Submitted:
30 October 2023
Posted:
31 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Functioning of Non-Enzymatic Antioxidants in Higher Plant Cells and the Ways of Boosting Their Biosynthesis
2.1. Isoprenoids
2.1.1. Biosynthesis of isoprenoids
2.1.1.1. Ubiquinone synthesis
2.1.1.2. Plastoquinone synthesis
2.1.1.3. Tococherol synthesis
2.1.1.4. Carotenoid synthesis
2.1.2. Antioxidant activity of isoprenoids
2.1.3. Genetic approaches for boosting isoprenoid production in plants
| Proteins | Species | Target genes | Anti-oxidants | Editing type | Result |
|---|---|---|---|---|---|
| Kaempferol 3-O-rhamnosyl transferase and kaempferol 3-O-glucosyltransferase | A. thaliana |
At1g30530, At5g17050 |
Ubiquinone | Knockout as a result of deletion and insertion | Ubiquinone content in the double knockout represented 160% of wild-type level [141] |
| PSY, phytoene synthase |
Oryza sativa | ZmPsy | Carotenoids | Marker-free targeted insertion at pre-determined plant genomic safe harbors (knock-in Erwinia uredovora carotenoid desaturase (SSU-crtI) and maize phytoene synthase (ZmPsy) both driven by the endosperm-specific glutelin promoter) |
High level of β-carotene in the endosperm [139] |
| SlCYC-B, lycopene-β-cyclase; SlDDB1, DNA damage UV binding protein 1; SlDET1, de-etiolated 1 |
Solanum lycoper-sycum |
DNA damage SlCYC-B, SlDDB1, SlDET1, | Carotenoids | Target activation-induced cytidine deaminase base-editing technology, substitution of a cytidine with a thymine | Variations in carotenoid accumulation with an additive effect for each single mutation [132,133] |
|
LCY-E, lycopene ε-cyclase; Blc, beta-lycopene cyclase; LCY-B1, lycopene β-cyclase 1; LCY-B2, lycopene β-cyclase 2; SGR1, Stay-green 1 |
S. lycopersycum |
DQ100158 (SGR1), EU533951 (LCY-E), XM_010313794 (Blc), EF650013 (LCY-B1), AF254793 (LCY-B2) |
Carotenoids | Knockout as a result of deletions, insertion, substitution | Lycopene content in tomato fruit was increased to about 5.1-fold [126] |
|
LCYE, lycopene ε-cyclase |
O. sativa (rice calli) |
LcyE | Gene replacement using HDR, substitution H523L | Orange-colored line, total carotenoid content was 6.8–9.6 times higher than that of wild-type calli, increased tolerance to salt stress [129] | |
| Nicotiana tabacum |
Ntε-LCY1, Ntε-LCY2 |
Knockout as a result of deletions, insertion, substitution | Increase of the total carotenoid and chlorophyll contents, photosynthetic efficiency, and levels of the stress response [131] | ||
| Musa sapientum (banana) | GN-LCYε | Knockout as a result of indels | Accumulation of β-carotene content up to 6-fold; absence or a drastic reduction in the levels of lutein and α-carotene [128] | ||
| EIL2, Ethylene-Insensitive 3/ Ethylene-Insensitive 3-Likes |
S. lycopersycum | EIL2 | Carotenoids, Ascorbate |
Knockout as a result of insertion | Yellow, orange fruits. 1.62-fold increase of ascorbate content via both the L-galactose and myoinositol pathways [61] |
| PDS, phytoene desaturases |
Malus domestica (apple) |
LC10183 (PDS) |
Carotenoids | Knockout as a result of deletions, insertion | Albino phenotypes of regenerated plantlets [44] |
| Fragaria sp. | PDS | Knockout as a result of deletions | Albino regenerants [45] | ||
| Daucus carota (Orange carrot ‘Kurodagosun’, ‘Deep purple’ carrot) | XM_017385289.1 (DcPDS and DcMYB113-like genes) | Knockout as a result of deletions, insertion, substitution | Albino plants and purple color depigmented plants [48] |
||
| Dioscorea rotundata | DrPDS | Knockout as a result of deletions, insertion | Phenotypes of variegated to complete albinism [46] | ||
| Allium cepa L. | AcPDS | Knockout as a result of deletions, indels | Regenerated shoots exhibited three distinct phenotypes: albino, chimeric, and pale green [47] | ||
| CCDs, carotenoid cleavage dioxygenases | Musa sapientum (banana) | CCDs | Carotenoids | Knockout as a result of deletions | Higher fold β-carotene accumulation in non-green tissue (roots) than in green tissue (leaf) [138] |
| βOHase2, β-carotene hydroxylase |
A. thaliana |
At5g52570 (BCH2) |
Xantho-phylls | Knockout as a result of deletions | Prevention of the negative effects of carotenoid overproduction on seed germination [140] |
| DnaJ, cysteine-rich zinc-binding domain |
O. sativa (rice calli) |
Orange gene (OsOr) | Chromoplast formation | Knockout as a result of deletions | Orange-colored line accumulated more lutein, β-carotene, and two β-carotene isomers; increased tolerance to salt stress [136] |
| F3H, flavanone 3-hydroxylases |
D. carota (Carrot calli, purple-colored) |
F3H | Dihydro-flavonols, leucoantho-cyanidins, proantho-cyanidins, antho-cyanidins, anthocyanins | Knockout as a result of deletions | Blockage of the anthocyanin biosynthesis, discoloration of calli [142] |
| F3′H, flavanone 3′-hydroxylase |
Oryza sativa L. (black rice) |
Os10g0320100 (OsF3′H) |
Flavan-3-oles | Knockout as a result of deletions, insertions | Ocher seeds, much lower anthocyanin content [143] |
| Euphorbia pulcherrima | F3'H | Increased ratio of pelargonidin to cyanidin, braigt color changed from vivid red to vivid reddish orange [144] | |||
| DFR, dihydroflavonol 4-reductase | Zea mays | GRMZM2G026930 (a1), MZM2G013726 (a4) | Leucoantho-cyanidins, proantho-cyanidins, antho-cyanidins, anthocyanins | Knockout as a result of deletions, insertions | Blockage of the anthocyanin biosynthesis [145] |
| S. lycopersycum | Solyc02g085020 (DFR) |
Blockage of the anthocyanin biosynthesis, hypocotyls and callus were green [146,147] | |||
|
Oryza sativa L. (black rice) |
Os01g0633500 (OsDFR) |
Much lower anthocyanin content, ocher seeds [143] |
|||
| Ipomoea nil |
AB006793 (InDFR-B) |
Anthocyanin-less white flowers [148] | |||
| S. lycopersycum | DFR | Green hypocotyl due to defective anthocyanin accumulation [147] | |||
| LDOX, leucoanthocyanidin dioxygenase |
Oryza sativa L. (black rice) |
Os01g0372500 (OsLDOX) |
Prontho-cyanidins, antho-cyanidins, anthocyanins | Knockout as a result of deletions and insertions | Brown seeds, much lower total anthocyanin content [143] |
| UGTs, UDP-glucosyltransferases | A. thaliana | UGT79B2 (At4g27560), UGT79B3, (At4g27570) | Anthocyanin | Knockout as a result of deletions and insertions | Reduced levels of flavonoids and increased susceptibility to abiotic stress [149] |
|
Gt5GT, anthocyanin 5-O-glucosyl transferase; Gt3′GT, anthocyanin 3′-O-glucosyl transferase; Gt5/3′AT, anthocyanin 5/3′-aromatic acyl transferase |
Gentian cv. Albireo (Gentiana-triflora × G. scabra) |
Gt5GT, Gt3′GT, Gt5/3′AT |
Anthocyanin | Knockout as a result of deletions and insertions | Transformants produced pale red violet, dull pink and pale mauve flowers [150] |
| PAP1, production of anthocyanin pigment 1 (MYB transcription factor (TF)) |
A. thaliana |
AT1G56650 (PAP1) |
Flavonoids | CRISPR/Cas9 activation system with the p65-HSF activators to increase endogenous transcriptional levels | Purple pigmentation of the leaves under a high light [151] |
|
ANT1, anthocyanin mutant 1 (Myb TFs) |
S. lycopersicum | ANT1 | Flavonoids | Gene targeting upstream of the ANT1 gene | Overexpression and ectopic accumulation of pigments in tomato tissues [152] |
| CRISPR/LbCpf1-based HDR, Gene targeting upstream of the ANT1 gene | Tomato purple phenotype with salinity tolerance [153] | ||||
| SlAN2-like, (R2R3-MYB TFs) |
Solyc10g086290 (SlAN2-like) |
Knockout as a result of deletion | Lower accumulation of anthocyanins, downregulation of multiple anthocyanin-related genes [154] | ||
| SlAN2 (R2R3-MYB TFs) |
SlAN2 | Knockout as a result of deletion and substitution | Flavonoid content and the relative expression levels of several anthocyanin-related genes in vegetative tissues were significantly lower [155] | ||
| DcPDS and DcMYB113-like (R2R3-MYB TFs) |
D. carota ('Deep Purple') |
DcPDS, DcMYB113-like | Knockout as a result of deletions | Regenerated albino shoots [48] |
|
| PtrMYB57 (R2R3-MYB TFs) |
Populus tomentosa Carr | PtrMYB57 | Anthocyanin and proantho-cyanidin | Knockout as a result of deletions | High anthocyanin and proanthocyanidin phenotype [156] |
| FtMYB45 (R2R3-MYB TFs) |
Fagopyrum tataricum | FtMYB45 | Flavonoids | Knockout as a result of deletions and insertion | Content of rutin, catechin, and other flavonoids was increased in hairy root mutants [157] |
| bZIP (basic region/leucine zipper TFs) |
Vitis vinifera | VvbZIP36 | Flavonoids | Knockout as a result of deletions and insertion | Accumulation of metabolites (naringenin chalcone, naringenin, dihydroflavonols and cyanidin-3-O-glucoside). Synthesis of stilbenes (α-viniferin), lignans and some flavonols (including quercetin-3-O-rhamnoside, kaempferol-3-O-rhamnoside and kaempferol-7-O-rhamnoside) was inhibited [158] |
| TTG1, Transparent Testa Glabra1 (MYB-bHLH-WD40 TFs) |
A. thaliana | TTG1 | Flavonoids | Knockout as a result of deletion | Mutants produce pale seeds and lack trichomes [159] |
| O. sativa L. | OsTTG1 | Decreased falvonoid accumulation in various rice organs [160] | |||
| TT, Transparent Testa (bHLH TFs) |
Brassica napus | BnTT8 | Proantho-cyanidin | Knockout as a result of deletion and insertion | The yellow-seeded phenotype, seeds with elevated seed oil and protein content and altered fatty acid composition [161,162] |
| N. tabacum L. |
NtAn1a, NtAn1b |
||||
| uORFGGP1 | Single nucleotide transversion from C to T in the 5′ UTR of the Solyc06g073320 sequence, leading to a change in the predicted amino acid sequence from serine to phenylalanine | Increased ascorbate content (2- to 5-fold higher), male sterility [163] | |||
| GST, Glutathione S-transferase | S. lycopersycum | SlGSTAA | Glutathione | Knockout as a result of deletions | Green hypocotyl owing to anthocyanin deficiency [164,165] |
| Gentian cv. Albireo (G. triflora × G. scabra) |
GST | Knockout as a result of deletions | Decreased anthocyanin accumulation in flower petals [150] | ||
| F. vesca | RAP, Reduced Anthocyanins in Petioles | Knockout as a result of deletions, insertion | Green stem and white-fruited phenotype [166] | ||
| Phosphorylase, GGP | Lactuca sativa | uORFAtVTC2LsGGP1 and LsGGP2 (homologs of AtVTC2) | Ascorbate | Knockout as a result of deletions and indels | Increased ascorbate content by ∼150% and oxidation stress tolerance [167] |
| S. lycopersicum | uORFAtVTC2LsGGP2 (homologs of AtVTC2) | Knockout, deletions, indels | Increased ascorbate content [126] |
2. Flavonoids
2.1. Biosynthesis of flavonoids
2.2. Antioxidant activity of flavonoids
2.3. Genetic approaches for boosting flavonoid production in plants
2.3.1. Regulation of the expression of individual genes encoding key enzymes in flavonoid biosynthesis
2.3.2. Regulation of transcription factor activity to enhance flavonoid biosynthesis
3. Ascorbate and Glutathione
3.1. Biosynthesis of ascorbate
| Genes | Enzymes | Functions | |
|---|---|---|---|
| At4g23100 | γ-glutamylcysteine synthetase, γ-ECS, ATECS1, ATGSH1, Cinnamyl Alcohol Dehydrogenase Homolog 2, Glutamate-Cysteine Ligase, GSH1, GSHA | Catalysis of the first, and rate-limiting, step of glutathione biosynthesis. | |
| At5g27380 | Glutathione Synthetase 2, ATGSH2, GSH2, GSHB | Binding γ-glutamylcysteine and glycine together to form glutathione | |
| At4g29130 | Hexokinase 1, HXK1, ATHXK1, GIN2 | Hexose phosphorylation activity | |
| At2g19860 | Hexokinase 2, HXK2, ATHXK2 | ||
| At1g47840 | Hexokinase 3, HXK3 | ||
| At4g24620 | Phosphoglucose Isomerase, PGI, Glucose-6-phosphate isomerase | Transformation of d-glucose-6-phosphate into d-fructose-6-phosphate | |
| At3g02570 At1g67070 | Man-6-phosphate Isomerase, Phosphomannose isomerase, PMI | D-mannose-6-P formation from d-fructose-6-phosphate [261] | |
| At2g45790 | Phosphomannomutase, PMM | Transformation of D-mannose 6-phosphate into D-mannose 1-phosphate [262,263] | |
| At2g39770 | GDP-D-mannose pyrophosphorylase, GMP1, Vitamin C Defective 1, VTC1, Cytokinesis Defective 1, CYT1, Embryo Defective 101, EMB101, Sensitive To Ozone 1, SOZ1, | Guanosine monophosphate transfer from GTP to GDP-D-Mannose [251,264,265] | |
| At5g28840 | GDP-D-mannose 3’,5’ Epimerase, GME | The conversion of GDP-D-mannose to GDP-L-galactose (Barber, 1979). GME is also able to catalyze the 3′ epimerization of GDP-mannose, giving GDP-l-gulose, which is the precursor of a possible side-branch biosynthetic pathway (the gulose pathway) for vitamin C synthesis [252,266]. Plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis. | |
| At5g55120 | VTC5 | GDP-L-Galactose Phosphorylase, GGP | Encodes a novel protein involved in ascorbate biosynthesis, which was shown to catalyze the transfer of GMP from GDP-galactose to a variety of hexose-1-phosphate acceptors [267] |
| At4g26850 | VTC2 | ||
| At3g02870 | L-Galactose 1-P-phosphatase, GPP, VTC4 |
The conversion of l-Galactose-1-phosphate into l-galactose [268,269,270] | |
| At3g07130 | Purple acid phosphatase with phytase activity, PAP15 | ||
