1. Introduction of Brain Diseases
Brain diseases encompass a broad spectrum of disorders that affect the structure or function of the brain, leading to a wide range of neurological and cognitive impairments. These conditions can be congenital, acquired, or degenerative in nature, and they pose significant challenges to both patients and healthcare providers. Understanding the complexities of brain diseases is essential for diagnosis, treatment, and ongoing research aimed at improving the quality of life for affected individuals.
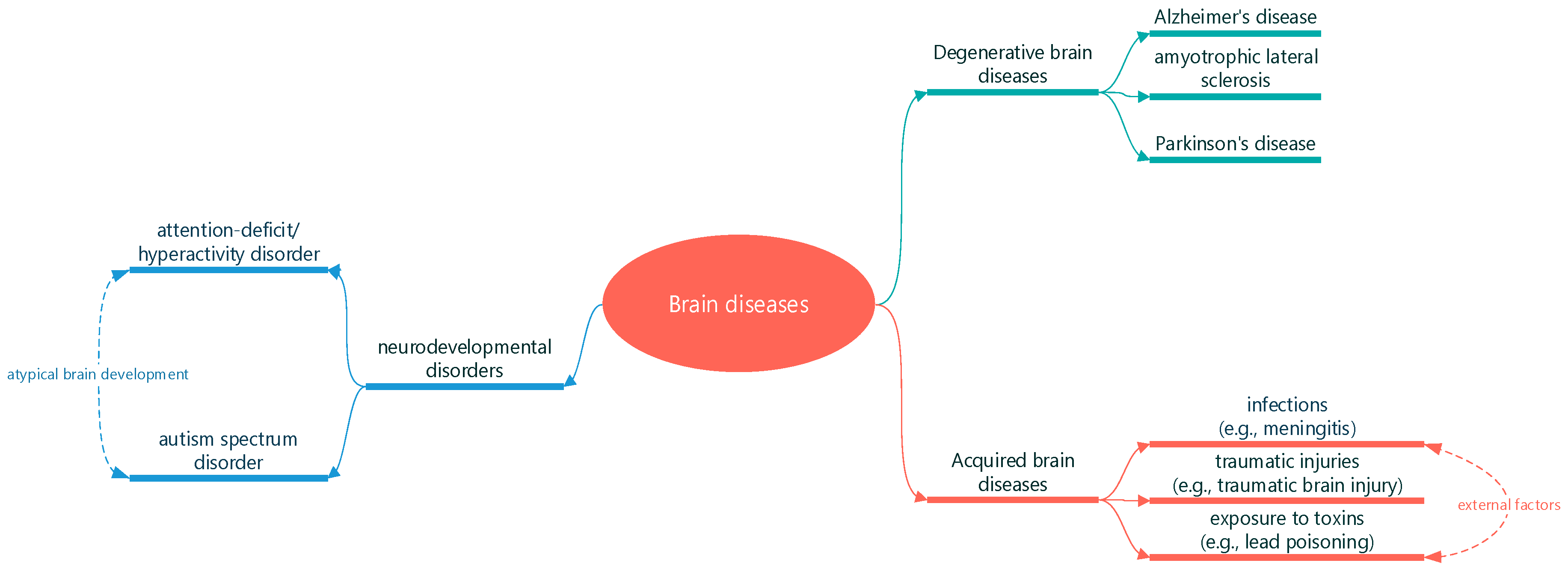
As shown in
Figure 1: one category of brain diseases is caused by neurodevelopmental disorders, such as autism spectrum disorder and attention-deficit/hyperactivity disorder (ADHD) [
1]. These conditions typically manifest early in life and are characterized by atypical brain development, leading to difficulties in social interaction, communication, and behavior regulation. While the exact causes remain under investigation, a combination of genetic and environmental factors is believed to contribute to their development [
2].
Acquired brain diseases, on the other hand, result from external factors such as infections (e.g., meningitis), traumatic injuries (e.g., traumatic brain injury), or exposure to toxins (e.g., lead poisoning) [
3]. These conditions often lead to sudden and severe neurological deficits and require prompt medical intervention. Treatment approaches may include antibiotics, surgery, or rehabilitation therapy, depending on the underlying cause and severity of the disease.
Degenerative brain diseases are a group of disorders characterized by the progressive deterioration of brain tissue. Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis (ALS) are well-known examples [
4]. These conditions are typically associated with aging and result in a gradual loss of cognitive or motor functions. While there is currently no cure for most degenerative brain diseases, ongoing research aims to better understand their underlying mechanisms and develop effective treatments to slow or halt their progression.
Brain diseases encompass a diverse array of conditions that affect the brain’s structure and function. They can be classified into neurodevelopmental disorders, acquired brain diseases, and degenerative brain diseases, each presenting unique challenges for diagnosis, treatment, and management. Advances in neuroscience and medical research continue to shed light on the underlying causes of these diseases and offer hope for improved therapies and interventions in the future [
5].
