Submitted:
30 October 2023
Posted:
31 October 2023
You are already at the latest version
Abstract
Keywords:
Introduction
- Traditional vaccine development techniques are time-consuming; conversely, in-silico epitope-based vaccines are developed quickly with high precision. [2]
- Reverse vaccinology, a branch of bioinformatics, can find novel antigens that conventional approaches can miss yet are essential for the immunogenicity of next-generation vaccines.
- Furthermore, unlike traditional vaccines that might require adjuvants to bolster the immune response, mRNA vaccines have been shown to elicit strong immunity without them [3].
- Additionally, mRNA vaccines don't have the risk of vector immunity, a concern with viral vector vaccines where prior exposure to the vector might reduce vaccine efficacy [4].
- Another notable advantage is the scalability of mRNA vaccine production, as they can be synthesized without the need for cell cultures, facilitating consistent and large-scale production [5].
- Importantly, mRNA does not integrate into the host genome, alleviating concerns associated with insertional mutagenesis [6].
- The first-generation COVID-19 mRNA vaccines posed challenges with ultra-cold storage requirements that are now resolved with improved formulations. [7]
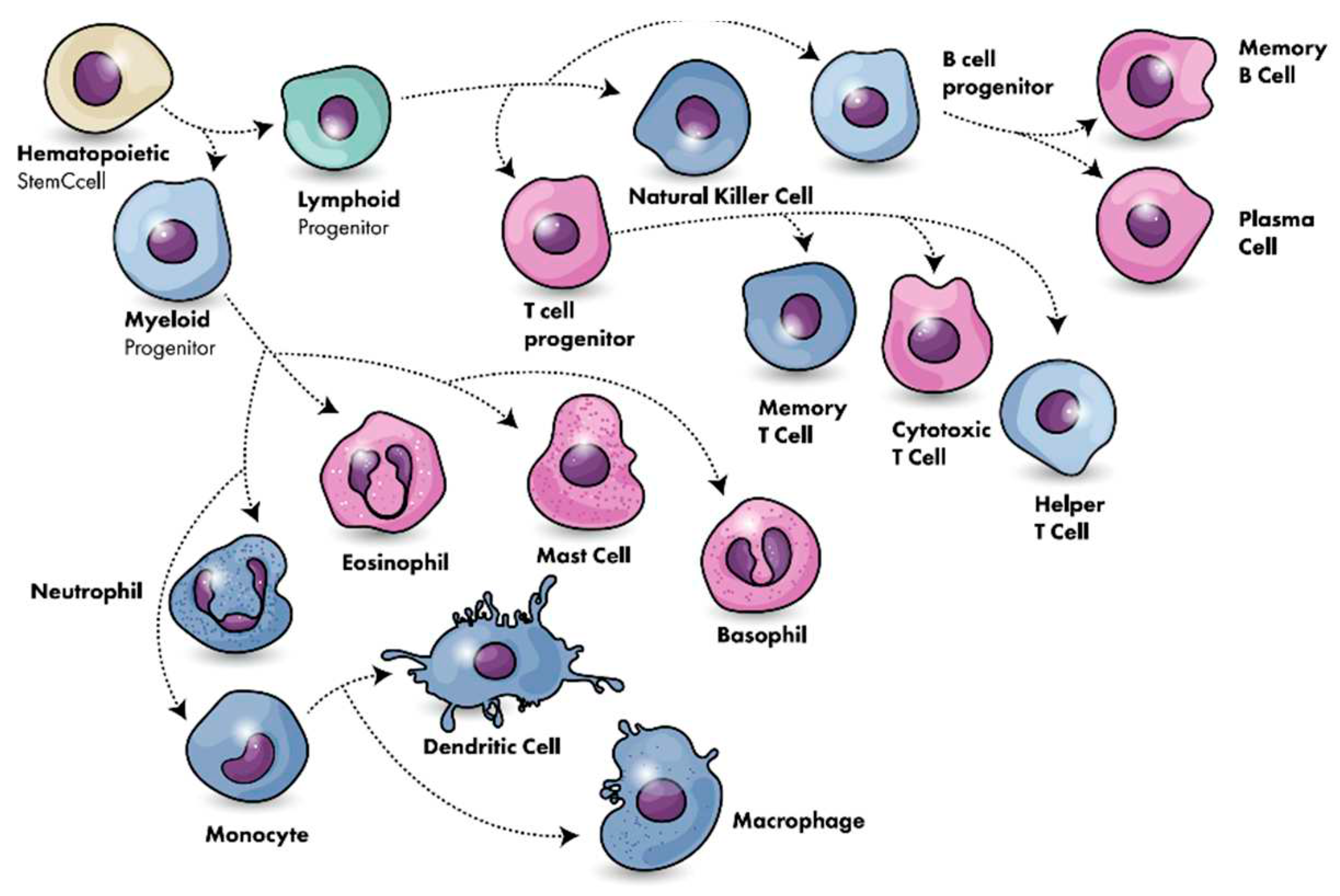
Immune System
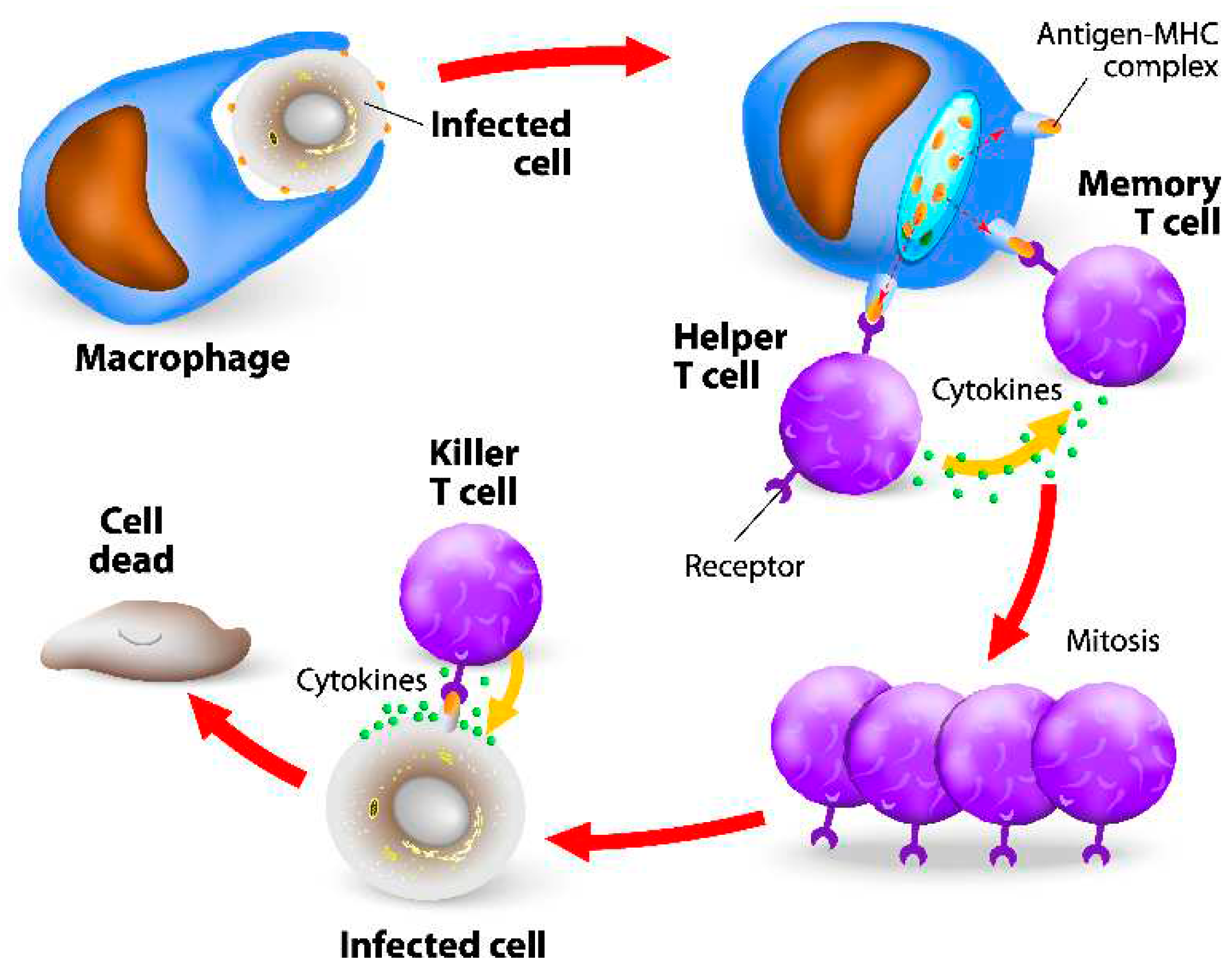
Cellular Immunity
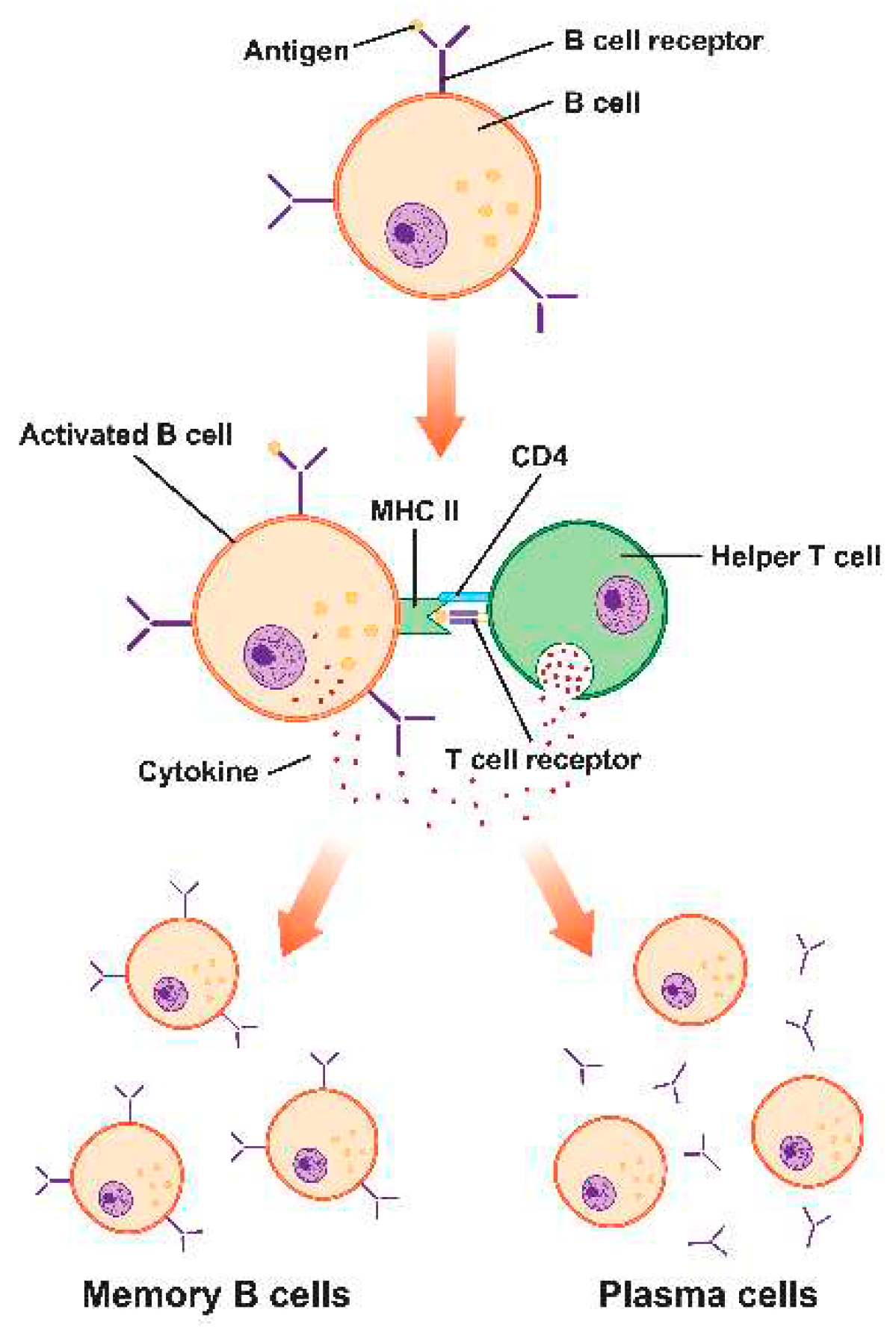
Humoral Immunity
Vaccine Types
Attenuated cell
Protein-based vaccines
Viral vector
Viral antigen
Nucleic acid
Dendritic cell
Adjuvants
Nucleoside Vaccines
DNA Vaccines
mRNA Vaccines
Longer-Term mRNA Vaccines
Specificity
Reverse Vaccinology
B-cell Epitopes
- disposal of short epitopes; [115]
- analysis of candidate antigens' fundamental properties (such as hydrophilicity, flexibility, surface exposure, and solvent availability); [116]