| At4g33670 | L-Galactose Dehydrogenase, GDH | The conversion of l-galactose into l-galactono-1,4-lactone [251] | |
| At3g47930 | L-Galactono 1,4-lactone Dehydrogenase, GLDH | Oxidation of L-galactono-1,4-lactone to AscA [264,271] | |
| At3g05620 At5g04970 At5g47500 At5g61680 | Methyl Esterases | The conversion of Methyl-D-Galacturonate into D-Galacturonate in D-Galacturonate pathway [272] | |
|
At1g14520 At4g26260 |
Myo-Inositol Oxygenase, MIOX1 | Convertion of Myo-inositol into L-Gulono-1,4-lactone Myo-inositol [254] | |
| At1g65770 | Ascorbic Acid Mannose Pathway Regulator 1, AMR1, ATFDA7, F-BOX/DUF295 ANCESTRAL 7 | Regulation of the mannose/L-galactose ascorbic acid biosynthetic pathway in response to developmental and environmental factors [273] | |
| At3g23230 | Ethylene Response Factor 98, ERF98, AtERF98, Transcriptional Regulator of Defense Response 1, TDR1, TTDR1 | Enhancement of the tolerance to salt through the transcriptional activation of ascorbic acid synthesis [274] | |
3.2. Biosynthesis of glutathione
2.3.3. Antioxidant activity of ascorbate and glutathione
2.3.4. The approaches for boosting ascorbate and glutathione production
3. Conclusion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Foyer, C.H.; Noctor, G. Redox Regulation in Photosynthetic Organisms: Signaling, Acclimation, and Practical Implications. Antioxid. Redox Signal. 2009, 11, 861–905. [Google Scholar] [CrossRef]
- Mubarakshina, M.M.; Ivanov, B.N.; Naydov, I.A.; Hillier, W.; Badger, M.R.; Krieger-Liszkay, A. Production and Diffusion of Chloroplastic H2O2 and Its Implication to Signalling. Journal of Experimental Botany 2010, 61, 3577–3587. [Google Scholar] [CrossRef] [PubMed]
- Borisova, M.M.M.; Kozuleva, M.A.; Rudenko, N.N.; Naydov, I.A.; Klenina, I.B.; Ivanov, B.N. Photosynthetic Electron Flow to Oxygen and Diffusion of Hydrogen Peroxide through the Chloroplast Envelope via Aquaporins. Biochimica et Biophysica Acta (BBA)-Bioenergetics 2012, 1817, 1314–1321. [Google Scholar] [CrossRef] [PubMed]
- Kozuleva, M.A.; Ivanov, B.N.; Vetoshkina, D.V.; Borisova-Mubarakshina, M.M. Minimizing an Electron Flow to Molecular Oxygen in Photosynthetic Electron Transfer Chain: An Evolutionary View. Frontiers in Plant Science 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- NOCTOR, G.; VELJOVIC-JOVANOVIC, S.; DRISCOLL, S.; NOVITSKAYA, L.; FOYER, C.H. Drought and Oxidative Load in the Leaves of C3 Plants: A Predominant Role for Photorespiration? Annals of Botany 2002, 89, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Mhamdi, A.; Foyer, C.H. The Roles of Reactive Oxygen Metabolism in Drought: Not So Cut and Dried. Plant Physiology 2014, 164, 1636–1648. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.M.; Mori, I.C.; Pei, Z.-M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.G.; Schroeder, J.I. NADPH Oxidase AtrbohD and AtrbohF Genes Function in ROS-Dependent ABA Signaling in Arabidopsis. The EMBO Journal 2003, 22, 2623–2633. [Google Scholar] [CrossRef]
- Mipeshwaree Devi, A.; Khedashwori Devi, K.; Premi Devi, P.; Lakshmipriyari Devi, M.; Das, S. Metabolic Engineering of Plant Secondary Metabolites: Prospects and Its Technological Challenges. Front Plant Sci 2023, 14, 1171154. [Google Scholar] [CrossRef]
- Liu, M.; Lu, S. Plastoquinone and Ubiquinone in Plants: Biosynthesis, Physiological Function and Metabolic Engineering. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Block, A.; Widhalm, J.R.; Fatihi, A.; Cahoon, R.E.; Wamboldt, Y.; Elowsky, C.; Mackenzie, S.A.; Cahoon, E.B.; Chapple, C.; Dudareva, N.; et al. The Origin and Biosynthesis of the Benzenoid Moiety of Ubiquinone (Coenzyme Q) in Arabidopsis. The Plant Cell 2014, 26, 1938–1948. [Google Scholar] [CrossRef]
- Soubeyrand, E.; Kelly, M.; Keene, S.A.; Bernert, A.C.; Latimer, S.; Johnson, T.S.; Elowsky, C.; Colquhoun, T.A.; Block, A.K.; Basset, G.J. Arabidopsis 4-COUMAROYL-COA LIGASE 8 Contributes to the Biosynthesis of the Benzenoid Ring of Coenzyme Q in Peroxisomes. Biochem J 2019, 476, 3521–3532. [Google Scholar] [CrossRef]
- Okada, K.; Kasahara, H.; Yamaguchi, S.; Kawaide, H.; Kamiya, Y.; Nojiri, H.; Yamane, H. Genetic Evidence for the Role of Isopentenyl Diphosphate Isomerases in the Mevalonate Pathway and Plant Development in Arabidopsis. Plant and Cell Physiology 2008, 49, 604–616. [Google Scholar] [CrossRef]
- Phillips, M.A.; D’Auria, J.C.; Gershenzon, J.; Pichersky, E. The Arabidopsis Thaliana Type I Isopentenyl Diphosphate Isomerases Are Targeted to Multiple Subcellular Compartments and Have Overlapping Functions in Isoprenoid Biosynthesis. The Plant Cell 2008, 20, 677–696. [Google Scholar] [CrossRef]
- Ducluzeau, A.-L.; Wamboldt, Y.; Elowsky, C.G.; Mackenzie, S.A.; Schuurink, R.C.; Basset, G.J.C. Gene Network Reconstruction Identifies the Authentic Trans-Prenyl Diphosphate Synthase That Makes the Solanesyl Moiety of Ubiquinone-9 in Arabidopsis. The Plant Journal 2012, 69, 366–375. [Google Scholar] [CrossRef]
- Okada, K.; Ohara, K.; Yazaki, K.; Nozaki, K.; Uchida, N.; Kawamukai, M.; Nojiri, H.; Yamane, H. The AtPPT1 Gene Encoding 4-Hydroxybenzoate Polyprenyl Diphosphate Transferase in Ubiquinone Biosynthesis Is Required for Embryo Development in Arabidopsis Thaliana. Plant Mol Biol 2004, 55, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Marbois, B.; Gin, P.; Gulmezian, M.; Clarke, C.F. The Yeast Coq4 Polypeptide Organizes a Mitochondrial Protein Complex Essential for Coenzyme Q Biosynthesis. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2009, 1791, 69–75. [Google Scholar] [CrossRef]
- Isaacson, T.; Ohad, I.; Beyer, P.; Hirschberg, J. Analysis in Vitro of the Enzyme CRTISO Establishes a Poly-Cis-Carotenoid Biosynthesis Pathway in Plants. Plant Physiology 2004, 136, 4246–4255. [Google Scholar] [CrossRef]
- Park, H.; Kreunen, S.S.; Cuttriss, A.J.; DellaPenna, D.; Pogson, B.J. Identification of the Carotenoid Isomerase Provides Insight into Carotenoid Biosynthesis, Prolamellar Body Formation, and Photomorphogenesis. The Plant Cell 2002, 14, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.X.; Gantt, E. GENES AND ENZYMES OF CAROTENOID BIOSYNTHESIS IN PLANTS. Annu Rev Plant Physiol Plant Mol Biol 1998, 49, 557–583. [Google Scholar] [CrossRef] [PubMed]
- Pogson, B.; McDonald, K.A.; Truong, M.; Britton, G.; DellaPenna, D. Arabidopsis Carotenoid Mutants Demonstrate That Lutein Is Not Essential for Photosynthesis in Higher Plants. Plant Cell 1996, 8, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Fiore, A.; Dall’Osto, L.; Fraser, P.D.; Bassi, R.; Giuliano, G. Elucidation of the β-Carotene Hydroxylation Pathway in Arabidopsis Thaliana. FEBS Letters 2006, 580, 4718–4722. [Google Scholar] [CrossRef]
- Kim, J.-E.; Cheng, K.M.; Craft, N.E.; Hamberger, B.; Douglas, C.J. Over-Expression of Arabidopsis Thaliana Carotenoid Hydroxylases Individually and in Combination with a Beta-Carotene Ketolase Provides Insight into in Vivo Functions. Phytochemistry 2010, 71, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Bugos, R.C.; Hieber, A.D.; Yamamoto, H.Y. Xanthophyll Cycle Enzymes Are Members of the Lipocalin Family, the First Identified from Plants*. Journal of Biological Chemistry 1998, 273, 15321–15324. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.Y.; Higashi, R.M. Violaxanthin De-Epoxidase: Lipid Composition and Substrate Specificity. Archives of Biochemistry and Biophysics 1978, 190, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Moran, G.R. 4-Hydroxyphenylpyruvate Dioxygenase. Archives of Biochemistry and Biophysics 2005, 433, 117–128. [Google Scholar] [CrossRef]
- Valentin, H.E.; Lincoln, K.; Moshiri, F.; Jensen, P.K.; Qi, Q.; Venkatesh, T.V.; Karunanandaa, B.; Baszis, S.R.; Norris, S.R.; Savidge, B.; et al. The Arabidopsis Vitamin E Pathway Gene5-1 Mutant Reveals a Critical Role for Phytol Kinase in Seed Tocopherol Biosynthesis. The Plant Cell 2006, 18, 212–224. [Google Scholar] [CrossRef]
- Wang, L.; Li, Q.; Zhang, A.; Zhou, W.; Jiang, R.; Yang, Z.; Yang, H.; Qin, X.; Ding, S.; Lu, Q.; et al. The Phytol Phosphorylation Pathway Is Essential for the Biosynthesis of Phylloquinone, Which Is Required for Photosystem I Stability in Arabidopsis. Molecular Plant 2017, 10, 183–196. [Google Scholar] [CrossRef]
- Hirooka, K.; Bamba, T.; Fukusaki, E.; Kobayashi, A. Cloning and Kinetic Characterization of Arabidopsis Thaliana Solanesyl Diphosphate Synthase. Biochem J 2003, 370, 679–686. [Google Scholar] [CrossRef]
- HIROOKA, K.; IZUMI, Y.; AN, C.-I.; NAKAZAWA, Y.; FUKUSAKI, E.; KOBAYASHI, A. Functional Analysis of Two Solanesyl Diphosphate Synthases from Arabidopsis Thaliana. Bioscience, Biotechnology, and Biochemistry 2005, 69, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Jun, L.; Saiki, R.; Tatsumi, K.; Nakagawa, T.; Kawamukai, M. Identification and Subcellular Localization of Two Solanesyl Diphosphate Synthases from Arabidopsis Thaliana. Plant and Cell Physiology 2004, 45, 1882–1888. [Google Scholar] [CrossRef] [PubMed]
- Grusak, M.A.; DellaPenna, D. Improving the Nutrient Composition of Plants to Enhance Human Nutrition and Health. Annual Review of Plant Physiology and Plant Molecular Biology 1999, 50, 133–161. [Google Scholar] [CrossRef]
- Ma, Y.; Yuan, L.; Wu, B.; Li, X.; Chen, S.; Lu, S. Genome-Wide Identification and Characterization of Novel Genes Involved in Terpenoid Biosynthesis in Salvia Miltiorrhiza. Journal of Experimental Botany 2012, 63, 2809–2823. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, S. Overview of Medicinally Important Diterpenoids Derived from Plastids. Mini Reviews in Medicinal Chemistry 2017, 17, 988–1001. [Google Scholar] [CrossRef]
- Kellogg, B.A.; Poulter, C.D. Chain Elongation in the Isoprenoid Biosynthetic Pathway. Current Opinion in Chemical Biology 1997, 1, 570–578. [Google Scholar] [CrossRef]
- Zhou, F.; Pichersky, E. More Is Better: The Diversity of Terpene Metabolism in Plants. Current Opinion in Plant Biology 2020, 55, 1–10. [Google Scholar] [CrossRef]
- Pu, X.; Dong, X.; Li, Q.; Chen, Z.; Liu, L. An Update on the Function and Regulation of Methylerythritol Phosphate and Mevalonate Pathways and Their Evolutionary Dynamics. Journal of Integrative Plant Biology 2021, 63, 1211–1226. [Google Scholar] [CrossRef] [PubMed]
- Closa, M.; Vranová, E.; Bortolotti, C.; Bigler, L.; Arró, M.; Ferrer, A.; Gruissem, W. The Arabidopsis Thaliana FPP Synthase Isozymes Have Overlapping and Specific Functions in Isoprenoid Biosynthesis, and Complete Loss of FPP Synthase Activity Causes Early Developmental Arrest. The Plant Journal 2010, 63, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.; Latimer, S.; Stutts, L.R.; Soubeyrand, E.; Block, A.K.; Basset, G.J. Kaempferol as a Precursor for Ubiquinone (Coenzyme Q) Biosynthesis: An Atypical Node between Specialized Metabolism and Primary Metabolism. Current Opinion in Plant Biology 2022, 66, 102165. [Google Scholar] [CrossRef]
- Bussell, J.D.; Reichelt, M.; Wiszniewski, A.A.G.; Gershenzon, J.; Smith, S.M. Peroxisomal ATP-Binding Cassette Transporter COMATOSE and the Multifunctional Protein ABNORMAL INFLORESCENCE MERISTEM Are Required for the Production of Benzoylated Metabolites in Arabidopsis Seeds. Plant Physiology 2014, 164, 48–54. [Google Scholar] [CrossRef]
- Xu, J.-J.; Zhang, X.-F.; Jiang, Y.; Fan, H.; Li, J.-X.; Li, C.-Y.; Zhao, Q.; Yang, L.; Hu, Y.-H.; Martin, C.; et al. A Unique Flavoenzyme Operates in Ubiquinone Biosynthesis in Photosynthesis-Related Eukaryotes. Science Advances 2021, 7, eabl3594. [Google Scholar] [CrossRef] [PubMed]
- Soubeyrand, E.; Johnson, T.S.; Latimer, S.; Block, A.; Kim, J.; Colquhoun, T.A.; Butelli, E.; Martin, C.; Wilson, M.A.; Basset, G.J. The Peroxidative Cleavage of Kaempferol Contributes to the Biosynthesis of the Benzenoid Moiety of Ubiquinone in Plants. The Plant Cell 2018, 30, 2910–2921. [Google Scholar] [CrossRef]
- Ksas, B.; Becuwe, N.; Chevalier, A.; Havaux, M. Plant Tolerance to Excess Light Energy and Photooxidative Damage Relies on Plastoquinone Biosynthesis. Sci Rep 2015, 5, 10919. [Google Scholar] [CrossRef]
- Collakova, E.; DellaPenna, D. The Role of Homogentisate Phytyltransferase and Other Tocopherol Pathway Enzymes in the Regulation of Tocopherol Synthesis during Abiotic Stress. Plant Physiol 2003, 133, 930–940. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, C.; Hirai, N.; Komori, S.; Wada, M.; Okada, K.; Osakabe, K.; Yamamoto, T.; Osakabe, Y. Efficient Genome Editing in Apple Using a CRISPR/Cas9 System. Sci Rep 2016, 6, 31481. [Google Scholar] [CrossRef] [PubMed]
- Wilson, F.M.; Harrison, K.; Armitage, A.D.; Simkin, A.J.; Harrison, R.J. CRISPR/Cas9-Mediated Mutagenesis of Phytoene Desaturase in Diploid and Octoploid Strawberry. Plant Methods 2019, 15, 45. [Google Scholar] [CrossRef] [PubMed]
- Syombua, E.D.; Zhang, Z.; Tripathi, J.N.; Ntui, V.O.; Kang, M.; George, O.O.; Edward, N.K.; Wang, K.; Yang, B.; Tripathi, L. A CRISPR/Cas9-Based Genome-Editing System for Yam (Dioscorea Spp.). Plant Biotechnology Journal 2021, 19, 645–647. [Google Scholar] [CrossRef] [PubMed]