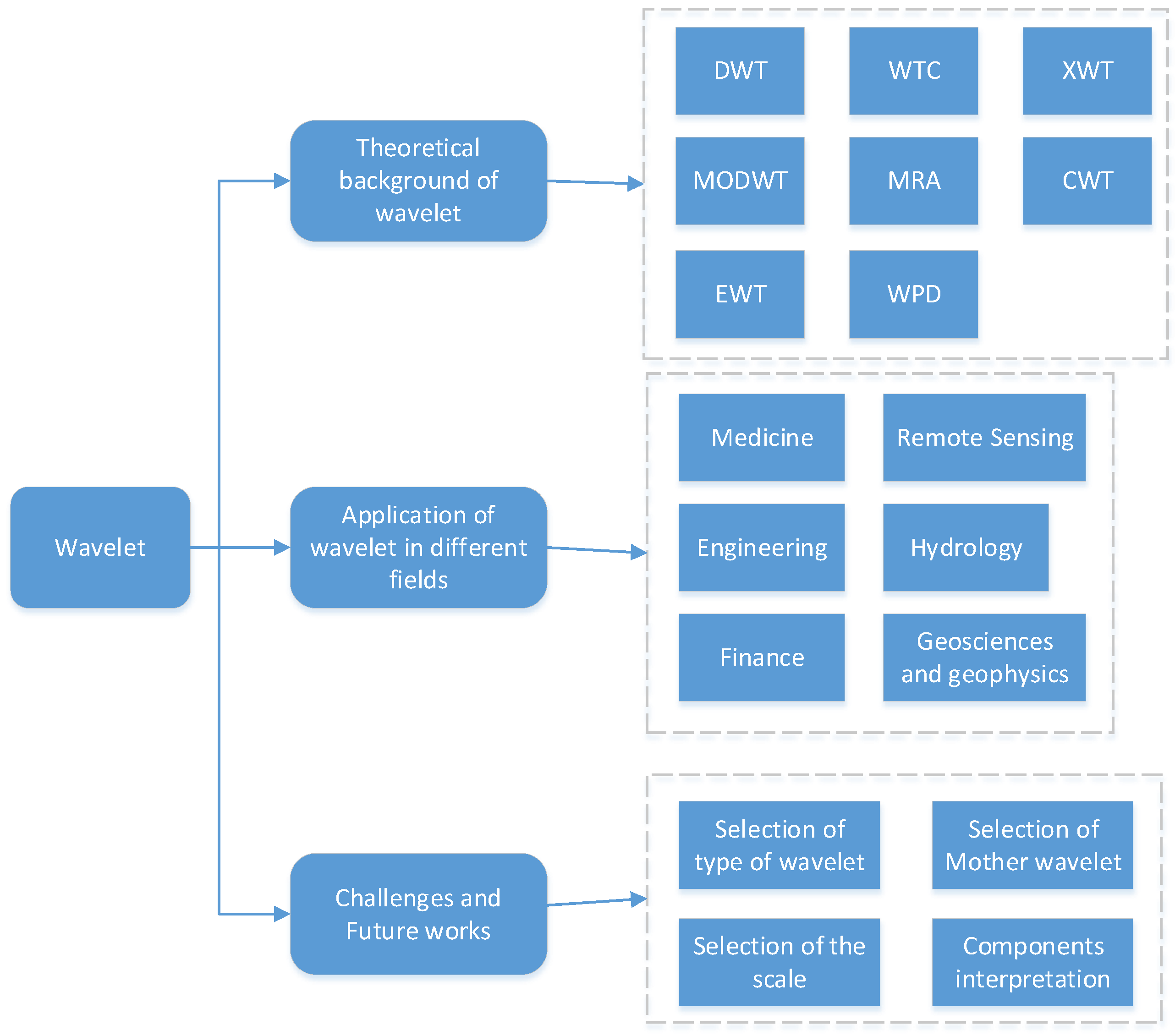
2. Introduction of Wavelet
Wavelet analysis is a powerful mathematical and signal processing technique that has gained prominence in various fields [
6], including mathematics, engineering, physics, and data analysis. It offers a unique way to analyze and represent signals or data in both the time and frequency domains [
7] simultaneously, making it a valuable tool for tasks like signal denoising, compression, and feature extraction. In this introduction, we will explore the fundamental concepts of wavelets, their origins, and their applications.
Wavelets are essentially mathematical functions or wave-like patterns that are used to transform data. Unlike traditional Fourier analysis, which decomposes a signal into a sum of sinusoidal functions with fixed frequencies [
8], wavelets allow for the decomposition of a signal into different wavelet functions, each associated with a specific scale or frequency [
9]. This flexibility is crucial because it enables wavelet analysis to capture both high-frequency and low-frequency components within a signal, making it well-suited for the analysis of complex signals with varying characteristics [
10].
The concept of wavelets can be traced back to the early 20th century, but it wasn’t until the late 20th century that wavelet analysis gained widespread recognition and use, thanks in part to the work of mathematicians like Jean Morlet and Ingrid Daubechies. Their contributions led to the development of discrete wavelet transforms (DWT) and continuous wavelet transforms (CWT), which are now essential tools in signal processing and data analysis [
11].
One of the key advantages of wavelets is their ability to perform multi-resolution analysis. This means that wavelet analysis can zoom in on fine details within a signal while also providing an overview of its coarse features, all in a single analysis. This makes wavelets particularly valuable in applications such as image processing, where different regions of an image may require different levels of detail [
12].
In practical terms, wavelet analysis finds applications in diverse fields. As shown in
Figure 2, In image processing, it is used for image compression and denoising. In data analysis, it is employed for feature extraction, time series analysis, and pattern recognition. In engineering, it is utilized for signal filtering and fault detection. Additionally, wavelets have found applications in the analysis of biological signals, such as electroencephalograms (EEGs) and electrocardiograms (ECGs) [
13], as well as in financial data analysis for tasks like stock market prediction.
Wavelet analysis is a versatile mathematical technique that has revolutionized the way we analyze and process data. Its ability to simultaneously capture both time and frequency information makes it invaluable in various fields, from signal processing to image analysis to finance. As technology advances and our understanding of wavelets deepens, their applications continue to expand, making them an essential tool in modern data analysis and signal processing. The following are four applications of wavelet analysis.
