting the role of thermodynamics in drug design, development, and optimization. Life 2022, 12, 1438. [Google Scholar] [CrossRef] [PubMed]
- Badavath, V.N.; Kumar, A.; Samanta, P.K.; Maji, S.; Das, A.; Blum, G.; Jha, A.; Sen, A. Determination of potential inhibitors based on isatin derivatives against SARS-CoV-2 main protease (mpro): a molecular docking, molecular dynamics and structure-activity relationship studies. Journal of Biomolecular Structure and Dynamics 2022, 40, 3110–3128. [Google Scholar] [CrossRef] [PubMed]
- Boike, L.; Henning, N.J.; Nomura, D.K. Advances in covalent drug discovery. Nature Reviews Drug Discovery 2022, 21, 881–898. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Banerjee, S.; Ghosh, K.; Gayen, S.; Jha, T. Protease targeted COVID-19 drug discovery and its challenges: Insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorganic & medicinal chemistry 2021, 29, 115860. [Google Scholar]
- Al-Karmalawy, A.A.; El-Gamil, D.S.; El-Shesheny, R.; Sharaky, M.; Alnajjar, R.; Kutkat, O.; Moatasim, Y.; Elagawany, M.; Al-Rashood, S.T.; Binjubair, F.A. Design and statistical optimisation of emulsomal nanoparticles for improved anti-SARS-CoV-2 activity of N-(5-nitrothiazol-2-yl)-carboxamido candidates: in vitro and in silico studies. Journal of Enzyme Inhibition and Medicinal Chemistry 2023, 38, 2202357. [Google Scholar] [CrossRef] [PubMed]
- de Souza Neto, L.R.; Moreira-Filho, J.T.; Neves, B.J.; Maidana, R.L.B.R.; Guimarães, A.C.R.; Furnham, N.; Andrade, C.H.; Silva, F.P., Jr. In silico strategies to support fragment-to-lead optimization in drug discovery. Frontiers in chemistry 2020, 8, 93. [Google Scholar] [CrossRef]
- Trisciuzzi, D.; Villoutreix, B.O.; Siragusa, L.; Baroni, M.; Cruciani, G.; Nicolotti, O. Targeting protein-protein interactions with low molecular weight and short peptide modulators: insights on disease pathways and starting points for drug discovery. Expert Opinion on Drug Discovery 2023, 1–16. [Google Scholar] [CrossRef]
- Kumar, H.M.S.; Herrmann, L.; Tsogoeva, S.B. Structural hybridization as a facile approach to new drug candidates. Bioorganic & Medicinal Chemistry Letters 2020, 30, 127514. [Google Scholar]
- Brogi, S.; Ramalho, T.C.; Kuca, K.; Medina-Franco, J.L.; Valko, M. In silico methods for drug design and discovery. 2020, 8, 612. [Google Scholar] [PubMed]
- Salehi, B.; Venditti, A.; Sharifi-Rad, M.; Kręgiel, D.; Sharifi-Rad, J.; Durazzo, A.; Lucarini, M.; Santini, A.; Souto, E.B.; Novellino, E. The therapeutic potential of apigenin. International journal of molecular sciences 2019, 20, 1305. [Google Scholar] [CrossRef] [PubMed]
- Pantaleão, S.Q.; Fernandes, P.O.; Gonçalves, J.E.; Maltarollo, V.G.; Honorio, K.M. Recent advances in the prediction of pharmacokinetics properties in drug design studies: a review. ChemMedChem 2022, 17, e202100542. [Google Scholar] [CrossRef]
- Tang, Z.; Zhang, X.; Shu, Y.; Guo, M.; Zhang, H.; Tao, W. Insights from nanotechnology in COVID-19 treatment. Nano today 2021, 36, 101019. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Li, X.; Lin, X. A review on applications of computational methods in drug screening and design. Molecules 2020, 25, 1375. [Google Scholar] [CrossRef] [PubMed]