- Mainkar, P.; Manape, T.K.; Satheesh, V.; Anandhan, S. CRISPR/Cas9-Mediated Editing of PHYTOENE DESATURASE Gene in Onion (Allium Cepa L.). Front Plant Sci 2023, 14, 1226911. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-S.; Feng, K.; Xiong, A.-S. CRISPR/Cas9-Mediated Multiply Targeted Mutagenesis in Orange and Purple Carrot Plants. Mol Biotechnol 2019, 61, 191–199. [Google Scholar] [CrossRef]
- Beyer, P.; Mayer, M.; Kleinig, H. Molecular Oxygen and the State of Geometric Isomerism of Intermediates Are Essential in the Carotene Desaturation and Cyclization Reactions in Daffodil Chromoplasts. European Journal of Biochemistry 1989, 184, 141–150. [Google Scholar] [CrossRef]
- Norris, S.R.; Barrette, T.R.; DellaPenna, D. Genetic Dissection of Carotenoid Synthesis in Arabidopsis Defines Plastoquinone as an Essential Component of Phytoene Desaturation. The Plant Cell 1995, 7, 2139–2149. [Google Scholar] [CrossRef]
- Carol, P.; Stevenson, D.; Bisanz, C.; Breitenbach, J.; Sandmann, G.; Mache, R.; Coupland, G.; Kuntz, M. Mutations in the Arabidopsis Gene IMMUTANS Cause a Variegated Phenotype by Inactivating a Chloroplast Terminal Oxidase Associated with Phytoene Desaturation. Plant Cell 1999, 11, 57–68. [Google Scholar] [CrossRef]
- DellaPenna, D.; Pogson, B.J. Vitamin Synthesis in Plants: Tocopherols and Carotenoids. Annu Rev Plant Biol 2006, 57, 711–738. [Google Scholar] [CrossRef]
- Pogson, B.J.; Rissler, H.M. Genetic Manipulation of Carotenoid Biosynthesis and Photoprotection. Philos Trans R Soc Lond B Biol Sci 2000, 355, 1395–1403. [Google Scholar] [CrossRef]
- Li, L.; Yuan, H. Chromoplast Biogenesis and Carotenoid Accumulation. Archives of Biochemistry and Biophysics 2013, 539, 102–109. [Google Scholar] [CrossRef]
- Park, S.; Kim, H.S.; Jung, Y.J.; Kim, S.H.; Ji, C.Y.; Wang, Z.; Jeong, J.C.; Lee, H.-S.; Lee, S.Y.; Kwak, S.-S. Orange Protein Has a Role in Phytoene Synthase Stabilization in Sweetpotato. Sci Rep 2016, 6, 33563. [Google Scholar] [CrossRef]
- Kang, L.; Kim, H.S.; Kwon, Y.S.; Ke, Q.; Ji, C.Y.; Park, S.-C.; Lee, H.-S.; Deng, X.; Kwak, S.-S. IbOr Regulates Photosynthesis under Heat Stress by Stabilizing IbPsbP in Sweetpotato. Frontiers in Plant Science 2017, 8. [Google Scholar] [CrossRef]
- Li, L.; Yang, Y.; Xu, Q.; Owsiany, K.; Welsch, R.; Chitchumroonchokchai, C.; Lu, S.; Van Eck, J.; Deng, X.-X.; Failla, M.; et al. The Or Gene Enhances Carotenoid Accumulation and Stability during Post-Harvest Storage of Potato Tubers. Mol Plant 2012, 5, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Crisp, P.; Walkey, D.G.A.; Bellman, E.; Roberts, E. A Mutation Affecting Curd Colour in Cauliflower (Brassica Oleracea L. Var. Botrytis DC). Euphytica 1975, 24, 173–176. [Google Scholar] [CrossRef]
- Lopez, A.B.; Van Eck, J.; Conlin, B.J.; Paolillo, D.J.; O’Neill, J.; Li, L. Effect of the Cauliflower Or Transgene on Carotenoid Accumulation and Chromoplast Formation in Transgenic Potato Tubers. Journal of Experimental Botany 2008, 59, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.-L.; Ho, T.-H.; Nguyen, D.-T. Overexpression of the IbOr Gene from Sweet Potato (Ipomea Batatas ’Hoang Long’) in Maize Increases Total Carotenoid and β-Carotene Contents. Turk J Biol 2017, 41, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhang, M.; Zhang, M.; Yang, M.; Dai, S.; Meng, Q.; Lv, W.; Zhuang, K. ETHYLENE-INSENSITIVE 3-LIKE 2 Regulates β-Carotene and Ascorbic Acid Accumulation in Tomatoes during Ripening. Plant Physiology 2023, 192, 2067–2080. [Google Scholar] [CrossRef]
- Niyogi, K.K. PHOTOPROTECTION REVISITED: Genetic and Molecular Approaches. Annu Rev Plant Physiol Plant Mol Biol 1999, 50, 333–359. [Google Scholar] [CrossRef] [PubMed]
- Newman, L.A.; Hadjeb, N.; Price, C.A. Synthesis of Two Chromoplast-Specific Proteins During Fruit Development in Capsicum Annuum. Plant Physiol 1989, 91, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Deruère, J.; Römer, S.; d’Harlingue, A.; Backhaus, R.A.; Kuntz, M.; Camara, B. Fibril Assembly and Carotenoid Overaccumulation in Chromoplasts: A Model for Supramolecular Lipoprotein Structures. The Plant Cell 1994, 6, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-H.; Lee, Y.; Kim, H.U. Fibrillin 5 Is Essential for Plastoquinone-9 Biosynthesis by Binding to Solanesyl Diphosphate Synthases in Arabidopsis. The Plant Cell 2015, 27, 2956–2971. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Sanchez, A.; Morelli, L.; Rodriguez-Concepcion, M. Arabidopsis FIBRILLIN6 Regulates Carotenoid Biosynthesis by Directly Promoting Phytoene Synthase Activity. 2022. 2022.06.30.498318. [Google Scholar] [CrossRef]
- Hundal, T.; Forsmark-Andrée, P.; Ernster, L.; Andersson, B. Antioxidant Activity of Reduced Plastoquinone in Chloroplast Thylakoid Membranes. Arch Biochem Biophys 1995, 324, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Maciejewska, U.; Polkowska-Kowalczyk, L.; Swiezewska, E.; Szkopinska, A. Plastoquinone: Possible Involvement in Plant Disease Resistance. Acta Biochim Pol 2002, 49, 775–780. [Google Scholar] [CrossRef]
- Kruk, J.; Jemioła-Rzemińska, M.; Burda, K.; Schmid, G.H.; Strzałka, K. Scavenging of Superoxide Generated in Photosystem I by Plastoquinol and Other Prenyllipids in Thylakoid Membranes. Biochemistry 2003, 42, 8501–8505. [Google Scholar] [CrossRef]
- Maroz, A.; Anderson, R.F.; Smith, R.A.J.; Murphy, M.P. Reactivity of Ubiquinone and Ubiquinol with Superoxide and the Hydroperoxyl Radical: Implications for in Vivo Antioxidant Activity. Free Radical Biology and Medicine 2009, 46, 105–109. [Google Scholar] [CrossRef]
- Borisova-Mubarakshina, M.M.; Vetoshkina, D.V.; Ivanov, B.N. Antioxidant and Signaling Functions of the Plastoquinone Pool in Higher Plants. Physiol Plant 2019, 166, 181–198. [Google Scholar] [CrossRef]
- Aikens, J.; Dix, T.A. Perhydroxyl Radical (HOO.) Initiated Lipid Peroxidation. The Role of Fatty Acid Hydroperoxides. Journal of Biological Chemistry 1991, 266, 15091–15098. [Google Scholar] [CrossRef] [PubMed]
- Kozuleva, M.A.; Petrova, A.A.; Mamedov, M.D.; Semenov, A.Yu.; Ivanov, B.N. O2 Reduction by Photosystem I Involves Phylloquinone under Steady-State Illumination. FEBS Letters 2014, 588, 4364–4368. [Google Scholar] [CrossRef] [PubMed]
- Kozuleva, M.A.; Ivanov, B.N. Superoxide Anion Radical Generation in Photosynthetic Electron Transport Chain. Biochemistry Moscow 2023, 88, 1045–1060. [Google Scholar] [CrossRef] [PubMed]
- Khorobrykh, S.A.; Ivanov, B.N. Oxygen Reduction in a Plastoquinone Pool of Isolated Pea Thylakoids. Photosynthesis Research 2002, 71, 209–219. [Google Scholar] [CrossRef]
- Mubarakshina, M.M.; Ivanov, B.N. The Production and Scavenging of Reactive Oxygen Species in the Plastoquinone Pool of Chloroplast Thylakoid Membranes. Physiologia Plantarum 2010, 140, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Grivennikova, V.G.; Vinogradov, A.D. Generation of Superoxide by the Mitochondrial Complex I. Biochim Biophys Acta 2006, 1757, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Muller, F.L.; Liu, Y.; Van Remmen, H. Complex III Releases Superoxide to Both Sides of the Inner Mitochondrial Membrane. J Biol Chem 2004, 279, 49064–49073. [Google Scholar] [CrossRef] [PubMed]
- Afanas’ev, I.B. Superoxide Ion: Chemistry and Biological Implications; CRC Press, 1991; ISBN 978-0-8493-5452-6. [Google Scholar]
- Khorobrykh, S.; Tyystjärvi, E. Plastoquinol Generates and Scavenges Reactive Oxygen Species in Organic Solvent: Potential Relevance for Thylakoids. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2018, 1859, 1119–1131. [Google Scholar] [CrossRef]
- Vetoshkina, D.V.; Ivanov, B.N.; Khorobrykh, S.A.; Proskuryakov, I.I.; Borisova-Mubarakshina, M.M. Involvement of the Chloroplast Plastoquinone Pool in the Mehler Reaction. Physiol Plant 2017, 161, 45–55. [Google Scholar] [CrossRef]
- Sanchez-Cruz, P.; Santos, A.; Diaz, S.; Alegría, A.E. Metal-Independent Reduction of Hydrogen Peroxide by Semiquinones. Chem. Res. Toxicol. 2014, 27, 1380–1386. [Google Scholar] [CrossRef] [PubMed]
- Borisova-Mubarakshina, M.M.; Naydov, I.A.; Ivanov, B.N. Oxidation of the Plastoquinone Pool in Chloroplast Thylakoid Membranes by Superoxide Anion Radicals. FEBS Letters 2018, 592, 3221–3228. [Google Scholar] [CrossRef]
- Neverov, K.V.; Krasnovsky Jr, A.A. Phosphorescence Analysis of the Chlorophyll Triplet States in Preparations of Photosystem II. Biophysics 2004, 49, 469–474. [Google Scholar]
- Rutherford, A.W.; Krieger-Liszkay, A. Herbicide-Induced Oxidative Stress in Photosystem II. Trends Biochem Sci 2001, 26, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Kruk, J.; Trebst, A. Plastoquinol as a Singlet Oxygen Scavenger in Photosystem II. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2008, 1777, 154–162. [Google Scholar] [CrossRef]
- Yadav, D.K.; Prasad, A.; Kruk, J.; Pospíšil, P. Evidence for the Involvement of Loosely Bound Plastosemiquinones in Superoxide Anion Radical Production in Photosystem II. PLOS ONE 2014, 9, e115466. [Google Scholar] [CrossRef] [PubMed]
- Gruszka, J.; Pawlak, A.; Kruk, J. Tocochromanols, Plastoquinol, and Other Biological Prenyllipids as Singlet Oxygen Quenchers—Determination of Singlet Oxygen Quenching Rate Constants and Oxidation Products. Free Radical Biology and Medicine 2008, 45, 920–928. [Google Scholar] [CrossRef]
- Ferretti, U.; Ciura, J.; Ksas, B.; Rác, M.; Sedlářová, M.; Kruk, J.; Havaux, M.; Pospíšil, P. Chemical Quenching of Singlet Oxygen by Plastoquinols and Their Oxidation Products in Arabidopsis. The Plant Journal 2018, 95, 848–861. [Google Scholar] [CrossRef]
- Rennenberg, A.P. Heinz Photooxidative Stress in Trees. In Causes of Photooxidative Stress and Amelioration of Defense Systems in Plants; CRC Press, 1993; ISBN 978-1-351-07045-4. [Google Scholar]
- Wang, S.Y.; Jiao, H. Scavenging Capacity of Berry Crops on Superoxide Radicals, Hydrogen Peroxide, Hydroxyl Radicals, and Singlet Oxygen. J. Agric. Food Chem. 2000, 48, 5677–5684. [Google Scholar] [CrossRef]
- Srivastava, S.; Phadke, R.S.; Govil, G.; Rao, C.N.R. Fluidity, Permeability and Antioxidant Behaviour of Model Membranes Incorporated with α-Tocopherol and Vitamin E Acetate. Biochimica et Biophysica Acta (BBA) - Biomembranes 1983, 734, 353–362. [Google Scholar] [CrossRef]
- Rajagopal, S.; Egorova, E.A.; Bukhov, N.G.; Carpentier, R. Quenching of Excited States of Chlorophyll Molecules in Submembrane Fractions of Photosystem I by Exogenous Quinones. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2003, 1606, 147–152. [Google Scholar] [CrossRef]
- Telfer, A. What Is β-Carotene Doing in the Photosystem II Reaction Centre? Philosophical Transactions: Biological Sciences 2002, 357, 1431–1440. [Google Scholar] [CrossRef]
- Mozzo, M.; Passarini, F.; Bassi, R.; van Amerongen, H.; Croce, R. Photoprotection in Higher Plants: The Putative Quenching Site Is Conserved in All Outer Light-Harvesting Complexes of Photosystem II. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2008, 1777, 1263–1267. [Google Scholar] [CrossRef] [PubMed]
- Alboresi, A.; Dall’Osto, L.; Aprile, A.; Carillo, P.; Roncaglia, E.; Cattivelli, L.; Bassi, R. Reactive Oxygen Species and Transcript Analysis upon Excess Light Treatment in Wild-Type Arabidopsis Thaliana vs a Photosensitive Mutant Lacking Zeaxanthin and Lutein. BMC Plant Biol 2011, 11, 62. [Google Scholar] [CrossRef]
- Krasnovsky Jr, A.; Paramonova, L.I. Interaction of Singlet Oxygen with Carotenoids: Rate Constants of Physical and Chemical Quenching. Biofizika 1983, 28, 725–729. [Google Scholar]
- Triantaphylidès, C.; Havaux, M. Singlet Oxygen in Plants: Production, Detoxification and Signaling. Trends in Plant Science 2009, 14, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Ashikhmin, A.A.; Benditkis, A.S.; Moskalenko, A.A.; Krasnovsky, A.A. Phytofluene as a Highly Efficient UVA Photosensitizer of Singlet Oxygen Generation. Biochemistry (Mosc) 2020, 85, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Manzano, D.; Fernández-Busquets, X.; Schaller, H.; González, V.; Boronat, A.; Arró, M.; Ferrer, A. The Metabolic Imbalance Underlying Lesion Formation in Arabidopsis Thaliana Overexpressing Farnesyl Diphosphate Synthase (Isoform 1S) Leads to Oxidative Stress and Is Triggered by the Developmental Decline of Endogenous HMGR Activity. Planta 2004, 219, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Tsegaye, Y.; Shintani, D.K.; DellaPenna, D. Overexpression of the Enzyme P-Hydroxyphenolpyruvate Dioxygenase in Arabidopsis and Its Relation to Tocopherol Biosynthesis. Plant Physiology and Biochemistry 2002, 40, 913–920. [Google Scholar] [CrossRef]
- Falk, J.; Andersen, G.; Kernebeck, B.; Krupinska, K. Constitutive Overexpression of Barley 4-Hydroxyphenylpyruvate Dioxygenase in Tobacco Results in Elevation of the Vitamin E Content in Seeds but Not in Leaves 1. FEBS Letters 2003, 540, 35–40. [Google Scholar] [CrossRef]