3. Application 1 – Feature Extraction
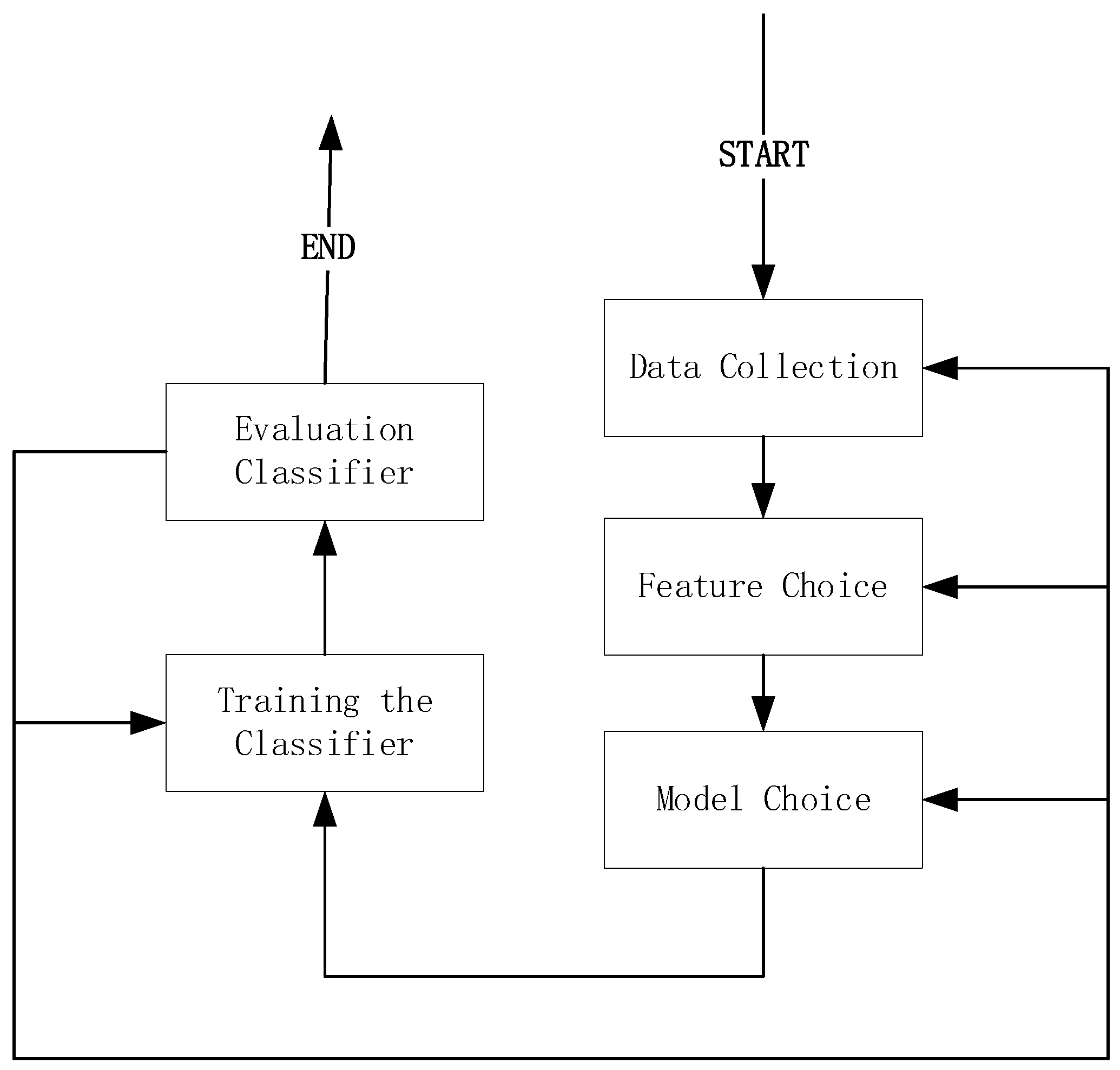
As shown in
Figure 3, the feature extraction method is mainly based on the relationship between attributes, such as combining different attributes to obtain new attributes, thus changing the original feature space. The main purpose is to try to reduce the number of attributes in the feature data set.
Feature extraction involves identifying relevant patterns or characteristics within brain signals that can differentiate between healthy individuals and those with neurological conditions. Here are four ways wavelet analysis facilitates this process: Wavelet analysis decomposes brain signals, such as EEG or fMRI data, into various frequency components at different scales [
14]. This decomposition enables the extraction of frequency-domain features, such as dominant frequency bands or spectral power, which can be indicative of specific brain diseases. For example, abnormalities in certain frequency bands, like alpha or beta rhythms, can be associated with conditions like epilepsy or Parkinson’s disease [
15]. By quantifying these features using wavelet analysis, researchers and clinicians can create discriminative metrics for disease detection.
Unlike traditional Fourier analysis, which provides a global view of frequency components, wavelet analysis offers time-frequency localization [
16]. This means that it can pinpoint when specific frequency components occur over time. In brain disease detection, this localization is crucial because it can identify transient events or irregularities in neural activity. For instance, epileptic seizures often manifest as sudden bursts of abnormal electrical activity [
17], and wavelet analysis can precisely capture and characterize these transient events, aiding in seizure detection and diagnosis.