- use of multimethod BCEs prediction strategies; [117]
- comparison of web-based BCEs prediction tool results with results from molecular interaction strategies like molecular docking and molecular dynamics simulations. [118]
T-cell Epitopes
Mutations
Targeted Vaccine Delivery
Multi-target Vaccines
Cancer Vaccines
Other Autoimmune Disorder Vaccines
Intellectual Property
10. Conclusions
Funding
Conflicts of Interest
References
- Jackson, N.A.C., Kester, K.E., Casimiro, D., Gurunathan, S., & DeRosa, F. (2020). The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vaccines, 5(1), 11. [CrossRef]
- Vivona S, Gardy JL, Ramachandran S, Brinkman FS, Raghava GP, Flower DR, Filippini F. Computer-aided biotechnology: from immuno-informatics to reverse vaccinology. Trends Biotechnol. 2008 Apr;26(4):190-200. [CrossRef]
- Pardi, N., Hogan, M.J., Porter, F.W., & Weissman, D. (2018). mRNA vaccines—a new era in vaccinology. Nature Reviews Drug Discovery, 17(4), 261-279. [CrossRef]
- Ewer, K.J., Lambe, T., Rollier, C.S., Spencer, A.J., Hill, A.V., & Dorrell, L. (2016). Viral vectors as vaccine platforms: from immunogenicity to impact. Current Opinion in Immunology, 41, 47-54. [CrossRef]
- Crommelin, D.J.A., Anchordoquy, T.J., Volkin, D.B., Jiskoot, W., & Mastrobattista, E. (2021). Addressing the Cold Reality of mRNA Vaccine Stability. Journal of Pharmaceutical Sciences, 110(3), 997-1001. Authors. [CrossRef]
- Kowalski, P.S., Rudra, A., Miao, L., & Anderson, D.G. (2019). Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Molecular Therapy, 27(4), 710-728. [CrossRef]
- Moderna. (2021). Moderna’s COVID-19 Vaccine Candidate Meets its Primary Efficacy Endpoint in the First Interim Analysis of the Phase 3 COVE Study. [Press release].