- Kim, S.-E.; Bian, X.; Lee, C.-J.; Park, S.-U.; Lim, Y.-H.; Kim, B.H.; Park, W.S.; Ahn, M.-J.; Ji, C.Y.; Yu, Y.; et al. Overexpression of 4-Hydroxyphenylpyruvate Dioxygenase (IbHPPD) Increases Abiotic Stress Tolerance in Transgenic Sweetpotato Plants. Plant Physiology and Biochemistry 2021, 167, 420–429. [Google Scholar] [CrossRef]
- Kanwischer, M.; Porfirova, S.; Bergmüller, E.; Dörmann, P. Alterations in Tocopherol Cyclase Activity in Transgenic and Mutant Plants of Arabidopsis Affect Tocopherol Content, Tocopherol Composition, and Oxidative Stress. Plant Physiology 2005, 137, 713–723. [Google Scholar] [CrossRef]
- Zbierzak, A.M.; Kanwischer, M.; Wille, C.; Vidi, P.-A.; Giavalisco, P.; Lohmann, A.; Briesen, I.; Porfirova, S.; Bréhélin, C.; Kessler, F.; et al. Intersection of the Tocopherol and Plastoquinol Metabolic Pathways at the Plastoglobule. Biochemical Journal 2009, 425, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hua, X.; Guo, J.; Qi, D.; Wang, L.; Liu, Z.; Jin, Z.; Chen, S.; Liu, G. Enhanced Tolerance to Drought Stress in Transgenic Tobacco Plants Overexpressing VTE1 for Increased Tocopherol Production from Arabidopsis Thaliana. Biotechnol Lett 2008, 30, 1275–1280. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.; He, S.; Liu, P.; Zhang, W.; Zhang, J.; Chen, S. The Role of Tocopherol Cyclase in Salt Stress Tolerance of Rice (Oryza Sativa). Sci. China Life Sci. 2011, 54, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Tavva, V.S.; Kim, Y.-H.; Kagan, I.A.; Dinkins, R.D.; Kim, K.-H.; Collins, G.B. Increased Alpha-Tocopherol Content in Soybean Seed Overexpressing the Perilla Frutescens Gamma-Tocopherol Methyltransferase Gene. Plant Cell Rep 2007, 26, 61–70. [Google Scholar] [CrossRef] [PubMed]
- YoungJin, K.; HongYul, S.; TaeIl, P.; SoHyeon, B.; WoonChul, S.; HyunSoon, K.; JungGon, K.; YongEui, C.; SongJoong, Y. Enhanced Biosynthesis of α-Tocopherol in Transgenic Soybean by Introducing γ-TMT Gene. Journal of Plant Biotechnology 2005, 7, 1–7. [Google Scholar]
- Li, Y.; Zhou, Y.; Wang, Z.; Sun, X.; Tang, K. Engineering Tocopherol Biosynthetic Pathway in Arabidopsis Leaves and Its Effect on Antioxidant Metabolism. Plant Science 2010, 178, 312–320. [Google Scholar] [CrossRef]
- Ghimire, B.K.; Seong, E.S.; Yu, C.Y.; Kim, S.-H.; Chung, I.-M. Evaluation of Phenolic Compounds and Antimicrobial Activities in Transgenic Codonopsis Lanceolata Plants via Overexpression of the γ-Tocopherol Methyltransferase (γ-Tmt) Gene. South African Journal of Botany 2017, 109, 25–33. [Google Scholar] [CrossRef]
- Ohara, K.; Kokado, Y.; Yamamoto, H.; Sato, F.; Yazaki, K. Engineering of Ubiquinone Biosynthesis Using the Yeast Coq2 Gene Confers Oxidative Stress Tolerance in Transgenic Tobacco. Plant J 2004, 40, 734–743. [Google Scholar] [CrossRef]
- Liu, M.; Ma, Y.; Du, Q.; Hou, X.; Wang, M.; Lu, S. Functional Analysis of Polyprenyl Diphosphate Synthase Genes Involved in Plastoquinone and Ubiquinone Biosynthesis in Salvia Miltiorrhiza. Front Plant Sci 2019, 10, 893. [Google Scholar] [CrossRef]
- Ksas, B.; Légeret, B.; Ferretti, U.; Chevalier, A.; Pospíšil, P.; Alric, J.; Havaux, M. The Plastoquinone Pool Outside the Thylakoid Membrane Serves in Plant Photoprotection as a Reservoir of Singlet Oxygen Scavengers. Plant, Cell & Environment 2018, 41, 2277–2287. [Google Scholar] [CrossRef]
- Ye, X.; Al-Babili, S.; Klöti, A.; Zhang, J.; Lucca, P.; Beyer, P.; Potrykus, I. Engineering the Provitamin A (Beta-Carotene) Biosynthetic Pathway into (Carotenoid-Free) Rice Endosperm. Science 2000, 287, 303–305. [Google Scholar] [CrossRef] [PubMed]
- Paine, J.A.; Shipton, C.A.; Chaggar, S.; Howells, R.M.; Kennedy, M.J.; Vernon, G.; Wright, S.Y.; Hinchliffe, E.; Adams, J.L.; Silverstone, A.L.; et al. Improving the Nutritional Value of Golden Rice through Increased Pro-Vitamin A Content. Nat Biotechnol 2005, 23, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Shewmaker, C.K.; Sheehy, J.A.; Daley, M.; Colburn, S.; Ke, D.Y. Seed-Specific Overexpression of Phytoene Synthase: Increase in Carotenoids and Other Metabolic Effects. Plant J 1999, 20, 401–412X. [Google Scholar] [CrossRef]
- Naqvi, S.; Zhu, C.; Farre, G.; Ramessar, K.; Bassie, L.; Breitenbach, J.; Perez Conesa, D.; Ros, G.; Sandmann, G.; Capell, T.; et al. Transgenic Multivitamin Corn through Biofortification of Endosperm with Three Vitamins Representing Three Distinct Metabolic Pathways. Proc Natl Acad Sci U S A 2009, 106, 7762–7767. [Google Scholar] [CrossRef]
- Paul, J.-Y.; Khanna, H.; Kleidon, J.; Hoang, P.; Geijskes, J.; Daniells, J.; Zaplin, E.; Rosenberg, Y.; James, A.; Mlalazi, B.; et al. Golden Bananas in the Field: Elevated Fruit pro-Vitamin A from the Expression of a Single Banana Transgene. Plant Biotechnol J 2017, 15, 520–532. [Google Scholar] [CrossRef]
- Pons, E.; Alquézar, B.; Rodríguez, A.; Martorell, P.; Genovés, S.; Ramón, D.; Rodrigo, M.J.; Zacarías, L.; Peña, L. Metabolic Engineering of β-Carotene in Orange Fruit Increases Its in Vivo Antioxidant Properties. Plant Biotechnol J 2014, 12, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Ahn, Y.O.; Ahn, M.-J.; Lee, H.-S.; Kwak, S.-S. Down-Regulation of β-Carotene Hydroxylase Increases β-Carotene and Total Carotenoids Enhancing Salt Stress Tolerance in Transgenic Cultured Cells of Sweetpotato. Phytochemistry 2012, 74, 69–78. [Google Scholar] [CrossRef]
- Davison, P.A.; Hunter, C.N.; Horton, P. Overexpression of Beta-Carotene Hydroxylase Enhances Stress Tolerance in Arabidopsis. Nature 2002, 418, 203–206. [Google Scholar] [CrossRef]
- Saeed, B.; Das, M.; Khurana, P. Overexpression of β-Carotene Hydroxylase1 (BCH1) in Indian Mulberry, Morus Indica Cv. K2, Confers Tolerance against UV, High Temperature and High Irradiance Stress Induced Oxidative Damage. Plant Cell Tiss Organ Cult 2015, 120, 1003–1014. [Google Scholar] [CrossRef]
- Wu, W.; Ji, J.; Wang, G.; Zhao, Q.; Jin, C.; Guan, C.; Josine, T.L. Overexpression of AtchyB in Eustoma Grandiflorum Shinn Enhances Its Tolerance to High-Light Via Zeaxanthin Accumulation. Plant Mol Biol Rep 2012, 30, 1433–1443. [Google Scholar] [CrossRef]
- Römer, S.; Lübeck, J.; Kauder, F.; Steiger, S.; Adomat, C.; Sandmann, G. Genetic Engineering of a Zeaxanthin-Rich Potato by Antisense Inactivation and Co-Suppression of Carotenoid Epoxidation. Metab Eng 2002, 4, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Chen, S.; Tian, H.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. Lycopene Is Enriched in Tomato Fruit by CRISPR/Cas9-Mediated Multiplex Genome Editing. Front Plant Sci 2018, 9, 559. [Google Scholar] [CrossRef]
- Diretto, G.; Tavazza, R.; Welsch, R.; Pizzichini, D.; Mourgues, F.; Papacchioli, V.; Beyer, P.; Giuliano, G. Metabolic Engineering of Potato Tuber Carotenoids through Tuber-Specific Silencing of Lycopene Epsilon Cyclase. BMC Plant Biol 2006, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Alok, A.; Shivani, null; Kumar, P.; Kaur, N.; Awasthi, P.; Chaturvedi, S.; Pandey, P.; Pandey, A.; Pandey, A.K.; et al. CRISPR/Cas9 Directed Editing of Lycopene Epsilon-Cyclase Modulates Metabolic Flux for β-Carotene Biosynthesis in Banana Fruit. Metab Eng 2020, 59, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Yu, J.; Kim, H.K.; Kim, J.Y.; Kim, M.-S.; Cho, Y.-G.; Bae, S.; Kang, K.K.; Jung, Y.J. Genome Editing of Golden SNP-Carrying Lycopene Epsilon-Cyclase (LcyE) Gene Using the CRSPR-Cas9/HDR and Geminiviral Replicon System in Rice. Int J Mol Sci 2022, 23, 10383. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, A.; Ohishi, K.; Yamada, T.; Shibata-Hatta, M.; Arai-Kichise, Y.; Watanabe, S.; Yoshikawa, H.; Wakasa, K. Biochemical and Molecular Characterization of Orange- and Tangerine-Colored Rice Calli. Plant Biotechnology 2015, 32, 193–203. [Google Scholar] [CrossRef]
- Song, W.; Wei, F.; Gao, S.; Dong, C.; Hao, J.; Jin, L.; Li, F.; Wei, P.; Guo, J.; Wang, R. Functional Characterization and Comparison of Lycopene Epsilon-Cyclase Genes in Nicotiana Tabacum. BMC Plant Biol 2022, 22, 252. [Google Scholar] [CrossRef]
- Hunziker, J.; Nishida, K.; Kondo, A.; Kishimoto, S.; Ariizumi, T.; Ezura, H. Multiple Gene Substitution by Target-AID Base-Editing Technology in Tomato. Sci Rep 2020, 10, 20471. [Google Scholar] [CrossRef]
- Hunziker, J.; Nishida, K.; Kondo, A.; Ariizumi, T.; Ezura, H. Phenotypic Characterization of High Carotenoid Tomato Mutants Generated by the Target-AID Base-Editing Technology. Front Plant Sci 2022, 13, 848560. [Google Scholar] [CrossRef]
- Wang, Z.; Ke, Q.; Kim, M.D.; Kim, S.H.; Ji, C.Y.; Jeong, J.C.; Lee, H.-S.; Park, W.S.; Ahn, M.-J.; Li, H.; et al. Transgenic Alfalfa Plants Expressing the Sweetpotato Orange Gene Exhibit Enhanced Abiotic Stress Tolerance. PLoS One 2015, 10, e0126050. [Google Scholar] [CrossRef] [PubMed]
- Berman, J.; Zorrilla-López, U.; Medina, V.; Farré, G.; Sandmann, G.; Capell, T.; Christou, P.; Zhu, C. The Arabidopsis ORANGE (AtOR) Gene Promotes Carotenoid Accumulation in Transgenic Corn Hybrids Derived from Parental Lines with Limited Carotenoid Pools. Plant Cell Rep 2017, 36, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Kim, J.Y.; Kim, J.H.; Go, J.Y.; Jung, Y.-S.; Lee, H.J.; Ahn, M.-J.; Yu, J.; Bae, S.; Kim, H.S.; et al. Biochemical Characterization of Orange-Colored Rice Calli Induced by Target Mutagenesis of OsOr Gene. Plants (Basel) 2022, 12, 56. [Google Scholar] [CrossRef]
- Zeng, J.; Wang, X.; Miao, Y.; Wang, C.; Zang, M.; Chen, X.; Li, M.; Li, X.; Wang, Q.; Li, K.; et al. Metabolic Engineering of Wheat Provitamin A by Simultaneously Overexpressing CrtB and Silencing Carotenoid Hydroxylase (TaHYD). J Agric Food Chem 2015, 63, 9083–9092. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, P.; Khan, S.; Lakhani, H.; Chaturvedi, S.; Shivani; Kaur, N.; Singh, J.; Kesarwani, A.K.; Tiwari, S. Transgene-Free Genome Editing Supports the Role of Carotenoid Cleavage Dioxygenase 4 as a Negative Regulator of β-Carotene in Banana. Journal of Experimental Botany 2022, 73, 3401–3416. [Google Scholar] [CrossRef]
- Dong, O.X.; Yu, S.; Jain, R.; Zhang, N.; Duong, P.Q.; Butler, C.; Li, Y.; Lipzen, A.; Martin, J.A.; Barry, K.W.; et al. Marker-Free Carotenoid-Enriched Rice Generated through Targeted Gene Insertion Using CRISPR-Cas9. Nat Commun 2020, 11, 1178. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Zhu, Q.; Wei, Z.; Owens, L.A.; Fish, T.; Kim, H.; Thannhauser, T.W.; Cahoon, E.B.; Li, L. Multi-Strategy Engineering Greatly Enhances Provitamin A Carotenoid Accumulation and Stability in Arabidopsis Seeds. aBIOTECH 2021, 2, 191–214. [Google Scholar] [CrossRef] [PubMed]
- Soubeyrand, E.; Latimer, S.; Bernert, A.C.; Keene, S.A.; Johnson, T.S.; Shin, D.; Block, A.K.; Colquhoun, T.A.; Schäffner, A.R.; Kim, J.; et al. 3-O-Glycosylation of Kaempferol Restricts the Supply of the Benzenoid Precursor of Ubiquinone (Coenzyme Q) in Arabidopsis Thaliana. Phytochemistry 2021, 186, 112738. [Google Scholar] [CrossRef]
- Klimek-Chodacka, M.; Oleszkiewicz, T.; Lowder, L.G.; Qi, Y.; Baranski, R. Efficient CRISPR/Cas9-Based Genome Editing in Carrot Cells. Plant Cell Rep 2018, 37, 575–586. [Google Scholar] [CrossRef]
- Jung, Y.J.; Lee, H.J.; Kim, J.H.; Kim, D.H.; Kim, H.K.; Cho, Y.-G.; Bae, S.; Kang, K.K. CRISPR/Cas9-Targeted Mutagenesis of F3′H, DFR and LDOX, Genes Related to Anthocyanin Biosynthesis in Black Rice (Oryza Sativa L.). Plant Biotechnol Rep 2019, 13, 521–531. [Google Scholar] [CrossRef]
- Nitarska, D.; Boehm, R.; Debener, T.; Lucaciu, R.C.; Halbwirth, H. First Genome Edited Poinsettias: Targeted Mutagenesis of Flavonoid 3’-Hydroxylase Using CRISPR/Cas9 Results in a Colour Shift. Plant Cell Tissue Organ Cult 2021, 147, 49–60. [Google Scholar] [CrossRef]
- Char, S.N.; Neelakandan, A.K.; Nahampun, H.; Frame, B.; Main, M.; Spalding, M.H.; Becraft, P.W.; Meyers, B.C.; Walbot, V.; Wang, K.; et al. An Agrobacterium-Delivered CRISPR/Cas9 System for High-Frequency Targeted Mutagenesis in Maize. Plant Biotechnol J 2017, 15, 257–268. [Google Scholar] [CrossRef]
- Danilo, B.; Perrot, L.; Botton, E.; Nogué, F.; Mazier, M. The DFR Locus: A Smart Landing Pad for Targeted Transgene Insertion in Tomato. PLoS One 2018, 13, e0208395. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Deng, L.; Yuan, G.; Zhao, W.; Ma, M.; Sun, C.; Du, M.; Li, C.; Li, C. Rapid Generation of a Tomato Male Sterility System and Its Feasible Application in Hybrid Seed Production. Theor Appl Genet 2023, 136, 197. [Google Scholar] [CrossRef]
- Watanabe, K.; Kobayashi, A.; Endo, M.; Sage-Ono, K.; Toki, S.; Ono, M. CRISPR/Cas9-Mediated Mutagenesis of the Dihydroflavonol-4-Reductase-B (DFR-B) Locus in the Japanese Morning Glory Ipomoea (Pharbitis) Nil. Sci Rep 2017, 7, 10028. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.-J.; Zhang, F.-J.; Zhang, G.-Z.; Jiang, X.-Y.; Yu, H.-M.; Hou, B.-K. The Arabidopsis UDP-Glycosyltransferases UGT79B2 and UGT79B3, Contribute to Cold, Salt and Drought Stress Tolerance via Modulating Anthocyanin Accumulation. Plant J 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Tasaki, K.; Higuchi, A.; Watanabe, A.; Sasaki, N.; Nishihara, M. Effects of Knocking out Three Anthocyanin Modification Genes on the Blue Pigmentation of Gentian Flowers. Sci Rep 2019, 9, 15831. [Google Scholar] [CrossRef]