Wavelet transformation results in a set of coefficients that represent the contribution of wavelets at various scales and positions in the signal. These coefficients can serve as valuable features for machine learning algorithms. By analyzing the magnitude and distribution of wavelet coefficients, one can create a feature vector that encapsulates the unique characteristics of a patient’s brain activity. This feature vector can then be used for classification tasks, distinguishing between individuals with different brain diseases or assessing disease progression.
Brain diseases often manifest as distinctive patterns in neuroimaging data. Wavelet analysis can enhance pattern recognition by isolating salient features within the data [
18]. This is particularly helpful when dealing with large and complex datasets. Machine learning algorithms can be trained on these wavelet-derived features to recognize specific disease-related patterns and make accurate diagnoses or predictions [
19].
4. Application 2 – Signal Denoising
Wavelet analysis decomposes the neuroimaging data into different scales, where each scale corresponds to a different level of detail or frequency component. Noise components typically occupy higher scales due to their high-frequency characteristics [
20]. By identifying and isolating these high-frequency scales, wavelet denoising effectively separates noise from the underlying brain signals [
10].
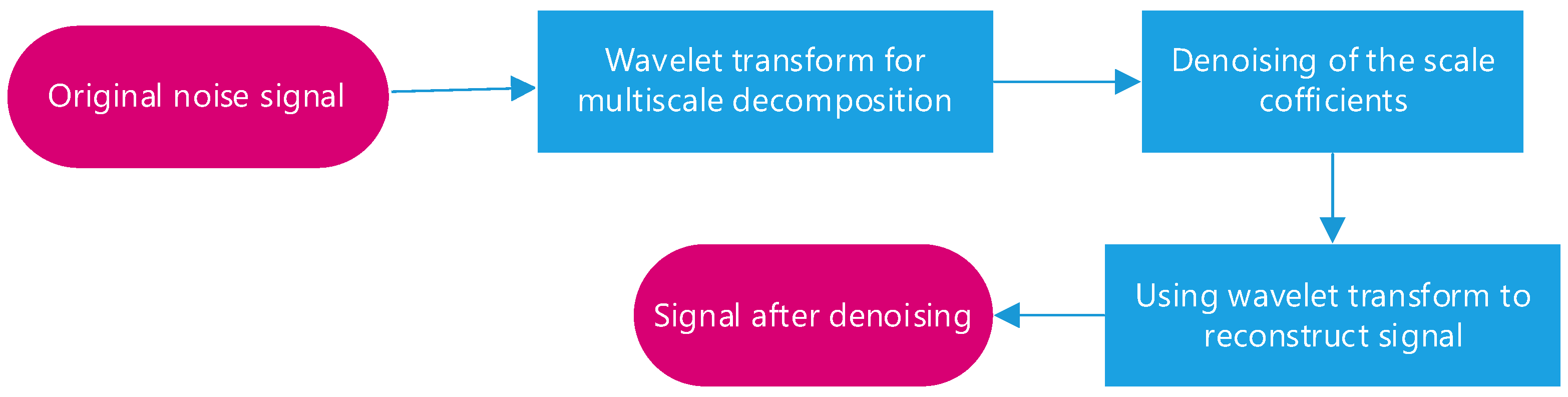
The flow chart of wavelet denoising is shown as
Figure 4. Wavelet denoising often employs a thresholding approach [
21]. After decomposing the signal, wavelet coefficients are compared to a predefined threshold. Coefficients below the threshold are considered noise and are set to zero, while coefficients above the threshold are retained as signal information [
22]. This thresholding process selectively removes noise while preserving the essential brain activity patterns. Adaptive thresholding methods, such as the Stein’s Unbiased Risk Estimate (SURE) threshold [
23], can optimize the denoising process for different types of data and noise characteristics.
Removing noise through wavelet denoising enhances the signal-to-noise ratio (SNR) of the neuroimaging data [
24]. This heightened SNR makes it easier to identify and analyze subtle abnormalities or disease-related patterns within the brain signals [
25]. For instance, in EEG data, wavelet denoising can sharpen the presence of epileptic spikes or abnormal oscillations, which are crucial for epilepsy diagnosis and treatment planning [
26].
Cleaned and denoised neuroimaging data provide a more accurate representation of the underlying brain activity, reducing the risk of false positives or misinterpretations during disease detection. Clinicians and researchers can rely on denoised data for more precise analysis, leading to improved diagnostics and a better understanding of the neurological condition. It also enhances the effectiveness of subsequent data analysis techniques, such as feature extraction or classification, which can be vital for disease detection and monitoring.
Wavelet analysis aids in the detection of brain diseases through denoising by separating noise from brain signals, employing thresholding techniques, enhancing the SNR, and ultimately improving the quality of the data for accurate diagnostics [
27]. This denoising process is invaluable in neuroimaging, as it allows for more reliable and sensitive detection of neurological abnormalities, facilitating early diagnosis and intervention for brain diseases.
5. Application 3 – Classification and Diagnosis
Brain diseases often manifest as distinct patterns or abnormalities in neuroimaging data. Wavelet analysis can enhance pattern recognition by isolating salient features within the data. Machine learning models can be trained on these wavelet-derived features to recognize specific patterns associated with different neurological conditions. For example, epileptic seizures may exhibit characteristic transient waveforms in EEG data, and wavelet-based features can help differentiate seizure events from normal brain activity, aiding in the diagnosis of epilepsy [
28].
Some brain diseases share common symptoms or characteristics, making accurate diagnosis challenging. Wavelet analysis can provide additional information that differentiates between similar conditions [
29]. For instance, in the case of neurodegenerative diseases like Alzheimer’s and Parkinson’s, wavelet analysis of brain imaging data can reveal distinct patterns of neural activity or connectivity that help distinguish between these conditions [
30]. This differentiation is critical for providing targeted treatment and interventions. Wavelet analysis is mainly used for classification and diagnosis through the process as shown in
Figure 5.