- Sabe, V.T.; Ntombela, T.; Jhamba, L.A.; Maguire, G.E.; Govender, T.; Naicker, T.; Kruger, H.G. Current trends in computer aided drug design and a highlight of drugs discovered via computational techniques: A review. European Journal of Medicinal Chemistry 2021, 224, 113705. [Google Scholar] [CrossRef] [PubMed]
- Zhan, P.; Pannecouque, C.; De Clercq, E.; Liu, X. Anti-HIV drug discovery and development: current innovations and future trends: miniperspective. Journal of medicinal chemistry 2016, 59, 2849–2878. [Google Scholar] [CrossRef] [PubMed]
- Rakib, A.; Nain, Z.; Sami, S.A.; Mahmud, S.; Islam, A.; Ahmed, S.; Siddiqui, A.B.F.; Babu, S.O.F.; Hossain, P.; Shahriar, A. A molecular modelling approach for identifying antiviral selenium-containing heterocyclic compounds that inhibit the main protease of SARS-CoV-2: An in silico investigation. Briefings in bioinformatics 2021, 22, 1476–1498. [Google Scholar] [CrossRef]
- Carracedo-Reboredo, P.; Liñares-Blanco, J.; Rodríguez-Fernández, N.; Cedrón, F.; Novoa, F.J.; Carballal, A.; Maojo, V.; Pazos, A.; Fernandez-Lozano, C. A review on machine learning approaches and trends in drug discovery. Computational and structural biotechnology journal 2021, 19, 4538–4558. [Google Scholar] [CrossRef]
- Huynh, T.; Cornell, W.; Luan, B. In silico Exploration of Inhibitors for SARS-CoV-2’s Papain-Like Protease. Front Chem 2020, 8, 624163. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Chakraborty, A.; Biswas, A.; Chowdhuri, S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors–an in silico docking and molecular dynamics simulation study. Journal of Biomolecular Structure and Dynamics 2021, 39, 4362–4374. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. Journal of chemical information and modeling 2020, 60, 3277–3286. [Google Scholar] [CrossRef] [PubMed]
- Yosri, N.; Abd El-Wahed, A.A.; Ghonaim, R.; Khattab, O.M.; Sabry, A.; Ibrahim, M.A.; Moustafa, M.F.; Guo, Z.; Zou, X.; Algethami, A.F. Anti-viral and immunomodulatory properties of propolis: Chemical diversity, pharmacological properties, preclinical and clinical applications, and in silico potential against SARS-CoV-2. Foods 2021, 10, 1776. [Google Scholar] [CrossRef] [PubMed]
- Chekroud, A.M.; Bondar, J.; Delgadillo, J.; Doherty, G.; Wasil, A.; Fokkema, M.; Cohen, Z.; Belgrave, D.; DeRubeis, R.; Iniesta, R. The promise of machine learning in predicting treatment outcomes in psychiatry. World Psychiatry 2021, 20, 154–170. [Google Scholar] [CrossRef] [PubMed]
- Pearce, J.M. A review of open source ventilators for COVID-19 and future pandemics. F1000Research 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, G.; Biagi, F.; Costa, P.; Karpiński, Z.; Mazza, J. The likely impact of COVID-19 on education: Reflections based on the existing literature and recent international datasets; Publications Office of the European Union Luxembourg, 2020; Volume 30275. [Google Scholar]
- Alanine, A.; Nettekoven, M.; Roberts, E.; Thomas, A.W. Lead generation-enhancing the success of drug discovery by investing in the hit to lead process. Combinatorial chemistry & high throughput screening 2003, 6, 51–66. [Google Scholar]
- Aftab, S.O.; Ghouri, M.Z.; Masood, M.U.; Haider, Z.; Khan, Z.; Ahmad, A.; Munawar, N. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. Journal of translational medicine 2020, 18, 1–15. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