- Park, J.-J.; Dempewolf, E.; Zhang, W.; Wang, Z.-Y. RNA-Guided Transcriptional Activation via CRISPR/dCas9 Mimics Overexpression Phenotypes in Arabidopsis. PLoS One 2017, 12, e0179410. [Google Scholar] [CrossRef]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-Frequency, Precise Modification of the Tomato Genome. Genome Biol 2015, 16, 232. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.V.; Sivankalyani, V.; Kim, E.-J.; Doan, D.T.H.; Tran, M.T.; Kim, J.; Sung, Y.W.; Park, M.; Kang, Y.J.; Kim, J.-Y. Highly Efficient Homology-Directed Repair Using CRISPR/Cpf1-Geminiviral Replicon in Tomato. Plant Biotechnol J 2020, 18, 2133–2143. [Google Scholar] [CrossRef]
- Yan, S.; Chen, N.; Huang, Z.; Li, D.; Zhi, J.; Yu, B.; Liu, X.; Cao, B.; Qiu, Z. Anthocyanin Fruit Encodes an R2R3-MYB Transcription Factor, SlAN2-like, Activating the Transcription of SlMYBATV to Fine-Tune Anthocyanin Content in Tomato Fruit. New Phytol 2020, 225, 2048–2063. [Google Scholar] [CrossRef] [PubMed]
- Zhi, J.; Liu, X.; Li, D.; Huang, Y.; Yan, S.; Cao, B.; Qiu, Z. CRISPR/Cas9-Mediated SlAN2 Mutants Reveal Various Regulatory Models of Anthocyanin Biosynthesis in Tomato Plant. Plant Cell Rep 2020, 39, 799–809. [Google Scholar] [CrossRef]
- Wan, S.; Li, C.; Ma, X.; Luo, K. PtrMYB57 Contributes to the Negative Regulation of Anthocyanin and Proanthocyanidin Biosynthesis in Poplar. Plant Cell Rep 2017, 36, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Wu, L.; Wang, M.; Yang, W.; Wang, X.; Ma, W.; Sun, W.; Chen, S.; Xiang, L.; Shi, Y. CRISPR/Cas9-Mediated Targeted Mutagenesis of FtMYB45 Promotes Flavonoid Biosynthesis in Tartary Buckwheat (Fagopyrum Tataricum). Front Plant Sci 2022, 13, 879390. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Fang, J.; Zhao, R.; Liu, X.; Yin, W.; Wang, Y.; Wang, X.; Wang, X.; Fang, Y. CRISPR/Cas9-Mediated Mutagenesis of VvbZIP36 Promotes Anthocyanin Accumulation in Grapevine (Vitis Vinifera). Horticulture Research 2022, 9, uhac022. [Google Scholar] [CrossRef]
- Ryder, P.; McHale, M.; Fort, A.; Spillane, C. Generation of Stable Nulliplex Autopolyploid Lines of Arabidopsis Thaliana Using CRISPR/Cas9 Genome Editing. Plant Cell Rep 2017, 36, 1005–1008. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Xia, X.; Zhang, Z.; He, J.; Nong, B.; Luo, T.; Feng, R.; Wu, Y.; Pan, Y.; et al. OsTTG1, a WD40 Repeat Gene, Regulates Anthocyanin Biosynthesis in Rice. Plant J 2021, 107, 198–214. [Google Scholar] [CrossRef]
- Zhai, Y.; Yu, K.; Cai, S.; Hu, L.; Amoo, O.; Xu, L.; Yang, Y.; Ma, B.; Jiao, Y.; Zhang, C.; et al. Targeted Mutagenesis of BnTT8 Homologs Controls Yellow Seed Coat Development for Effective Oil Production in Brassica Napus L. Plant Biotechnol J 2020, 18, 1153–1168. [Google Scholar] [CrossRef]
- Tian, Y.; Liu, X.; Fan, C.; Li, T.; Qin, H.; Li, X.; Chen, K.; Zheng, Y.; Chen, F.; Xu, Y. Enhancement of Tobacco (Nicotiana Tabacum L.) Seed Lipid Content for Biodiesel Production by CRISPR-Cas9-Mediated Knockout of NtAn1. Front Plant Sci 2020, 11, 599474. [Google Scholar] [CrossRef]
- Deslous, P.; Bournonville, C.; Decros, G.; Okabe, Y.; Mauxion, J.-P.; Jorly, J.; Gadin, S.; Brès, C.; Mori, K.; Ferrand, C.; et al. Overproduction of Ascorbic Acid Impairs Pollen Fertility in Tomato. Journal of Experimental Botany 2021, 72, 3091–3107. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, S.; Wang, H.; Luo, B.; Cai, Y.; Li, X.; Zhang, Y.; Wang, X. Rapid Generation of Tomato Male-Sterile Lines with a Marker Use for Hybrid Seed Production by CRISPR/Cas9 System. Mol Breed 2021, 41, 25. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Deng, L.; Yuan, G.; Zhao, W.; Ma, M.; Sun, C.; Du, M.; Li, C.; Li, C. A CRISPR-Cas9-Derived Male Sterility System for Tomato Breeding. Agronomy 2023, 13, 1785. [Google Scholar] [CrossRef]
- Gao, Q.; Luo, H.; Li, Y.; Liu, Z.; Kang, C. Genetic Modulation of RAP Alters Fruit Coloration in Both Wild and Cultivated Strawberry. Plant Biotechnol J 2020, 18, 1550–1561. [Google Scholar] [CrossRef]
- Zhang, H.; Si, X.; Ji, X.; Fan, R.; Liu, J.; Chen, K.; Wang, D.; Gao, C. Genome Editing of Upstream Open Reading Frames Enables Translational Control in Plants. Nat Biotechnol 2018, 36, 894–898. [Google Scholar] [CrossRef]
- Samanta, A.; Das, G.; Das, S. Roles of Flavonoids in Plants. International Journal of pharmaceutical science and technology 2011, 6, 12–35. [Google Scholar]
- Takahashi, A.; Ohnishi, T. The Significance of the Study about the Biological Effects of Solar Ultraviolet Radiation Using the Exposed Facility on the International Space Station. Biol Sci Space 2004, 18, 255–260. [Google Scholar] [CrossRef]
- Cheng, G.W.; Breen, P.J. Activity of Phenylalanine Ammonia-Lyase (PAL) and Concentrations of Anthocyanins and Phenolics in Developing Strawberry Fruit. Journal of the American Society for Horticultural Science 1991, 116, 865–869. [Google Scholar] [CrossRef]
- Forkmann, G.; Heller, W. 1.26 - Biosynthesis of Flavonoids. In Comprehensive Natural Products Chemistry; Barton, S.D., Nakanishi, K., Meth-Cohn, O., Eds.; Pergamon: Oxford, 1999; pp. 713–748. ISBN 978-0-08-091283-7. [Google Scholar]
- Ohl, S.; Hedrick, S.A.; Chory, J.; Lamb, C.J. Functional Properties of a Phenylalanine Ammonia-Lyase Promoter from Arabidopsis. The Plant Cell 1990, 2, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Shufflebottom, D.; Edwards, K.; Schuch, W.; Bevan, M. Transcription of Two Members of a Gene Family Encoding Phenylalanine Ammonia-Lyase Leads to Remarkably Different Cell Specificities and Induction Patterns. Plant J 1993, 3, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Werck-Reichhart, D.; Bak, S.; Paquette, S. Cytochromes P450. Arabidopsis Book 2002, 1, e0028. [Google Scholar] [CrossRef] [PubMed]
- Wohl, J.; Petersen, M. Functional Expression and Characterization of Cinnamic Acid 4-Hydroxylase from the Hornwort Anthoceros Agrestis in Physcomitrella Patens. Plant Cell Rep 2020, 39, 597–607. [Google Scholar] [CrossRef]
- Ehlting, J.; Büttner, D.; Wang, Q.; Douglas, C.J.; Somssich, I.E.; Kombrink, E. Three 4-Coumarate:Coenzyme A Ligases in Arabidopsis Thaliana Represent Two Evolutionarily Divergent Classes in Angiosperms. The Plant Journal 1999, 19, 9–20. [Google Scholar] [CrossRef]
- Shirley, B.W.; Kubasek, W.L.; Storz, G.; Bruggemann, E.; Koornneef, M.; Ausubel, F.M.; Goodman, H.M. Analysis of Arabidopsis Mutants Deficient in Flavonoid Biosynthesis. Plant J 1995, 8, 659–671. [Google Scholar] [CrossRef]
- Schoenbohm, C.; Martens, S.; Eder, C.; Forkmann, G.; Weisshaar, B. Identification of the Arabidopsis Thaliana Flavonoid 3’-Hydroxylase Gene and Functional Expression of the Encoded P450 Enzyme. Biol Chem 2000, 381, 749–753. [Google Scholar] [CrossRef]
- Falcone Ferreyra, M.L.; Emiliani, J.; Rodriguez, E.J.; Campos-Bermudez, V.A.; Grotewold, E.; Casati, P. The Identification of Maize and Arabidopsis Type I FLAVONE SYNTHASEs Links Flavones with Hormones and Biotic Interactions. Plant Physiol 2015, 169, 1090–1107. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E.; Chamberlin, M.; Snook, M.; Siame, B.; Butler, L.; Swenson, J.; Maddock, S.; St Clair, G.; Bowen, B. Engineering Secondary Metabolism in Maize Cells by Ectopic Expression of Transcription Factors. Plant Cell 1998, 10, 721–740. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.-Y.; Sharma, S.B.; Paiva, N.L.; Ferreira, D.; Dixon, R.A. Role of Anthocyanidin Reductase, Encoded by BANYULS in Plant Flavonoid Biosynthesis. Science 2003, 299, 396–399. [Google Scholar] [CrossRef]
- Devic, M.; Guilleminot, J.; Debeaujon, I.; Bechtold, N.; Bensaude, E.; Koornneef, M.; Pelletier, G.; Delseny, M. The BANYULS Gene Encodes a DFR-like Protein and Is a Marker of Early Seed Coat Development. Plant J 1999, 19, 387–398. [Google Scholar] [CrossRef]
- Jones, P.; Messner, B.; Nakajima, J.-I.; Schäffner, A.R.; Saito, K. UGT73C6 and UGT78D1, Glycosyltransferases Involved in Flavonol Glycoside Biosynthesis in Arabidopsis Thaliana. J Biol Chem 2003, 278, 43910–43918. [Google Scholar] [CrossRef]
- Tohge, T.; Nishiyama, Y.; Hirai, M.Y.; Yano, M.; Nakajima, J.; Awazuhara, M.; Inoue, E.; Takahashi, H.; Goodenowe, D.B.; Kitayama, M.; et al. Functional Genomics by Integrated Analysis of Metabolome and Transcriptome of Arabidopsis Plants Over-Expressing an MYB Transcription Factor. Plant J 2005, 42, 218–235. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Tohge, T.; Niida, R.; Saito, K. Identification of a Flavonol 7-O-Rhamnosyltransferase Gene Determining Flavonoid Pattern in Arabidopsis by Transcriptome Coexpression Analysis and Reverse Genetics. J Biol Chem 2007, 282, 14932–14941. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Fukushima, A.; Nakabayashi, R.; Hanada, K.; Matsuda, F.; Sugawara, S.; Inoue, E.; Kuromori, T.; Ito, T.; Shinozaki, K.; et al. Two Glycosyltransferases Involved in Anthocyanin Modification Delineated by Transcriptome Independent Component Analysis in Arabidopsis Thaliana. Plant J 2012, 69, 154–167. [Google Scholar] [CrossRef]
- Stracke, R.; Jahns, O.; Keck, M.; Tohge, T.; Niehaus, K.; Fernie, A.R.; Weisshaar, B. Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-Dependent Flavonol Glycoside Accumulation in Arabidopsis Thaliana Plants Reveals MYB11-, MYB12- and MYB111-Independent Flavonol Glycoside Accumulation. New Phytologist 2010, 188, 985–1000. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, L.; Wang, H.; Chen, X.; Wang, Y.; Yang, H.; Wei, C.; Wan, X.; Xia, T. The R2R3-MYB, bHLH, WD40, and Related Transcription Factors in Flavonoid Biosynthesis. Funct Integr Genomics 2013, 13, 75–98. [Google Scholar] [CrossRef]
- Deng, X.; Bashandy, H.; Ainasoja, M.; Kontturi, J.; Pietiäinen, M.; Laitinen, R.A.E.; Albert, V.A.; Valkonen, J.P.T.; Elomaa, P.; Teeri, T.H. Functional Diversification of Duplicated Chalcone Synthase Genes in Anthocyanin Biosynthesis of Gerbera Hybrida. New Phytologist 2014, 201, 1469–1483. [Google Scholar] [CrossRef] [PubMed]
- Schijlen, E.G.W.M.; de Vos, C.H.R.; Martens, S.; Jonker, H.H.; Rosin, F.M.; Molthoff, J.W.; Tikunov, Y.M.; Angenent, G.C.; van Tunen, A.J.; Bovy, A.G. RNA Interference Silencing of Chalcone Synthase, the First Step in the Flavonoid Biosynthesis Pathway, Leads to Parthenocarpic Tomato Fruits. Plant Physiol 2007, 144, 1520–1530. [Google Scholar] [CrossRef]
- Parage, C.; Tavares, R.; Réty, S.; Baltenweck-Guyot, R.; Poutaraud, A.; Renault, L.; Heintz, D.; Lugan, R.; Marais, G.A.B.; Aubourg, S.; et al. Structural, Functional, and Evolutionary Analysis of the Unusually Large Stilbene Synthase Gene Family in Grapevine. Plant Physiology 2012, 160, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Poutaraud, A.; Hugueney, P. Metabolism and Roles of Stilbenes in Plants. Plant Science 2009, 177, 143–155. [Google Scholar] [CrossRef]
- Wang, X.; Hu, H.; Wu, Z.; Fan, H.; Wang, G.; Chai, T.; Wang, H. Tissue-Specific Transcriptome Analyses Reveal Candidate Genes for Stilbene, Flavonoid and Anthraquinone Biosynthesis in the Medicinal Plant Polygonum Cuspidatum. BMC Genomics 2021, 22, 353. [Google Scholar] [CrossRef] [PubMed]
- Bomati, E.K.; Austin, M.B.; Bowman, M.E.; Dixon, R.A.; Noel, J.P. Structural Elucidation of Chalcone Reductase and Implications for Deoxychalcone Biosynthesis. J Biol Chem 2005, 280, 30496–30503. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Singh, R.K.; Moehninsi; Navarre, D.A. R2R3-MYB Transcription Factors, StmiR858 and Sucrose Mediate Potato Flavonol Biosynthesis. Horticulture Research 2021, 8, 25. [Google Scholar] [CrossRef]
- Casas, M.I.; Falcone-Ferreyra, M.L.; Jiang, N.; Mejía-Guerra, M.K.; Rodríguez, E.; Wilson, T.; Engelmeier, J.; Casati, P.; Grotewold, E. Identification and Characterization of Maize Salmon Silks Genes Involved in Insecticidal Maysin Biosynthesis. The Plant Cell 2016, 28, 1297–1309. [Google Scholar] [CrossRef] [PubMed]
- Alsayari, A.; Muhsinah, A.B.; Hassan, M.Z.; Ahsan, M.J.; Alshehri, J.A.; Begum, N. Aurone: A Biologically Attractive Scaffold as Anticancer Agent. European Journal of Medicinal Chemistry 2019, 166, 417–431. [Google Scholar] [CrossRef]
- Nakayama, T.; Yonekura-Sakakibara, K.; Sato, T.; Kikuchi, S.; Fukui, Y.; Fukuchi-Mizutani, M.; Ueda, T.; Nakao, M.; Tanaka, Y.; Kusumi, T.; et al. Aureusidin Synthase: A Polyphenol Oxidase Homolog Responsible for Flower Coloration. Science 2000, 290, 1163–1166. [Google Scholar] [CrossRef]
- Stich, K.; Eidenberger, T.; Wurst, F.; Forkmann, G. Flavonol Synthase Activity and the Regulation of Flavonol and Anthocyanin Biosynthesis during Flower Development in Dianthus Caryophyllus L. (Carnation). Zeitschrift für Naturforschung C 1992, 47, 553–560. [Google Scholar] [CrossRef]