Wavelet-based features provide quantitative measures of brain activity, enabling clinicians to objectively assess the severity and progression of brain diseases [
31]. This is particularly valuable for conditions like multiple sclerosis or traumatic brain injury, where changes in brain activity over time can be indicative of disease progression. By quantifying these changes using wavelet-derived features, clinicians can make more informed decisions regarding treatment and patient management [
32].
Wavelet analysis supports the detection of brain diseases through classification and diagnosis by extracting informative features [
33], enhancing pattern recognition, aiding in differential diagnosis, and providing quantitative assessments of brain activity. This approach not only helps identify neurological conditions but also contributes to more accurate and tailored treatment strategies, ultimately improving patient outcomes and quality of life.
6. Application 4 – Time-Frequency Analysis
One of the key advantages of wavelet analysis is its ability to provide time-frequency localization of neural signals [
34]. Unlike traditional Fourier analysis [
35], which offers a global view of signal frequencies, wavelet analysis pinpoints when specific frequency components occur over time [
36]. In the context of brain diseases, this localization is crucial for detecting transient events or irregularities in neural activity. For instance, epileptic seizures often manifest as sudden bursts of abnormal electrical activity, and wavelet analysis can precisely capture and characterize these transient events, aiding in seizure detection and diagnosis [
37]. The main steps of time-frequency analysis are shown in
Figure 6.
Many brain diseases, such as Alzheimer’s disease or Parkinson’s disease, are progressive and involve changes in brain activity patterns over time. Wavelet analysis allows for the tracking of these changes by continuously assessing the time-varying frequency content of neuroimaging data [
38]. Researchers can identify shifts in dominant frequency bands or evolving patterns of neural synchronization associated with disease progression. This information is invaluable for monitoring the advancement of the disease and assessing the effectiveness of treatments.
Time-frequency analysis [
39] using wavelets can help identify specific biomarkers associated with brain diseases. Biomarkers are measurable indicators of disease presence or progression [
40]. By examining the time-varying spectral content of neural signals, researchers can discover unique patterns or oscillatory behaviors that are characteristic of a particular neurological condition. These biomarkers can serve as early diagnostic tools and provide insights into disease mechanisms, aiding in the development of targeted therapies [
41].
For brain diseases that require ongoing treatment or intervention, wavelet-based time-frequency analysis can be used to assess the effects of therapies on brain activity [
42]. By comparing pre- and post-treatment neuroimaging data, clinicians can determine whether the treatment has resulted in changes in time-varying neural patterns. This information helps in tailoring treatment plans and making necessary adjustments to improve patient outcomes.
Wavelet analysis enhances the detection of brain diseases through time-frequency analysis by providing precise time-frequency localization, facilitating disease progression monitoring, identifying biomarkers, and evaluating the effectiveness of treatments [
43]. These insights aid in early diagnosis, personalized treatment strategies, and a better understanding of the dynamic nature of neurological conditions, ultimately improving patient care and outcomes [
44].
7. Other Analysis Tools
There are several other analysis tools and techniques used in the field of neuroscience and the study of brain diseases. These tools complement wavelet analysis and provide additional methods for analyzing and understanding brain-related data. Here are a few notable ones: FFT is a widely used technique for transforming time-domain signals into the frequency domain [
45]. It helps identify the dominant frequency components in neuroimaging data, which is particularly valuable in studying brain rhythms and oscillations. FFT is often used alongside wavelet analysis to provide complementary frequency information. ICA is a blind source separation technique that can separate mixed signals into their original sources [
46]. In neuroimaging, ICA is used to identify independent components in data, allowing researchers to isolate and study specific brain networks or sources of activity. PCA is a dimensionality reduction technique that is used to extract the most significant features or components from complex datasets [
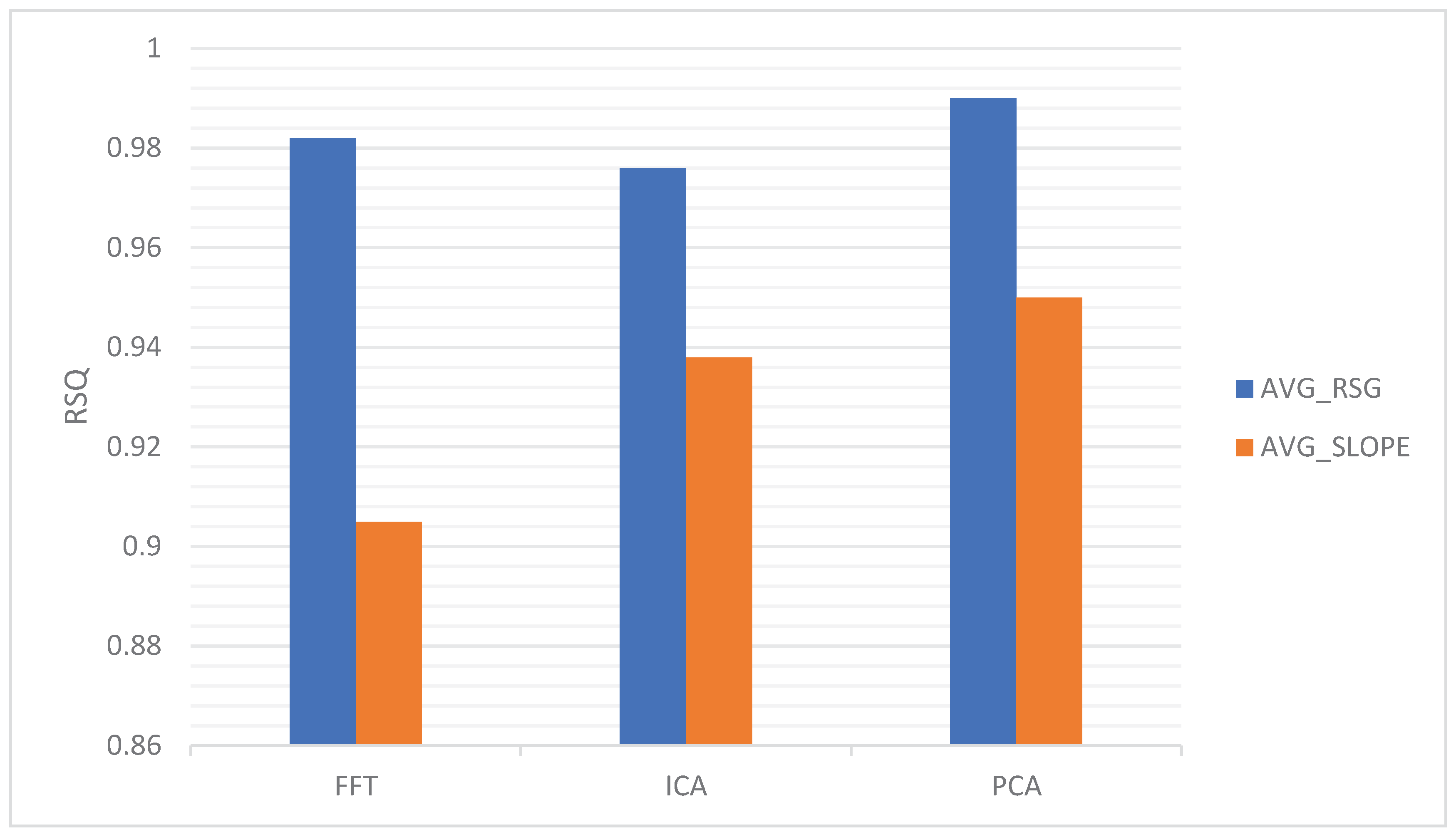
47]. It can be applied to reduce the dimensionality of neuroimaging data, making it more manageable for subsequent analysis or visualization. The analysis tools of FFT, ICA, and PCA exhibit variations in the data of AVG_RSG and AVG_SLOPE, as illustrated in
Figure 7.
8. Discussion and Conclusion
In conclusion, wavelet theories and techniques have emerged as invaluable tools in the realm of neuroscience and the study of brain diseases. Wavelet analysis provides a versatile framework for the analysis of neuroimaging data, offering benefits such as time-frequency localization, noise reduction, and feature extraction. These capabilities aid in the identification, characterization, and monitoring of various neurological conditions.
Wavelet analysis’s ability to extract meaningful features from brain signals plays a pivotal role in disease detection. By capturing frequency components, wavelet analysis can highlight abnormalities or patterns associated with specific brain diseases [
48]. This feature extraction, combined with machine learning algorithms, enables the development of robust diagnostic tools that enhance early detection and personalized treatment strategies.
Furthermore, wavelet-based time-frequency analysis provides a dynamic view of brain activity [
49], facilitating the monitoring of disease progression and the identification of biomarkers. This information is vital for understanding the evolving nature of neurological disorders and evaluating the effectiveness of therapeutic interventions. Overall, wavelet theories and techniques have greatly enriched our understanding of brain diseases, leading to improved diagnosis, treatment, and patient care in the field of neuroscience.
Funding
This research did not receive any grants.
Acknowledgment
We thank all the anonymous reviewers for their hard reviewing work.
References
- J. K. Kern, D. A. Geier, P. G. King, L. K. Sykes, J. A. Mehta, and M. R. Geier, “Shared brain connectivity issues, symptoms, and comorbidities in autism spectrum disorder, attention deficit/hyperactivity disorder, and Tourette syndrome,” Brain connectivity, vol. 5, pp. 321-335, 2015. [CrossRef]
- R. J. Boyd, D. Avramopoulos, L. L. Jantzie, and A. S. McCallion, “Neuroinflammation represents a common theme amongst genetic and environmental risk factors for Alzheimer and Parkinson diseases,” Journal of Neuroinflammation, vol. 19, p. 223, 2022. [CrossRef]
- L. Goldman, E. M. Siddiqui, A. Khan, S. Jahan, M. U. Rehman, S. Mehan, et al., “Understanding acquired brain injury: A review,” Biomedicines, vol. 10, p. 2167, 2022. [CrossRef]
- E. Gaggelli, H. Kozlowski, D. Valensin, and G. Valensin, “Copper homeostasis and neurodegenerative disorders (Alzheimer’s, prion, and Parkinson’s diseases and amyotrophic lateral sclerosis),” Chemical reviews, vol. 106, pp. 1995-2044, 2006. [CrossRef]
- D. Gitler, P. Dhillon, and J. Shorter, “Neurodegenerative disease: models, mechanisms, and a new hope,” Disease models & mechanisms, vol. 10, pp. 499-502, 2017. [CrossRef]
- S.-H. Wang, “Diagnosis of COVID-19 by Wavelet Renyi Entropy and Three-Segment Biogeography-Based Optimization,” International Journal of Computational Intelligence Systems, vol. 13, pp. 1332-1344, 2020. [CrossRef]
- Y. Zhang, “A Comprehensive Survey on Fractional Fourier Transform,” Fundamenta Informaticae, vol. 151, pp. 1-48, 2017. [CrossRef]
- W. Zhou, Z. Feng, Y. Xu, X. Wang, and H. Lv, “Empirical Fourier decomposition: An accurate signal decomposition method for nonlinear and non-stationary time series analysis,” Mechanical Systems and Signal Processing, vol. 163, p. 108155, 2022. [CrossRef]
- S.-H. Wang, “Unilateral sensorineural hearing loss identification based on double-density dual-tree complex wavelet transform and multinomial logistic regression,” Integrated Computer-Aided Engineering, vol. 26, pp. 411-426, 2019. [CrossRef]