- Jiang, X.; Shi, Y.; Fu, Z.; Li, W.-W.; Lai, S.; Wu, Y.; Wang, Y.; Liu, Y.; Gao, L.; Xia, T. Functional Characterization of Three Flavonol Synthase Genes from Camellia Sinensis: Roles in Flavonol Accumulation. Plant Science 2020, 300, 110632. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, D.-H.; Yang, J.-H.; Lee, J.-Y.; Lim, S.-H. Increased Flavonol Levels in Tobacco Expressing AcFLS Affect Flower Color and Root Growth. International Journal of Molecular Sciences 2020, 21, 1011. [Google Scholar] [CrossRef]
- Cao, Y.; Xing, M.; Xu, C.; Li, X. Biosynthesis of Flavonol and Its Regulation in Plants. Acta Horticulturae Sinica 2018, 45, 177–192. [Google Scholar] [CrossRef]
- Yun, C.-S.; Yamamoto, T.; Nozawa, A.; Tozawa, Y. Expression of Parsley Flavone Synthase I Establishes the Flavone Biosynthetic Pathway in Arabidopsis Thaliana. Biosci Biotechnol Biochem 2008, 72, 968–973. [Google Scholar] [CrossRef]
- Wei, S.; Zhang, W.; Fu, R.; Zhang, Y. Genome-Wide Characterization of 2-Oxoglutarate and Fe(II)-Dependent Dioxygenase Family Genes in Tomato during Growth Cycle and Their Roles in Metabolism. BMC Genomics 2021, 22, 126. [Google Scholar] [CrossRef]
- Li, H.; Li, D.; Yang, Z.; Zeng, Q.; Luo, Y.; He, N. Flavones Produced by Mulberry Flavone Synthase Type I Constitute a Defense Line against the Ultraviolet-B Stress. Plants (Basel) 2020, 9, 215. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Yang, J.; Cui, M.-Y.; Liu, J.; Fang, Y.; Yan, M.; Qiu, W.; Shang, H.; Xu, Z.; Yidiresi, R.; et al. The Reference Genome Sequence of Scutellaria Baicalensis Provides Insights into the Evolution of Wogonin Biosynthesis. Molecular Plant 2019, 12, 935–950. [Google Scholar] [CrossRef]
- Winefield, C.S.; Lewis, D.H.; Swinny, E.E.; Zhang, H.; Arathoon, H.S.; Fischer, T.C.; Halbwirth, H.; Stich, K.; Gosch, C.; Forkmann, G.; et al. Investigation of the Biosynthesis of 3-Deoxyanthocyanins in Sinningia Cardinalis. Physiologia Plantarum 2005, 124, 419–430. [Google Scholar] [CrossRef]
- Bruce, W.; Folkerts, O.; Garnaat, C.; Crasta, O.; Roth, B.; Bowen, B. Expression Profiling of the Maize Flavonoid Pathway Genes Controlled by Estradiol-Inducible Transcription Factors CRC and P. The Plant Cell 2000, 12, 65–79. [Google Scholar] [CrossRef]
- Styles, E.D.; Ceska, O. Genetic Control of 3-Hydroxy- and 3-Deoxy- Flavonoids in Zea Mays. Phytochemistry 1975, 14, 413–415. [Google Scholar] [CrossRef]
- Schijlen, E.G.W.M.; Ric de Vos, C.H.; van Tunen, A.J.; Bovy, A.G. Modification of Flavonoid Biosynthesis in Crop Plants. Phytochemistry 2004, 65, 2631–2648. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, S.; Lee, E.; Walker, A.R.; Tanner, G.J.; Larkin, P.J.; Ashton, A.R. The Arabidopsis TDS4 Gene Encodes Leucoanthocyanidin Dioxygenase (LDOX) and Is Essential for Proanthocyanidin Synthesis and Vacuole Development. Plant J 2003, 35, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.-M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and Biochemistry of Seed Flavonoids. Annu Rev Plant Biol 2006, 57, 405–430. [Google Scholar] [CrossRef]
- Tanner, G.J.; Francki, K.T.; Abrahams, S.; Watson, J.M.; Larkin, P.J.; Ashton, A.R. Proanthocyanidin Biosynthesis in Plants. Purification of Legume Leucoanthocyanidin Reductase and Molecular Cloning of Its cDNA. J Biol Chem 2003, 278, 31647–31656. [Google Scholar] [CrossRef]
- Bogs, J.; Downey, M.O.; Harvey, J.S.; Ashton, A.R.; Tanner, G.J.; Robinson, S.P. Proanthocyanidin Synthesis and Expression of Genes Encoding Leucoanthocyanidin Reductase and Anthocyanidin Reductase in Developing Grape Berries and Grapevine Leaves. Plant Physiol 2005, 139, 652–663. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Wang, L.; Han, Z.; Jiang, Y.; Zhao, L.; Liu, H.; Yang, L.; Luo, K. Molecular Cloning and Characterization of PtrLAR3, a Gene Encoding Leucoanthocyanidin Reductase from Populus Trichocarpa, and Its Constitutive Expression Enhances Fungal Resistance in Transgenic Plants. J Exp Bot 2012, 63, 2513–2524. [Google Scholar] [CrossRef]
- Grotewold, E. The Genetics and Biochemistry of Floral Pigments. Annu Rev Plant Biol 2006, 57, 761–780. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, H.; Wu, R. Recent Advances on Blue Flower Formation. Chinese Bulletin of Botany 2020, 55, 216. [Google Scholar] [CrossRef]
- Yamada, Y.; Sato, F. Chapter Eight - Transcription Factors in Alkaloid Biosynthesis. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Academic Press, 2013; Vol. 305, pp. 339–382. [Google Scholar]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB Transcription Factors in Arabidopsis. Trends Plant Sci 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Meraj, T.A.; Fu, J.; Raza, M.A.; Zhu, C.; Shen, Q.; Xu, D.; Wang, Q. Transcriptional Factors Regulate Plant Stress Responses Through Mediating Secondary Metabolism. Genes 2020, 11, 346. [Google Scholar] [CrossRef]
- Cao, Y.; Jia, H.; Xing, M.; Jin, R.; Grierson, D.; Gao, Z.; Sun, C.; Chen, K.; Xu, C.; Li, X. Genome-Wide Analysis of MYB Gene Family in Chinese Bayberry (Morella Rubra) and Identification of Members Regulating Flavonoid Biosynthesis. Front Plant Sci 2021, 12, 691384. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential Regulation of Closely Related R2R3-MYB Transcription Factors Controls Flavonol Accumulation in Different Parts of the Arabidopsis Thaliana Seedling. The Plant Journal 2007, 50, 660–677. [Google Scholar] [CrossRef]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as Antioxidants in Plants: Location and Functional Significance. Plant Science 2012, 196, 67–76. [Google Scholar] [CrossRef]
- Lien, E.J.; Ren, S.; Bui, H.H.; Wang, R. Quantitative Structure-Activity Relationship Analysis of Phenolic Antioxidants. Free Radic Biol Med 1999, 26, 285–294. [Google Scholar] [CrossRef]
- Bors, W.; Saran, M. Radical Scavenging by Flavonoid Antioxidants. Free Radic Res Commun 1987, 2, 289–294. [Google Scholar] [CrossRef]
- Jovanovic, S.V.; Steenken, S.; Hara, Y.; Simic, M.G. Reduction Potentials of Flavonoid and Model Phenoxyl Radicals. Which Ring in Flavonoids Is Responsible for Antioxidant Activity? J. Chem. Soc., Perkin Trans. 2 1996, 2497–2504. [Google Scholar] [CrossRef]
- Sekher Pannala, A.; Chan, T.S.; O’Brien, P.J.; Rice-Evans, C.A. Flavonoid B-Ring Chemistry and Antioxidant Activity: Fast Reaction Kinetics. Biochemical and Biophysical Research Communications 2001, 282, 1161–1168. [Google Scholar] [CrossRef]
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-Antioxidant Activity Relationships of Flavonoids and Phenolic Acids. Free Radic Biol Med 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Nagai, S.; Ohara, K.; Mukai, K. Kinetic Study of the Quenching Reaction of Singlet Oxygen by Flavonoids in Ethanol Solution. J Phys Chem B 2005, 109, 4234–4240. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of Oxidative and Drought Tolerance in Arabidopsis by Overaccumulation of Antioxidant Flavonoids. Plant J 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Wolfenden, B.S.; Willson, R.L. Radical-Cations as Reference Chromogens in Kinetic Studies of Ono-Electron Transfer Reactions: Pulse Radiolysis Studies of 2,2′-Azinobis-(3-Ethylbenzthiazoline-6-Sulphonate). J. Chem. Soc., Perkin Trans. 2 1982, 805–812. [Google Scholar] [CrossRef]
- Agati, G.; Matteini, P.; Goti, A.; Tattini, M. Chloroplast-Located Flavonoids Can Scavenge Singlet Oxygen. New Phytol 2007, 174, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Pandey, A.K. Chemistry and Biological Activities of Flavonoids: An Overview. The Scientific World Journal 2013, 2013, e162750. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine; Oxford University Press, 2015; ISBN 978-0-19-871748-5. [Google Scholar]
- Colliver, S.; Bovy, A.; Collins, G.; Muir, S.; Robinson, S.; de Vos, C.H.R.; Verhoeyen, M.E. Improving the Nutritional Content of Tomatoes through Reprogramming Their Flavonoid Biosynthetic Pathway. Phytochemistry Reviews 2002, 1, 113–123. [Google Scholar] [CrossRef]
- Lukaszewicz, M.; Matysiak-Kata, I.; Skala, J.; Fecka, I.; Cisowski, W.; Szopa, J. Antioxidant Capacity Manipulation in Transgenic Potato Tuber by Changes in Phenolic Compounds Content. J Agric Food Chem 2004, 52, 1526–1533. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, L.-J.; Wang, Y.; Liu, S.; Geng, Z.; Song, A.; Jiang, J.; Chen, S.; Chen, F. Functional Identification of a Flavone Synthase and a Flavonol Synthase Genes Affecting Flower Color Formation in Chrysanthemum Morifolium. Plant Physiol Biochem 2021, 166, 1109–1120. [Google Scholar] [CrossRef]
- Wang, M.; Ren, T.; Huang, R.; Li, Y.; Zhang, C.; Xu, Z. Overexpression of an Apocynum Venetum Flavonols Synthetase Gene Confers Salinity Stress Tolerance to Transgenic Tobacco Plants. Plant Physiol Biochem 2021, 162, 667–676. [Google Scholar] [CrossRef]
- Wang, M.; Qin, L.; Xie, C.; Li, W.; Yuan, J.; Kong, L.; Yu, W.; Xia, G.; Liu, S. Induced and Constitutive DNA Methylation in a Salinity-Tolerant Wheat Introgression Line. Plant and Cell Physiology 2014, 55, 1354–1365. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Kim, J.H.; Kwon, J.; Jeong, C.Y.; Lee, W.; Lee, D.; Hong, S.-W.; Lee, H. Characterization of Arabidopsis Thaliana FLAVONOL SYNTHASE 1 (FLS1) -Overexpression Plants in Response to Abiotic Stress. Plant Physiology and Biochemistry 2016, 103, 133–142. [Google Scholar] [CrossRef]
- Kumar, V.; Nadda, G.; Kumar, S.; Yadav, S.K. Transgenic Tobacco Overexpressing Tea cDNA Encoding Dihydroflavonol 4-Reductase and Anthocyanidin Reductase Induces Early Flowering and Provides Biotic Stress Tolerance. PLOS ONE 2013, 8, e65535. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, S.; Wang, T.; Liu, H.; Xu, X.; Chen, K.; Zhang, P. The Moss Flavone Synthase I Positively Regulates the Tolerance of Plants to Drought Stress and UV-B Radiation. Plant Sci 2020, 298, 110591. [Google Scholar] [CrossRef] [PubMed]
- Mehrtens, F.; Kranz, H.; Bednarek, P.; Weisshaar, B. The Arabidopsis Transcription Factor MYB12 Is a Flavonol-Specific Regulator of Phenylpropanoid Biosynthesis. Plant Physiol 2005, 138, 1083–1096. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Kong, W.; Wong, G.; Fu, L.; Peng, R.; Li, Z.; Yao, Q. AtMYB12 Regulates Flavonoids Accumulation and Abiotic Stress Tolerance in Transgenic Arabidopsis Thaliana. Mol Genet Genomics 2016, 291, 1545–1559. [Google Scholar] [CrossRef] [PubMed]
- Misra, P.; Pandey, A.; Tiwari, M.; Chandrashekar, K.; Sidhu, O.P.; Asif, M.H.; Chakrabarty, D.; Singh, P.K.; Trivedi, P.K.; Nath, P.; et al. Modulation of Transcriptome and Metabolome of Tobacco by Arabidopsis Transcription Factor, AtMYB12, Leads to Insect Resistance. Plant Physiol 2010, 152, 2258–2268. [Google Scholar] [CrossRef] [PubMed]
- Mitsunami, T.; Nishihara, M.; Galis, I.; Alamgir, K.M.; Hojo, Y.; Fujita, K.; Sasaki, N.; Nemoto, K.; Sawasaki, T.; Arimura, G. Overexpression of the PAP1 Transcription Factor Reveals a Complex Regulation of Flavonoid and Phenylpropanoid Metabolism in Nicotiana Tabacum Plants Attacked by Spodoptera Litura. PLoS One 2014, 9, e108849. [Google Scholar] [CrossRef]
- Bovy, A.; de Vos, R.; Kemper, M.; Schijlen, E.; Almenar Pertejo, M.; Muir, S.; Collins, G.; Robinson, S.; Verhoeyen, M.; Hughes, S.; et al. High-Flavonol Tomatoes Resulting from the Heterologous Expression of the Maize Transcription Factor Genes LC and C1. Plant Cell 2002, 14, 2509–2526. [Google Scholar] [CrossRef]
- Khusnutdinov, E.; Sukhareva, A.; Panfilova, M.; Mikhaylova, E. Anthocyanin Biosynthesis Genes as Model Genes for Genome Editing in Plants. Int J Mol Sci 2021, 22, 8752. [Google Scholar] [CrossRef]
- Mackon, E.; Jeazet Dongho Epse Mackon, G.C.; Guo, Y.; Ma, Y.; Yao, Y.; Liu, P. Development and Application of CRISPR/Cas9 to Improve Anthocyanin Pigmentation in Plants: Opportunities and Perspectives. Plant Sci 2023, 333, 111746. [Google Scholar] [CrossRef]
- Castillejo, C.; Waurich, V.; Wagner, H.; Ramos, R.; Oiza, N.; Muñoz, P.; Triviño, J.C.; Caruana, J.; Liu, Z.; Cobo, N.; et al. Allelic Variation of MYB10 Is the Major Force Controlling Natural Variation in Skin and Flesh Color in Strawberry (Fragaria Spp.) Fruit. Plant Cell 2020, 32, 3723–3749. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, G.L.; Jones, M.A.; Smirnoff, N. The Biosynthetic Pathway of Vitamin C in Higher Plants. Nature 1998, 393, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Wolucka, B.A.; Van Montagu, M. GDP-Mannose 3’,5’-Epimerase Forms GDP-L-Gulose, a Putative Intermediate for the de Novo Biosynthesis of Vitamin C in Plants. J Biol Chem 2003, 278, 47483–47490. [Google Scholar] [CrossRef] [PubMed]
- Lorence, A.; Chevone, B.I.; Mendes, P.; Nessler, C.L. Myo-Inositol Oxygenase Offers a Possible Entry Point into Plant Ascorbate Biosynthesis. Plant Physiol 2004, 134, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Agius, F.; González-Lamothe, R.; Caballero, J.L.; Muñoz-Blanco, J.; Botella, M.A.; Valpuesta, V. Engineering Increased Vitamin C Levels in Plants by Overexpression of a D-Galacturonic Acid Reductase. Nat Biotechnol 2003, 21, 177–181. [Google Scholar] [CrossRef]