- Pavlova, G. Guyo, and A. Pavlov, “Multiresolution wavelet analysis of noisy datasets with different measures for decomposition coefficients,” Physica A: Statistical Mechanics and its Applications, vol. 585, p. 126406, 2022. [CrossRef]
- R. C. Guido, F. Pedroso, A. Furlan, R. C. Contreras, L. G. Caobianco, and J. S. Neto, “CWT× DWT× DTWT× SDTWT: Clarifying terminologies and roles of different types of wavelet transforms,” International Journal of Wavelets, Multiresolution and Information Processing, vol. 18, p. 2030001, 2020. [CrossRef]
- K. Sui and H.-G. Kim, “Research on application of multimedia image processing technology based on wavelet transform,” EURASIP Journal on Image and Video Processing, vol. 2019, pp. 1-9, 2019. [CrossRef]
- N. Bajaj, “Wavelets for EEG Analysis,” Wavelet Theory, pp. 1-16, 2020. [CrossRef]
- Khosla, P. Khandnor, and T. Chand, “A comparative analysis of signal processing and classification methods for different applications based on EEG signals,” Biocybernetics and Biomedical Engineering, vol. 40, pp. 649-690, 2020. [CrossRef]
- R. Cohan, K. A. Bearss, and J. F. DeSouza, “Frequency-specific biomarkers in neurodegenerative disorders: Implications of alpha and beta oscillations in motor behaviour,” Journal of Neurology & Neuromedicine, vol. 4, 2019. [CrossRef]
- Silik, M. Noori, W. A. Altabey, R. Ghiasi, and Z. Wu, “Comparative analysis of wavelet transform for time-frequency analysis and transient localization in structural health monitoring,” Structural Durability & Health Monitoring, vol. 15, p. 1, 2021. [CrossRef]
- Z. ElSayed, M. Ozer, and N. Elsayed, “Epilepsy Seizure Detection: Anatomy and Analysis,” arXiv preprint arXiv:2305.19347, 2023.
- S. Pittner and S. V. Kamarthi, “Feature extraction from wavelet coefficients for pattern recognition tasks,” IEEE Transactions on pattern analysis and machine intelligence, vol. 21, pp. 83-88, 1999. [CrossRef]
- J. R. Malgheet, N. B. Manshor, L. S. Affendey, and A. B. Abdul Halin, “Iris recognition development techniques: a comprehensive review,” Complexity, vol. 2021, pp. 1-32, 2021. [CrossRef]
- S. Wang, “Texture Analysis Method Based on Fractional Fourier Entropy and Fitness-scaling Adaptive Genetic Algorithm for Detecting Left-sided and Right-sided Sensorineural Hearing Loss,” Fundamenta Informaticae, vol. 151, pp. 505-521, 2017. [CrossRef]
- M. Srivastava, C. L. Anderson, and J. H. Freed, “A new wavelet denoising method for selecting decomposition levels and noise thresholds,” IEEE access, vol. 4, pp. 3862-3877, 2016. [CrossRef]
- H. Hu, L. Zhang, H. Yan, Y. Bai, and P. Wang, “Denoising and baseline drift removal method of MEMS hydrophone signal based on VMD and wavelet threshold processing,” Ieee Access, vol. 7, pp. 59913-59922, 2019. [CrossRef]
- Z. Apostolakis, J. S. Shin, C. Meral, J.-L. Robert, A. Sadeghi, and F. Vignon, “Adaptive Slow-time Singular Value Thresholding (SVT) based on Stein’s Unbiased Risk Estimate (SURE) for Ultrasound Image Random Noise Reduction,” in 2020 IEEE International Ultrasonics Symposium (IUS), 2020, pp. 1-4. [CrossRef]
- Z. A. A. Alyasseri, A. T. Khader, M. A. Al-Betar, A. K. Abasi, and S. N. Makhadmeh, “EEG signals denoising using optimal wavelet transform hybridized with efficient metaheuristic methods,” IEEE Access, vol. 8, pp. 10584-10605, 2019. [CrossRef]
- Y. Zhang, “A Multilayer Perceptron Based Smart Pathological Brain Detection System by Fractional Fourier Entropy,” Journal of Medical Systems, vol. 40, p. 173, 2016. [CrossRef]
- N. Mäkinen, “Deep learning approach for epileptic seizure detection,” N. Mäkinen, 2020. [CrossRef]
- P. S. Chaitanya and S. K. Satpathy, “A Multilevel De-Noising Approach for Precision Edge-Based Fragmentation in MRI Brain Tumor Segmentation,” Traitement du Signal, vol. 40, 2023. [CrossRef]
- V. Grigorovsky, U. Tufa, D. Jacobs, and B. L. Bardakjian, “Machine Intelligence-Based Epileptic Seizure Forecasting,” Neural Engineering, pp. 535-565, 2020. [CrossRef]
- M. Aljalal, S. A. Aldosari, M. Molinas, K. AlSharabi, and F. A. Alturki, “Detection of Parkinson’s disease from EEG signals using discrete wavelet transform, different entropy measures, and machine learning techniques,” Scientific Reports, vol. 12, p. 22547, 2022. [CrossRef]
- A.-M. Tăuţan, B. Ionescu, and E. Santarnecchi, “Artificial intelligence in neurodegenerative diseases: A review of available tools with a focus on machine learning techniques,” Artificial Intelligence in Medicine, vol. 117, p. 102081, 2021. [CrossRef]