- Smirnoff, N. Ascorbic Acid: Metabolism and Functions of a Multi-Facetted Molecule. Curr Opin Plant Biol 2000, 3, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Tyapkina, D.Y.; Kochieva, E.Z.; Slugina, M.A. Vitamin C in Fleshy Fruits: Biosynthesis, Recycling, Genes, and Enzymes. Vavilov Journal of Genetics and Breeding 2019, 23, 270–280. [Google Scholar] [CrossRef]
- Maruta, T.; Yonemitsu, M.; Yabuta, Y.; Tamoi, M.; Ishikawa, T.; Shigeoka, S. Arabidopsis Phosphomannose Isomerase 1, but Not Phosphomannose Isomerase 2, Is Essential for Ascorbic Acid Biosynthesis. J Biol Chem 2008, 283, 28842–28851. [Google Scholar] [CrossRef]
- Badejo, A.A.; Jeong, S.T.; Goto-Yamamoto, N.; Esaka, M. Cloning and Expression of GDP-d-Mannose Pyrophosphorylase Gene and Ascorbic Acid Content of Acerola (Malpighia Glabra L.) Fruit at Ripening Stages. Plant Physiology and Biochemistry 2007, 45, 665–672. [Google Scholar] [CrossRef]
- Badejo, A.A.; Tanaka, N.; Esaka, M. Analysis of GDP-d-Mannose Pyrophosphorylase Gene Promoter from Acerola (Malpighia Glabra) and Increase in Ascorbate Content of Transgenic Tobacco Expressing the Acerola Gene. Plant and Cell Physiology 2008, 49, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Badejo, A.A.; Fujikawa, Y.; Esaka, M. Gene Expression of Ascorbic Acid Biosynthesis Related Enzymes of the Smirnoff-Wheeler Pathway in Acerola (Malpighia Glabra). J Plant Physiol 2009, 166, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, J.; Park, S.W. Role of L-Ascorbate in Alleviating Abiotic Stresses in Crop Plants. Bot Stud 2014, 55, 38. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Yu, C.; Qin, H.; Liu, X.; Zhang, A.; Johansen, I.E.; Wang, D. Molecular and Functional Analysis of Phosphomannomutase (PMM) from Higher Plants and Genetic Evidence for the Involvement of PMM in Ascorbic Acid Biosynthesis in Arabidopsis and Nicotiana Benthamiana. Plant J 2007, 49, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Hoeberichts, F.A.; Vaeck, E.; Kiddle, G.; Coppens, E.; van de Cotte, B.; Adamantidis, A.; Ormenese, S.; Foyer, C.H.; Zabeau, M.; Inzé, D.; et al. A Temperature-Sensitive Mutation in the Arabidopsis Thaliana Phosphomannomutase Gene Disrupts Protein Glycosylation and Triggers Cell Death. J Biol Chem 2008, 283, 5708–5718. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N.; Conklin, P.L.; Loewus, F.A. BIOSYNTHESIS OF ASCORBIC ACID IN PLANTS: A Renaissance. Annu Rev Plant Physiol Plant Mol Biol 2001, 52, 437–467. [Google Scholar] [CrossRef]
- Fenech, M.; Amaya, I.; Valpuesta, V.; Botella, M.A. Vitamin C Content in Fruits: Biosynthesis and Regulation. Front Plant Sci 2018, 9, 2006. [Google Scholar] [CrossRef]
- Maruta, T.; Ichikawa, Y.; Mieda, T.; Takeda, T.; Tamoi, M.; Yabuta, Y.; Ishikawa, T.; Shigeoka, S. The Contribution of Arabidopsis Homologs of L-Gulono-1,4-Lactone Oxidase to the Biosynthesis of Ascorbic Acid. Biosci Biotechnol Biochem 2010, 74, 1494–1497. [Google Scholar] [CrossRef]
- Dowdle, J.; Ishikawa, T.; Gatzek, S.; Rolinski, S.; Smirnoff, N. Two Genes in Arabidopsis Thaliana Encoding GDP-L-Galactose Phosphorylase Are Required for Ascorbate Biosynthesis and Seedling Viability. Plant J 2007, 52, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Conklin, P.L.; Gatzek, S.; Wheeler, G.L.; Dowdle, J.; Raymond, M.J.; Rolinski, S.; Isupov, M.; Littlechild, J.A.; Smirnoff, N. Arabidopsis Thaliana VTC4 Encodes L-Galactose-1-P Phosphatase, a Plant Ascorbic Acid Biosynthetic Enzyme. J Biol Chem 2006, 281, 15662–15670. [Google Scholar] [CrossRef]
- Torabinejad, J.; Donahue, J.L.; Gunesekera, B.N.; Allen-Daniels, M.J.; Gillaspy, G.E. VTC4 Is a Bifunctional Enzyme That Affects Myoinositol and Ascorbate Biosynthesis in Plants. Plant Physiol 2009, 150, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gruszewski, H.A.; Chevone, B.I.; Nessler, C.L. An Arabidopsis Purple Acid Phosphatase with Phytase Activity Increases Foliar Ascorbate. Plant Physiol 2008, 146, 431–440. [Google Scholar] [CrossRef]
- Leferink, N.G.H.; van den Berg, W.A.M.; van Berkel, W.J.H. L-Galactono-Gamma-Lactone Dehydrogenase from Arabidopsis Thaliana, a Flavoprotein Involved in Vitamin C Biosynthesis. FEBS J 2008, 275, 713–726. [Google Scholar] [CrossRef]
- Louvet, R.; Cavel, E.; Gutierrez, L.; Guénin, S.; Roger, D.; Gillet, F.; Guerineau, F.; Pelloux, J. Comprehensive Expression Profiling of the Pectin Methylesterase Gene Family during Silique Development in Arabidopsis Thaliana. Planta 2006, 224, 782–791. [Google Scholar] [CrossRef]
- Zhang, W.; Lorence, A.; Gruszewski, H.A.; Chevone, B.I.; Nessler, C.L. AMR1, an Arabidopsis Gene That Coordinately and Negatively Regulates the Mannose/l-Galactose Ascorbic Acid Biosynthetic Pathway. Plant Physiology 2009, 150, 942–950. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Zhang, R.; Huang, R. The Ethylene Response Factor AtERF98 Enhances Tolerance to Salt through the Transcriptional Activation of Ascorbic Acid Synthesis in Arabidopsis. Plant J 2012, 71, 273–287. [Google Scholar] [CrossRef]
- Roberts, R.M. The Metabolism of D-Mannose- 14 C to Polysaccharide in Corn Roots. Specific Labeling of L-Galactose, D-Mannose, and L-Fucose. Arch Biochem Biophys 1971, 145, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Baydoun, E.A.-H.; Fry, S.C. [2-3H]Mannose Incorporation in Cultured Plant Cells: Investigation of L-Galactose Residues of the Primary Cell Wall. Journal of Plant Physiology 1988, 132, 484–490. [Google Scholar] [CrossRef]
- Bulley, S.; Laing, W. The Regulation of Ascorbate Biosynthesis. Curr Opin Plant Biol 2016, 33, 15–22. [Google Scholar] [CrossRef]
- Hancock, R.D.; Viola, R. Biosynthesis and Catabolism of L-Ascorbic Acid in Plants. Critical Reviews in Plant Sciences 2005, 24, 167–188. [Google Scholar] [CrossRef]
- Laing, W.A.; Bulley, S.; Wright, M.; Cooney, J.; Jensen, D.; Barraclough, D.; MacRae, E. A Highly Specific L-Galactose-1-Phosphate Phosphatase on the Path to Ascorbate Biosynthesis. Proc Natl Acad Sci U S A 2004, 101, 16976–16981. [Google Scholar] [CrossRef]
- Gatzek, S.; Wheeler, G.L.; Smirnoff, N. Antisense Suppression of L-Galactose Dehydrogenase in Arabidopsis Thaliana Provides Evidence for Its Role in Ascorbate Synthesis and Reveals Light Modulated l-Galactose Synthesis. Plant J 2002, 30, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.H.; Mittova, V.; Kiddle, G.; Heazlewood, J.L.; Bartoli, C.G.; Theodoulou, F.L.; Foyer, C.H. Control of Ascorbate Synthesis by Respiration and Its Implications for Stress Responses. Plant Physiol 2003, 133, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Schertl, P.; Sunderhaus, S.; Klodmann, J.; Grozeff, G.E.G.; Bartoli, C.G.; Braun, H.-P. L-Galactono-1,4-Lactone Dehydrogenase (GLDH) Forms Part of Three Subcomplexes of Mitochondrial Complex I in Arabidopsis Thaliana. J Biol Chem 2012, 287, 14412–14419. [Google Scholar] [CrossRef]
- Pineau, B.; Layoune, O.; Danon, A.; De Paepe, R. L-Galactono-1,4-Lactone Dehydrogenase Is Required for the Accumulation of Plant Respiratory Complex I. J Biol Chem 2008, 283, 32500–32505. [Google Scholar] [CrossRef]
- Davey, M.W.; Gilot, C.; Persiau, G.; Østergaard, J.; Han, Y.; Bauw, G.C.; Van Montagu, M.C. Ascorbate Biosynthesis in Arabidopsis Cell Suspension Culture. Plant Physiology 1999, 121, 535–544. [Google Scholar] [CrossRef]
- Loewus, F.A.; Kelly, S. The Metabolism of P-Galacturonic Acid and Its Methyl Ester in the Detached Ripening Strawberry. Arch Biochem Biophys 1961, 95, 483–493. [Google Scholar] [CrossRef]
- Smirnoff, N. Chapter 4 - Vitamin C: The Metabolism and Functions of Ascorbic Acid in Plants. In Advances in Botanical Research; Rébeillé, F., Douce, R., Eds.; Biosynthesis of Vitamins in Plants Part B; Academic Press, 2011; Vol. 59, pp. 107–177. [Google Scholar]
- Akram, N.A.; Shafiq, F.; Ashraf, M. Ascorbic Acid-A Potential Oxidant Scavenger and Its Role in Plant Development and Abiotic Stress Tolerance. Front Plant Sci 2017, 8, 613. [Google Scholar] [CrossRef]
- Jain, A.K.; Nessler, C.L. Metabolic Engineering of an Alternative Pathway for Ascorbic Acid Biosynthesis in Plants. Molecular Breeding 2000, 6, 73–78. [Google Scholar] [CrossRef]
- Endres, S.; Tenhaken, R. Myoinositol Oxygenase Controls the Level of Myoinositol in Arabidopsis, but Does Not Increase Ascorbic Acid. Plant Physiol 2009, 149, 1042–1049. [Google Scholar] [CrossRef]
- Tóth, S.Z.; Nagy, V.; Puthur, J.T.; Kovács, L.; Garab, G. The Physiological Role of Ascorbate as Photosystem II Electron Donor: Protection against Photoinactivation in Heat-Stressed Leaves. Plant Physiology 2011, 156, 382–392. [Google Scholar] [CrossRef]
- Nepal, N.; Yactayo-Chang, J.P.; Medina-Jiménez, K.; Acosta-Gamboa, L.M.; González-Romero, M.E.; Arteaga-Vázquez, M.A.; Lorence, A. Mechanisms Underlying the Enhanced Biomass and Abiotic Stress Tolerance Phenotype of an Arabidopsis MIOX Over-Expresser. Plant Direct 2019, 3, e00165. [Google Scholar] [CrossRef]
- Zechmann, B.; Müller, M. Subcellular Compartmentation of Glutathione in Dicotyledonous Plants. Protoplasma 2010, 246, 15–24. [Google Scholar] [CrossRef]
- Zechmann, B. Compartment-Specific Importance of Glutathione during Abiotic and Biotic Stress. Front Plant Sci 2014, 5, 566. [Google Scholar] [CrossRef] [PubMed]
- Hell, R.; Bergmann, L. λ-Glutamylcysteine Synthetase in Higher Plants: Catalytic Properties and Subcellular Localization. Planta 1990, 180, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.; Oliver, D.J. Glutathione Metabolic Genes Coordinately Respond to Heavy Metals and Jasmonic Acid in Arabidopsis. Plant Cell 1998, 10, 1539–1550. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, Q.; Ahmad, R.; Kwak, S.-S.; Rashid, A.; Anjum, N. Ascorbate and Glutathione: Protectors of Plants in Oxidative Stress. In Ascorbate-Glutathione Pathway and Stress Tolerance in Plants; 1970; pp. 209–229. ISBN 978-90-481-9403-2. [Google Scholar]
- Richman, P.G.; Meister, A. Regulation of Gamma-Glutamyl-Cysteine Synthetase by Nonallosteric Feedback Inhibition by Glutathione. J Biol Chem 1975, 250, 1422–1426. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Foyer, C.H. ASCORBATE AND GLUTATHIONE: Keeping Active Oxygen Under Control. Annu Rev Plant Physiol Plant Mol Biol 1998, 49, 249–279. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.J. The Integration of Glutathione Homeostasis and Redox Signaling. J Plant Physiol 2008, 165, 1390–1403. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, A.; Hernandez, J.A.; Pastori, G.; del Rio LA, null; Sevilla, F. Role of the Ascorbate-Glutathione Cycle of Mitochondria and Peroxisomes in the Senescence of Pea Leaves. Plant Physiol 1998, 118, 1327–1335. [Google Scholar] [CrossRef] [PubMed]
- Asada, K. The Water-Water Cycle as Alternative Photon and Electron Sinks. Philos Trans R Soc Lond B Biol Sci 2000, 355, 1419–1431. [Google Scholar] [CrossRef] [PubMed]
- Miyake, C.; Asada, K. Thylakoid-Bound Ascorbate Peroxidase in Spinach Chloroplasts and Photoreduction of Its Primary Oxidation Product Monodehydroascorbate Radicals in Thylakoids. Plant and Cell Physiology 1992, 33, 541–553. [Google Scholar] [CrossRef]
- Kaiser, W. The Effect of Hydrogen Peroxide on CO2 Fixation of Isolated Intact Chloroplasts. Biochimica et Biophysica Acta (BBA) - Bioenergetics 1976, 440, 476–482. [Google Scholar] [CrossRef]
- Ivanov, B.; Kozuleva, M.; Mubarakshina, M.; Ivanov, B.; Kozuleva, M.; Mubarakshina, M. Oxygen Metabolism in Chloroplast. In Cell Metabolism - Cell Homeostasis and Stress Response; IntechOpen, 2012; ISBN 978-953-307-978-3. [Google Scholar]
- Gotoh, N.; Niki, E. Rates of Interactions of Superoxide with Vitamin E, Vitamin C and Related Compounds as Measured by Chemiluminescence. Biochimica et Biophysica Acta (BBA) - General Subjects 1992, 1115, 201–207. [Google Scholar] [CrossRef]
- Winterbourn, C.C.; Metodiewa, D. The Reaction of Superoxide with Reduced Glutathione. Archives of Biochemistry and Biophysics 1994, 314, 284–290. [Google Scholar] [CrossRef]
- Allen, J.F. A Two-Step Mechanism for the Photosynthetic Reduction of Oxygen by Ferredoxin. Biochemical and Biophysical Research Communications 1975, 66, 36–43. [Google Scholar] [CrossRef]
- Kramarenko, G.G.; Hummel, S.G.; Martin, S.M.; Buettner, G.R. Ascorbate Reacts with Singlet Oxygen to Produce Hydrogen Peroxide. phot 2006, 82, 1634–1637. [Google Scholar] [CrossRef]
- Devasagayam, T.P.A.; Sundquist, A.R.; Di Mascio, P.; Kaiser, S.; Sies, H. Activity of Thiols as Singlet Molecular Oxygen Quenchers. Journal of Photochemistry and Photobiology B: Biology 1991, 9, 105–116. [Google Scholar] [CrossRef]
- Jain, P.; Bhatla, S.C. Signaling Role of Phospholipid Hydroperoxide Glutathione Peroxidase (PHGPX) Accompanying Sensing of NaCl Stress in Etiolated Sunflower Seedling Cotyledons. Plant Signaling & Behavior 2014, 9, e977746. [Google Scholar] [CrossRef]
- Broad, R.C.; Bonneau, J.P.; Hellens, R.P.; Johnson, A.A.T. Manipulation of Ascorbate Biosynthetic, Recycling, and Regulatory Pathways for Improved Abiotic Stress Tolerance in Plants. Int J Mol Sci 2020, 21, 1790. [Google Scholar] [CrossRef]
- Eltelib, H.A.; Fujikawa, Y.; Esaka, M. Overexpression of the Acerola (Malpighia Glabra) Monodehydroascorbate Reductase Gene in Transgenic Tobacco Plants Results in Increased Ascorbate Levels and Enhanced Tolerance to Salt Stress. South African Journal of Botany 2012, 78, 295–301. [Google Scholar] [CrossRef]
- Shin, S.-Y.; Kim, M.-H.; Kim, Y.-H.; Park, H.-M.; Yoon, H.-S. Co-Expression of Monodehydroascorbate Reductase and Dehydroascorbate Reductase from Brassica Rapa Effectively Confers Tolerance to Freezing-Induced Oxidative Stress. Mol Cells 2013, 36, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Souriau, N.; Perret, S.; Lelandais, M.; Kunert, K.J.; Pruvost, C.; Jouanin, L. Overexpression of Glutathione Reductase but Not Glutathione Synthetase Leads to Increases in Antioxidant Capacity and Resistance to Photoinhibition in Poplar Trees. Plant Physiol 1995, 109, 1047–1057. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Liang, Y.; Zeng, S.; Yan, J.; Li, K.; Xu, H. Overexpression of Tomato Glutathione Reductase (SlGR) in Transgenic Tobacco Enhances Salt Tolerance Involving the S-Nitrosylation of GR. Plant Physiol Biochem 2023, 196, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Mano, J.; Tanaka, K.; Wang, S.; Zhang, M.; Deng, X.; Zhang, S. High Level of Reduced Glutathione Contributes to Detoxification of Lipid Peroxide-Derived Reactive Carbonyl Species in Transgenic Arabidopsis Overexpressing Glutathione Reductase under Aluminum Stress. Physiol Plant 2017, 161, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Raja, V.; Wani, U.M.; Wani, Z.A.; Jan, N.; Kottakota, C.; Reddy, M.K.; Kaul, T.; John, R. Pyramiding Ascorbate-Glutathione Pathway in Lycopersicum Esculentum Confers Tolerance to Drought and Salinity Stress. Plant Cell Rep 2022, 41, 619–637. [Google Scholar] [CrossRef] [PubMed]
- Bashir, S.; Jan, N.; Wani, U.M.; Raja, V.; John, R. Co-over Expression of Ascorbate Glutathione Pathway Enzymes Improve Mercury Tolerance in Tomato. Plant Physiol Biochem 2022, 186, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Borgohain, P.; Saha, B.; Agrahari, R.; Chowardhara, B.; Sahoo, S.; van der Vyver, C.; Panda, S.K. SlNAC2 Overexpression in Arabidopsis Results in Enhanced Abiotic Stress Tolerance with Alteration in Glutathione Metabolism. Protoplasma 2019, 256, 1065–1077. [Google Scholar] [CrossRef] [PubMed]
- Liang Zhu Y, null; Pilon-Smits, E.A.; Jouanin, L.; Terry, N. Overexpression of Glutathione Synthetase in Indian Mustard Enhances Cadmium Accumulation and Tolerance. Plant Physiol 1999, 119, 73–80. [Google Scholar] [CrossRef]
- Yang, Y.; Li, J.; Li, H.; Ding, Y.; Wu, W.; Qin, R.; Ni, J.; Xu, R.; Wei, P.; Yang, J. OsGSTU5 and OsGSTU37 Encoding Glutathione Reductases Are Required for Cadmium Tolerance in Rice. Int. J. Environ. Sci. Technol. 2023, 20, 10253–10260. [Google Scholar] [CrossRef]
- Qi, Y.C.; Liu, W.Q.; Qiu, L.Y.; Zhang, S.M.; Ma, L.; Zhang, H. Overexpression of Glutathione S-Transferase Gene Increases Salt Tolerance of Arabidopsis. Russ J Plant Physiol 2010, 57, 233–240. [Google Scholar] [CrossRef]
- Roxas, V.P.; Smith, R.K.; Allen, E.R.; Allen, R.D. Overexpression of Glutathione S-Transferase/Glutathione Peroxidase Enhances the Growth of Transgenic Tobacco Seedlings during Stress. Nat Biotechnol 1997, 15, 988–991. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wu, M.; Teng, Y.; Jia, S.; Yu, D.; Wei, T.; Chen, C.; Song, W. Overexpression of the Glutathione Peroxidase 5 (RcGPX5) Gene From Rhodiola Crenulata Increases Drought Tolerance in Salvia Miltiorrhiza. Frontiers in Plant Science 2019, 9. [Google Scholar] [CrossRef]
- Yoshimura, K.; Miyao, K.; Gaber, A.; Takeda, T.; Kanaboshi, H.; Miyasaka, H.; Shigeoka, S. Enhancement of Stress Tolerance in Transgenic Tobacco Plants Overexpressing Chlamydomonas Glutathione Peroxidase in Chloroplasts or Cytosol. Plant J 2004, 37, 21–33. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Bela, K.; Horváth, E.; Rigó, G.; Gallé, Á.; Szabados, L.; Fehér, A.; Csiszár, J. Overexpression of the Arabidopsis Glutathione Peroxidase-like 5 Gene (AtGPXL5) Resulted in Altered Plant Development and Redox Status. Environmental and Experimental Botany 2019, 167, 103849. [Google Scholar] [CrossRef]
- Tasaki, K.; Yoshida, M.; Nakajima, M.; Higuchi, A.; Watanabe, A.; Nishihara, M. Molecular Characterization of an Anthocyanin-Related Glutathione S-Transferase Gene in Japanese Gentian with the CRISPR/Cas9 System. BMC Plant Biol 2020, 20, 370. [Google Scholar] [CrossRef]
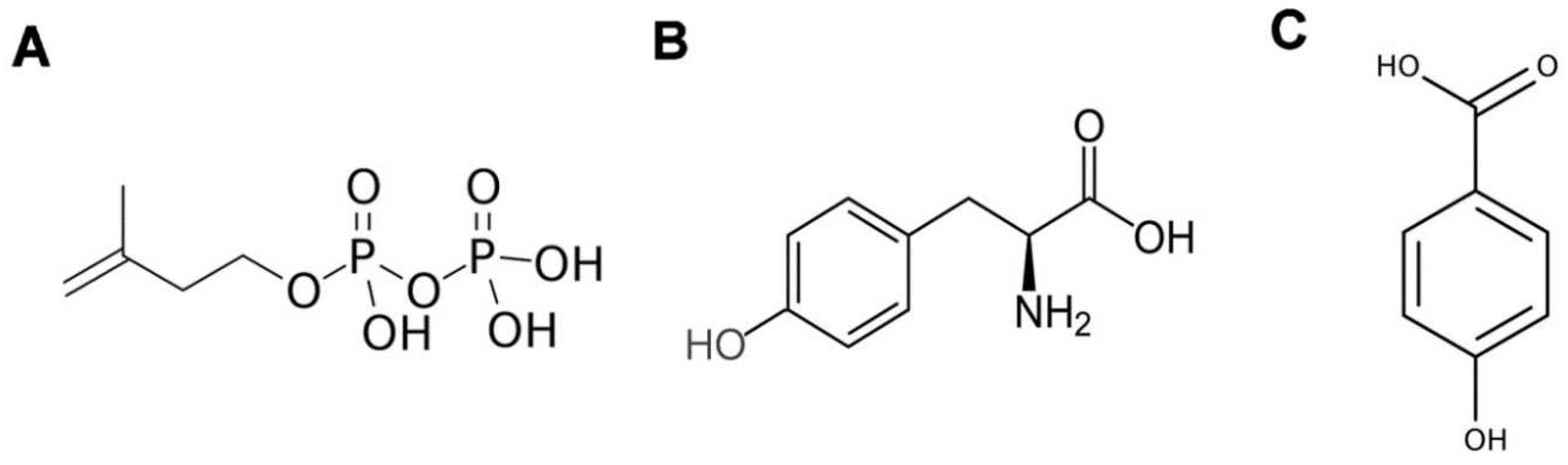
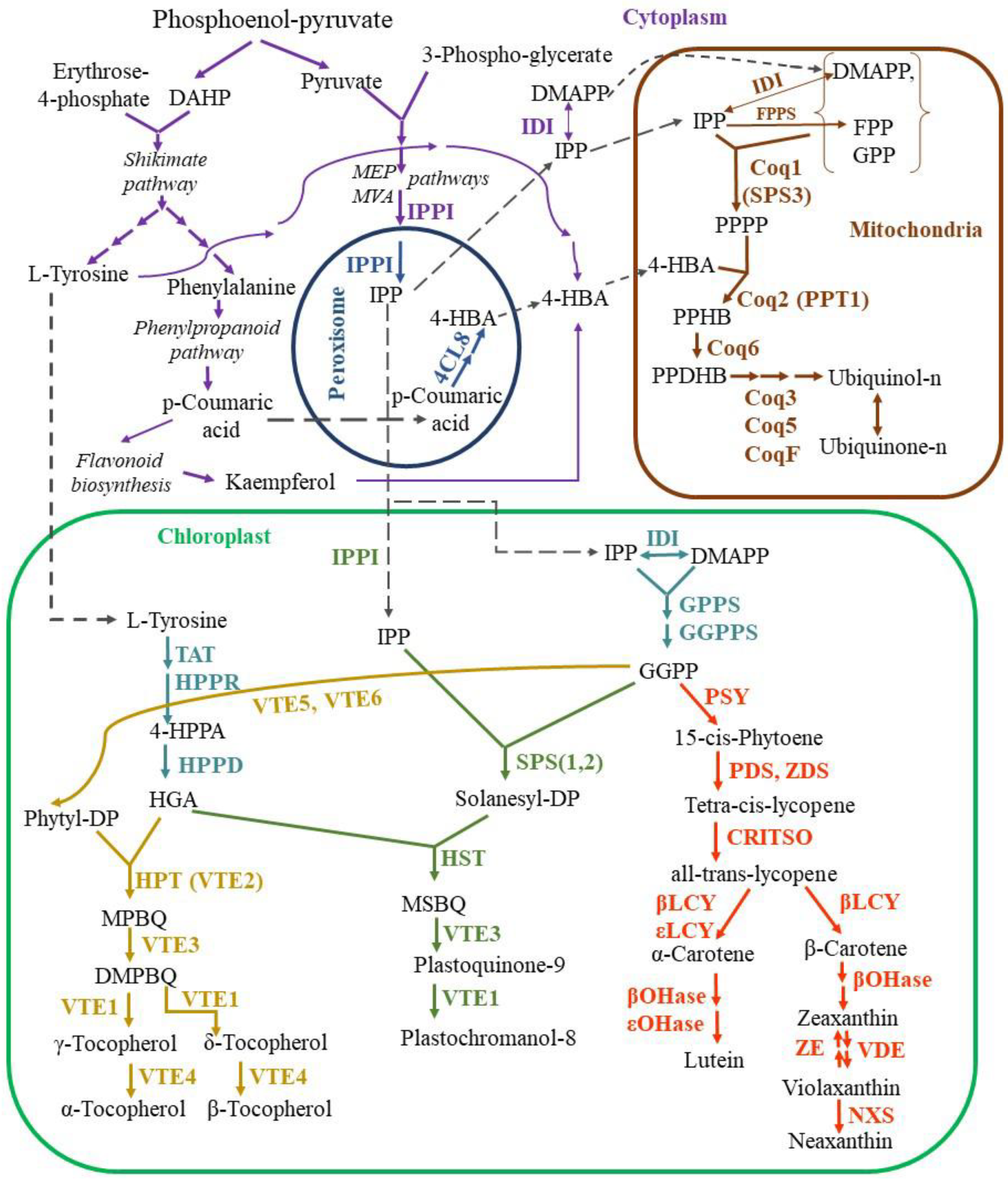
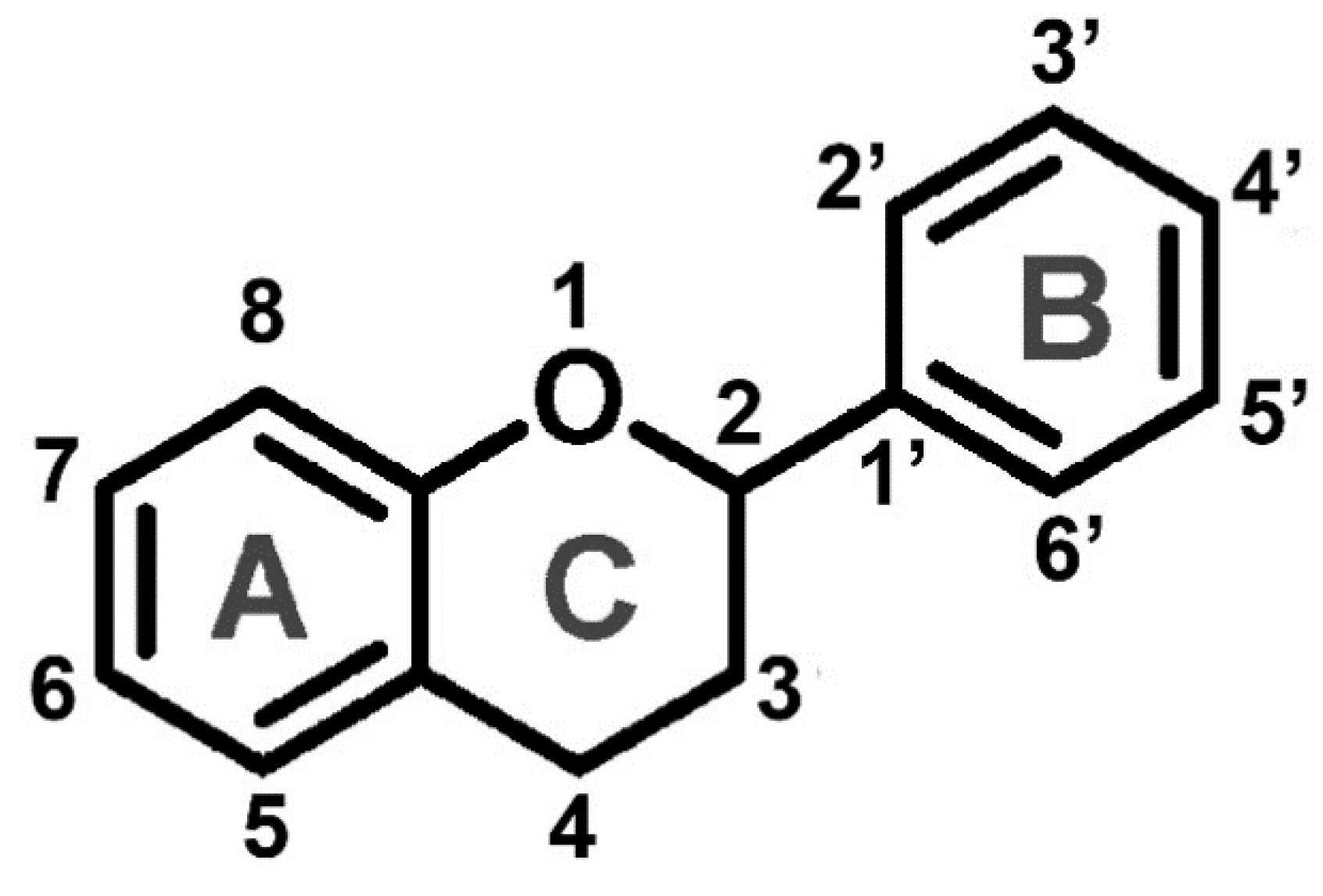
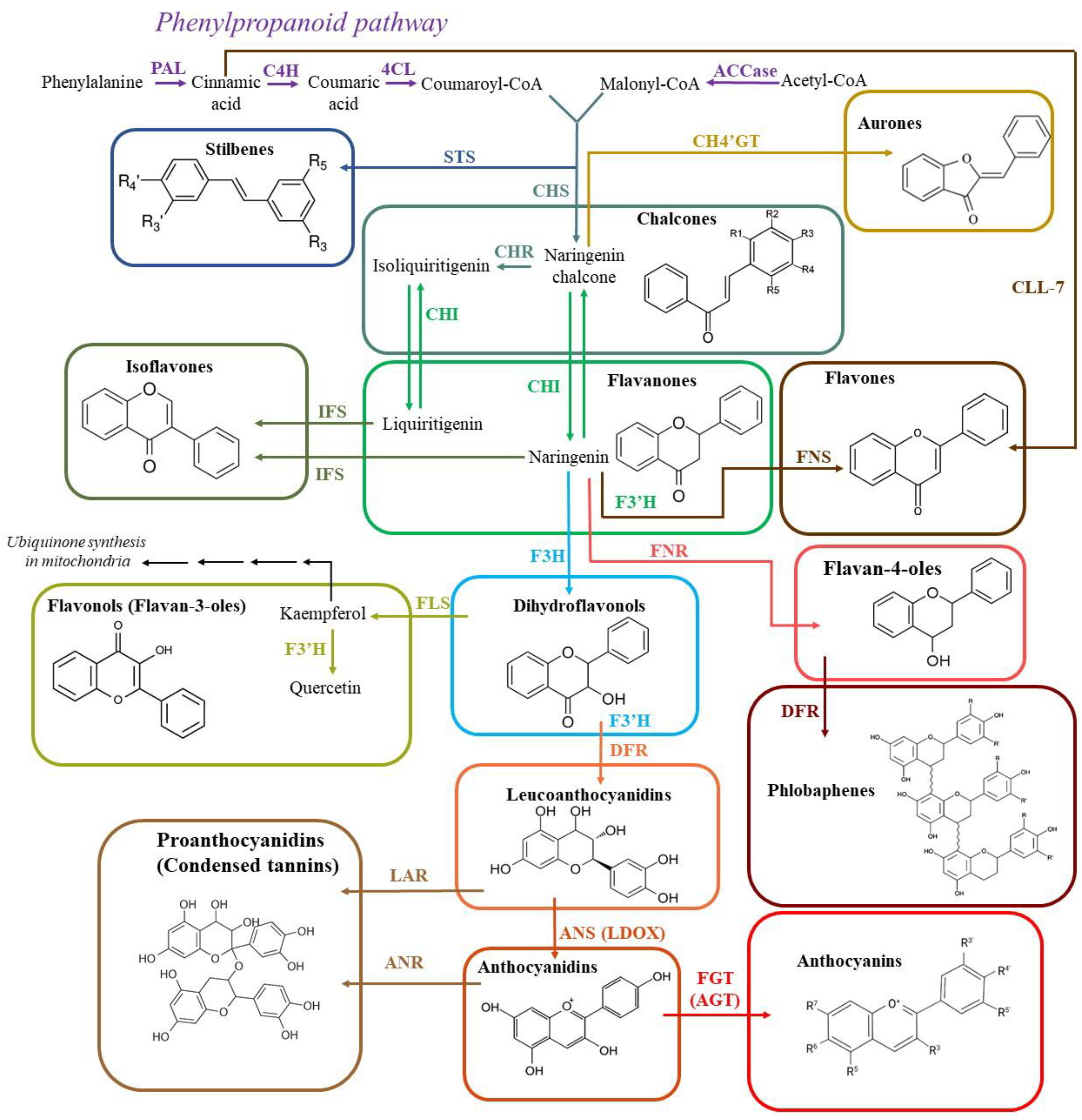
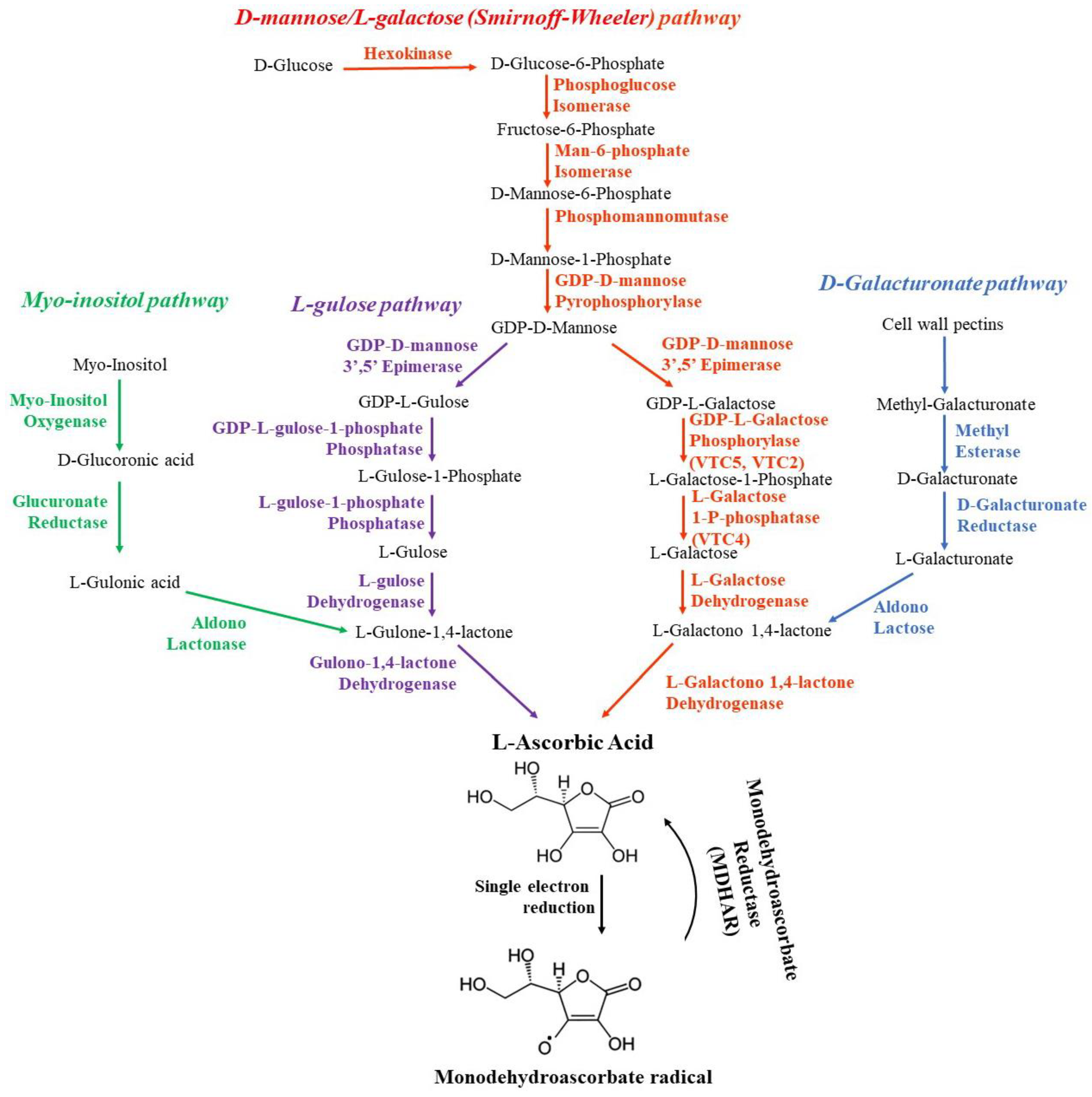
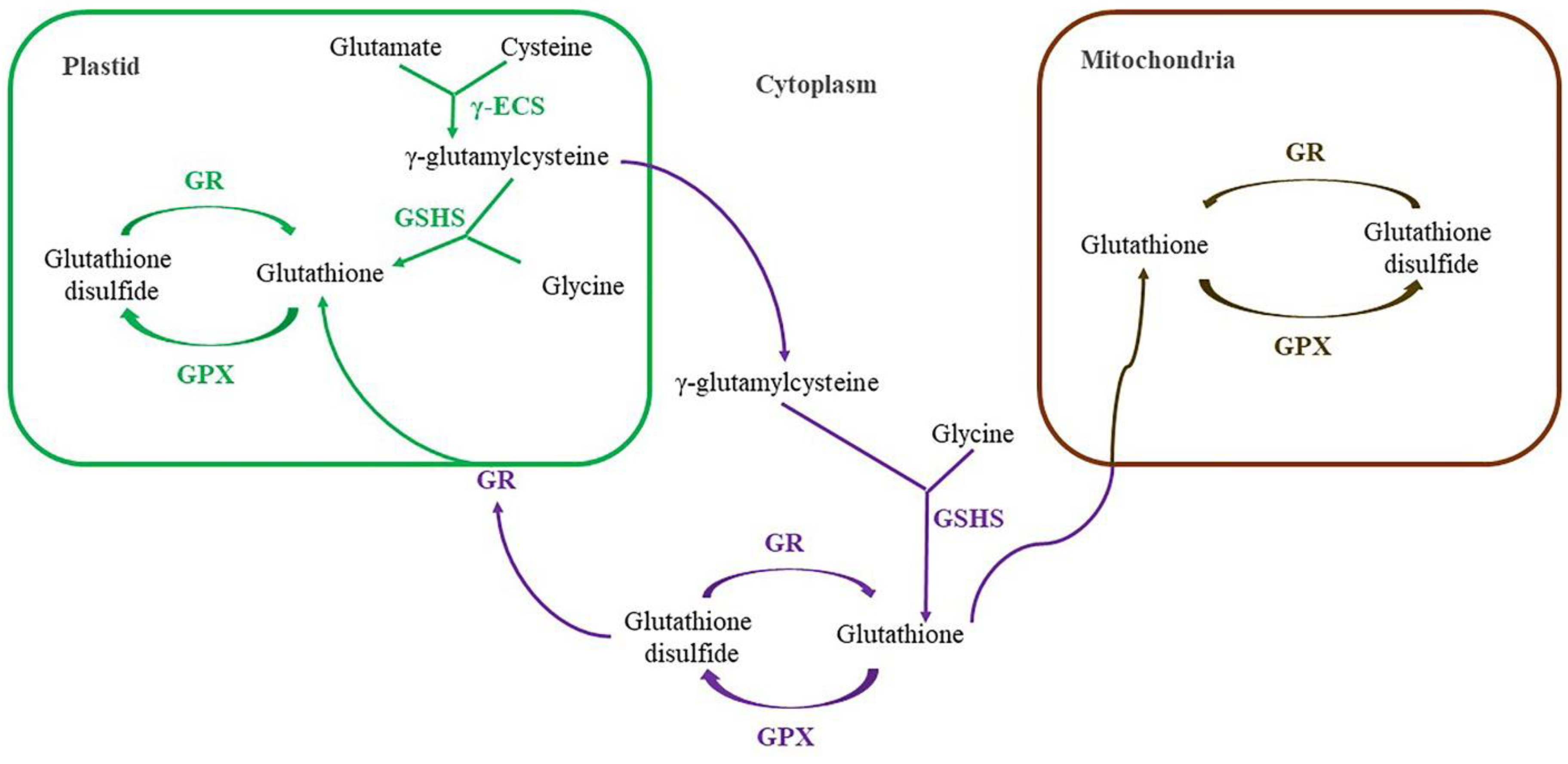
| Genes | Enzymes | Functions | |
|---|---|---|---|
| At5g16440 | IPPI, isopentenyl diphosphate isomerases | Enzymes with dimethyl allyl diphosphate isomerase activity. It is involved in the biosynthesis of IPP, isopentenyl diphosphate. IPP is a subject for further condensation reactions to form intermediates in the synthesis of plastidic and mitochondrial isoprenoids (carotenoids, tocopherols, PQ, PCr8, UQ) | |
| At3g02780 | |||
| At4g19010 | AT4G 19010 |
Peroxisomal 4-coumarate CoA ligases | 4-HBA (hydroxybezoic acid) biosynthesis from phenylalanine in peroxisomes for further UQ biosynthesis [10,11] |
| At5g38120 | 4CL8 | ||
| At5g47770 | FPS1 | Farnesyl diphosphate synthases | Isoprenoid farnesyl diphosphate (FPP) biosynthesis for further UQ biosynthesis [12,13] |
| At4g17190 | FPS2 | ||
| At2g34630 | CoQ1, SPS3, solanesyl diphosphate synthase | Isoprene polymerization for further UQ biosynthesis [14] | |
| At4g23660 | Coq2, PPT1, 4-hydroxybenzoate polyprenyl diphosphate transferase | Rate-limiting enzyme in UQ biosynthesis. Catalysis of benzoquinone ring of 4-HB condensation with polyisoprenoid side chain of polyprenyl-pyrophosphate to form 3-polyprenyl-4-hydroxybenzoate [15] |
|
| At3g24200 | Coq6 | Flavin-dependent monooxygenases (Toda et al., 2014; Latimer et al., 2021) | Aromatic hydroxylation of C-H in different positions in UQ biosynthesis |
| At1g24340 | CoqF | ||
| At2g30920 | Coq3 | S -adenosyl- l -methionine (SAM)-dependent methyl transferases | |
| At5g57300 | Coq5 | ||
| At2g03690 | Coq4 | Presumably, a scaffold protein, which is responsible for organization of UQ biosynthetic complex [16] | |
| At5g17230 | PSY, phytoene synthase | Condensation of two molecules of GGDP to produce phytoene for further carotenoid biosynthesis |
|
| At4g14210 | PDS, phytoene desaturase | Desaturation of phytoene to ζ-carotene by introduction of four double bonds into phytoene for further carotenoid biosynthesis | |
| At3g04870 | ZDS, ζ-carotene desaturase | Reduction ζ-carotene to lycopene by introduction of four double bonds for further carotenoid biosynthesis | |
| At1g06820 | CRTISO, carotenoid isomerase | Catalyzes cis-trans isomerization of poly-cis-carotenoids to all-trans-lycopene. Together with PDS and ZDS, CRTiso is required to complete the synthesis of lycopene from phytoene for further carotenoid biosynthesis [17,18] | |
| At3g10230 | βLCY1, β-carotene cyclase, ATLCY, LYC, Lycopene cyclase | Introduction of a ring at both ends of symmetrical lycopene to form the bicyclic β-carotene [19] | |
| At5g57030 | εLCY, ε- carotene cyclase | Required to form lutein [20] | |
| At4g25700 | βOHase1 | β-carotene hydroxylases | Conversion of beta-carotene to zeaxanthin via cryptoxanthin [21] |
| At5g52570 | βOHase2 | ||
| At3g53130 | εOHase, ε-carotene hydroxylase | Involved in epsilon ring hydroxylation to carotene for lutein biosynthesis [22] | |
| At5g67030 | ZE, zeaxanthin epoxidase | Introduction of epoxide groups into both rings of zeaxanthin to form violaxanthin [23] | |
| At1g08550 | VDE, | De-epoxidation of violaxanthin to zeaxanthin [24] | |
| At1g06570 | HPPD, 4-hydroxyphenylpyruvate dioxygenase (α-ketoisocaproate dioxygenase, KIC dioxygenase) |
Homogentisate (HGA) synthesis from hydroxyphenylpyruvate for further biosynthesis of PQ, PCr and tocopherols [25] | |
| At5g04490 | VTE5, phytol kinase | Phosphorylation of free phytol for further biosynthesis of tocopherols [26] | |
| At1g78620 | VTE6, phytyl-phosphate kinase | A key enzyme for phytol phosphorylation for further biosynthesis of tocopherols and phylloquinone [27] | |
| At1g78510 | SPS1 | Solanesyl diphosphate synthases | Solanesyl diphosphate condensation from geranylgeranyl diphosphate (GGDP) and isopentenyl phosphate (IPP) for further biosynthesis of PQ and PCrs [28,29,30] |
| At1g17050 | SPS2 | ||
| At5g09820 | FBN5-B | Fibrillins | Specifically interacted with solanesyl SPS1 and SPS2 |
| At3g11945 | HST, homogentisate solanesyl diphosphate transferase | Condensation of homogentisate (HGA) with solanesyl diphosphate with formation of methyl-solanesyl-benzoquinone (MSBQ) for further biosynthesis of PQ and PCrs | |
| At2g18950 | HPT, homogentisate phytyl transferase (VTE2) | Catalysis of condensation of HGA and Phytyl-DP to form dimethyl-phytyl-benzoquinone (MPBQ) for further biosynthesis of tocopherols | |
| At3g63410 | VTE3 2-methyl-6-phytyl-1,4-benzoquinol methyltransferase |
Methyl-solanesyl-benzoquinone (MSBQ) conversion to PQ9 and methyl-phytyl-benzoquinone (MPBQ) conversion to DMPBQ for further biosynthesis of tocopherols [31] | |
| At4g32770 | VTE1, tocopherol cyclase | Plastochromanol-8 synthesis from reduced plastoquinone-9; α-tocopherol biosynthesis from γ -tocopherol [31] | |
| At1g64970 | VTE4, G-TMT, γ-tocopherol methyltransferase |
Conversion of δ- and γ-tocopherols (and tocotrienols) to β- and α-tocopherols [31] | |
| Genes | Enzymes | Functions |
|---|---|---|
| At2g37040 | PAL, phenylalanine ammonia lyase | The deamination of phenylalanine to trans-cinnamic acid [172,173] |
| At3g53260 | ||
| At2g30490 | C4H, Cinnamic acid 4-hydroxylase | The hydroxylation of trans-cinnamic acid [174,175] |
| At1g65060 | 4CL, 4-coumarate CoA ligase | Coumaric acid conversion to coumaroyl-CoA, which is the last step of phenylpropanoid pathway [176] |
| At5g13930 | CHS, chalcone synthase (ATCHS, Transparent Testa 4, TT4) | The condensation of activated coumaric acid with three molecules of activated malonic acid in the form of malonyl-CoA to the formation of naringenin-chalcone. A key enzyme involved in the biosynthesis of flavonoids [177] |
| At3g55120 | CHI, chalcone isomerase (A11, ATCHI, Chalcone flavanone isomerase, Transparent Testa 5, TT5) | Catalysis of the conversion of chalcones into flavanones [177]. At3g55120 is co-expressed with CHS encoding gene [54] |
| At3g51240 | F3H, flavanone 3-hydroxylase | Encodes flavanone 3-hydroxylase that is coordinately expressed with CHSs and CHIs and involved in flavonoid biosynthesis [178]. |
| At5g07990 | F3’H, flavanone 3’-hydroxylase (CYP75B1, Cytochrome P450 75B1, D501, Transparent Testa 7, TT7) | Hydroxylation of 3’-position of B-ring of flavonoids with catalysis of dihydroquercetin and quercetin formation from dihydrokaempferol and kaempferol, respectively [178] |
| At5g24530 | FNS, flavone synthase (AtDMR6, Downy Mildew Resistant6) | The conversion of the flavanones into flavones. This class is also shown to comprise soluble Fe2+/2-oxoglutarate-dependent dioxygenases, which are oxygen- and NADPH-dependent cytochrome P450 membrane-bound monooxygenases [179] |
| At5g08640 | FLS, flavonol synthase (ATFLS1) | Encodes a flavonol synthase that catalyzes formation of flavonols from dihydroflavonols. Co-expressed with CHI and CHS (qRT-PCR) |
| At5g63590 | ||
| At5g42800 | DFR, dihydroflavonol 4-reductase | The reduction of the 4-keto group of dihydroflavonol to the corresponding leucoanthocyanidin (Shirley et al., 1998). Synthesis of phlobaphenes from flavan-4-oles in Zea mays [180] |
| At1g61720 | ANR, anthocyanidin reductase | Synthesis of proanthocyanidins (condensed tannins) from leukoanthocyanidins and anthocyanidins [181] |
| At4g22880 | ANS, anthocyanidin synthase (LDOX, Leucoanthocyanidin dioxygenase) | Convertion leucoanthocyanidins to anthocyanins [182] |
| At5g17050 | FGT, flavonoid glycosyltransferases | Glycosylation of anthocyanidins to anthocyanins [183,184,185,186] |
| At1g30530 | ||
| At5g17030 | ||
| At2g36790 | ||
| At1g06000 | ||
| At4g14090 | ||
| At5g54060 | ||
| At2g47460 | MYB domain protein 12, MYB12, ATMYB12, PFG1 | Flavonol synthesis regulators. Strongly activate the promoters of CHS, F3H, FLS and CHI [187] |
| At3g62610 | AtMYB11, PFG2 | |
| At5g49330 | AtMYB111, PFG3 | |
| At2g46510 | bHLH17 (ABA-inducible bHLH-type transcription factor, AIB, ATAIB, JA-associated MYC2-like 1, JAM1 | Positive regulator of flavonoid biosynthesis [188] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