- Guerra, V. D’Onofrio, F. Ferreri, M. Bologna, and A. Antonini, “Objective measurement versus clinician-based assessment for Parkinson’s disease,” Expert Review of Neurotherapeutics, vol. 23, pp. 689-702, 2023. [CrossRef]
- Li, Q. Wang, M. Zou, P. Cai, X. Li, K. Feng, et al., “A radiomics model based on preoperative gadoxetic acid–enhanced magnetic resonance imaging for predicting post-hepatectomy liver failure in patients with hepatocellular carcinoma,” Frontiers in Oncology, vol. 13, p. 1164739, 2023. [CrossRef]
- F. A. Alturki, K. AlSharabi, A. M. Abdurraqeeb, and M. Aljalal, “EEG signal analysis for diagnosing neurological disorders using discrete wavelet transform and intelligent techniques,” Sensors, vol. 20, p. 2505, 2020. [CrossRef]
- Rasheed, M. Osama, M. Rafique, A. D. K. Tareen, K. J. Lone, S. A. Qureshi, et al., “Time-frequency analysis of radon and thoron data using continuous wavelet transform,” Physica Scripta, vol. 98, p. 105008, 2023. [CrossRef]
- Y. D. Zhang and S. C. Satapathy, “Fruit category classification by fractional Fourier entropy with rotation angle vector grid and stacked sparse autoencoder,” Expert Systems, vol. 39, 2022. [CrossRef]
- Wirsing, “Time frequency analysis of wavelet and Fourier transform,” Wavelet theory, 2020.
- E. Guillén-García, L. Morales-Velazquez, A. L. Zorita-Lamadrid, O. Duque-Perez, R. A. Osornio-Rios, and R. d. J. Romero-Troncoso, “Accurate identification and characterisation of transient phenomena using wavelet transform and mathematical morphology,” IET Generation, Transmission & Distribution, vol. 13, pp. 4021-4028, 2019. [CrossRef]
- D. Savva, G. K. Matsopoulos, and G. D. Mitsis, “A wavelet-based approach for estimating time-varying connectivity in resting-state functional magnetic resonance imaging,” Brain Connectivity, vol. 12, pp. 285-298, 2022. [CrossRef]
- Y. Karaca, “Secondary pulmonary tuberculosis recognition by rotation angle vector grid-based fractional Fourier entropy,” Fractals, vol. 30, Article ID: 2240047, 2022. [CrossRef]
- C. Restrepo, D. Dueñas, Z. Corredor, and Y. Liscano, “Advances in Genomic Data and Biomarkers: Revolutionizing NSCLC Diagnosis and Treatment,” Cancers, vol. 15, p. 3474, 2023. [CrossRef]
- F. L. Guest, H. Rahmoune, and P. C. Guest, “Early diagnosis and targeted treatment strategy for improved therapeutic outcomes in Alzheimer’s disease,” Reviews on New Drug Targets in Age-Related Disorders, pp. 175-191, 2020. [CrossRef]
- S.-H. Wang, “DSSAE: Deep Stacked Sparse Autoencoder Analytical Model for COVID-19 Diagnosis by Fractional Fourier Entropy,” ACM Transactions on Management Information Systems, vol. 13, Article ID: 2, 2021. [CrossRef]
- C. Yen, C.-L. Lin, and M.-C. Chiang, “Exploring the Frontiers of Neuroimaging: A Review of Recent Advances in Understanding Brain Functioning and Disorders,” Life, vol. 13, p. 1472, 2023. [CrossRef]
- J. Sun, “Preliminary study on angiosperm genus classification by weight decay and combination of most abundant color index with fractional Fourier entropy,” Multimedia Tools and Applications, vol. 77, pp. 22671-22688, 2018. [CrossRef]
- Lu, W.-X. Ren, and S.-D. Wang, “Fractional Fourier transform: Time-frequency representation and structural instantaneous frequency identification,” Mechanical Systems and Signal Processing, vol. 178, p. 109305, 2022. [CrossRef]
- Tripathi, R. Singh, and U. Pandey, “Effective Independent Component Analysis Algorithm (EICA) for Blind Source Separation of Mixed Images for Biomedical Applications,” in Proceedings of Trends in Electronics and Health Informatics: TEHI 2021, ed: Springer, 2022, pp. 311-327. [CrossRef]
- B. M. S. Hasan and A. M. Abdulazeez, “A review of principal component analysis algorithm for dimensionality reduction,” Journal of Soft Computing and Data Mining, vol. 2, pp. 20-30, 2021. [CrossRef]
- Sharma, S. Patel, and U. R. Acharya, “Automated detection of abnormal EEG signals using localized wavelet filter banks,” Pattern Recognition Letters, vol. 133, pp. 188-194, 2020. [CrossRef]
- E. Miller, K. Dos Santos, R. Marshall, and I. Kougioumtzoglou, “Joint time-frequency analysis of dynamic cerebral autoregulation using generalized harmonic wavelets,” Physiological measurement, vol. 41, p. 024002, 2020. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).