- Freyn, A.W., Ramos da Silva, J., Rosado, V.C., Bliss, C.M., Pine, M., Mui, B.L., Sahin, U. (2020). A multi-targeting, nucleoside-modified mRNA influenza virus vaccine provides broad protection in mice. Molecular Therapy, 28(7), 1569-1584. [CrossRef]
- Polack, F.P., Thomas, S.J., Kitchin, N., Absalon, J., Gurtman, A., Lockhart, S., Perez, J.L. (2020). Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. New England Journal of Medicine, 383(27), 2603-2615. [CrossRef]
- Dolgin, E. (2021). The tangled history of mRNA vaccines. Nature, 597(7874), 318-324. [CrossRef]
- Alameh, M.G., Weissman, D., & Pardi, N. (2021). Messenger RNA-Based Vaccines Against Infectious Diseases. Current Topics in Microbiology and Immunology, 429, 1-40. [CrossRef]
- Vaccine Tracker. https://covid19.trackvaccines.org (accessed on 1 September 2023).
- WHO Covid-19 Vaccine Landscape. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 1 September 2023).
- WHO Covid Dashboard. https://covid19.who.int (accessed on 1 September 2023).
- https://clinicaltrials.gov/search?cond=SARS-CoV2&intr=Vaccine (accessed on 1 September 2023).
- https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 1 September 2023).
- Nobel Prize 2023 Medicine or Physiology. https://www.nobelprize.org/prizes/medicine/2023/press-release/ (accessed on 12 October 2023).
- Sahin, U., & Türeci, Ö. (2018). Personalized vaccines for cancer immunotherapy. Science, 359(6382), 1355-1360. [CrossRef]
- Pardi, N., Tuyishime, S., Muramatsu, H., Kariko, K., & Weissman, D. (2015). Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. Journal of Controlled Release, 217, 345-351. [CrossRef]
- Rojas LA, Sethna Z, Soares KC, Olcese C, Pang N, Patterson E, Lihm J, Ceglia N, Guasp P, Chu A, et al., Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature. 2023 Jun;618(7963):144-150. [CrossRef]
- Holtkamp S, Kreiter S, Selmi A, Simon P, Koslowski M, Huber C, Türeci O, Sahin U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006 Dec 15;108(13):4009-17. [CrossRef]
- Barbier AJ, Jiang AY, Zhang P, Wooster R, Anderson DG. The clinical progress of mRNA vaccines and immunotherapies. Nat Biotechnol. 2022 Jun;40(6):840-854. [CrossRef]
- Liu C, Shi Q, Huang X, Koo S, Kong N, Tao W. mRNA-based cancer therapeutics. Nat Rev Cancer. 2023 Aug;23(8):526-543. [CrossRef]
- Pubmed Search. https://pubmed.ncbi.nlm.nih.gov/?term=mrna+vaccine&filter=datesearch.y_5&sort=pubdate (accessed on 1 September 2023).
- Rajpurkar P, Chen E, Banerjee O, Topol EJ. AI in health and medicine. Nat Med. 2022 Jan;28(1):31-38. [CrossRef]
- Brenden K. Petersen, Jiachen Yang, Will S. Grathwohl, Chase Cockrell, Claudio Santiago, Gary An, and Daniel M. Faissol, Deep Reinforcement Learning and Simulation as a Path Toward Precision Medicine Journal of Computational Biology 2019 26:6, 597-604. [CrossRef]
- Goudsmit, J., van den Biggelaar, A.H.J., Koudstaal, W. et al. Immune age and biological age as determinants of vaccine responsiveness among elderly populations: the Human Immunomics Initiative research program. Eur J Epidemiol 36, 753–762 (2021). [CrossRef]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al.,. Highly accurate protein structure prediction with AlphaFold. Nature. 2021 Aug;596(7873):583-589. [CrossRef]
- Hie, B.L., Shanker, V.R., Xu, D. Efficient evolution of human antibodies from general protein language models. Nat Biotechnol (2023). [CrossRef]
- Ong, E., He, Y. (2022). Vaccine Design by Reverse Vaccinology and Machine Learning. Methods in molecular biology (Clifton, N.J.), 2414, 1–16. [CrossRef] [PubMed]
- Parihar, A., Malviya, S., Khan, R. (2022). Immunoinformatics and reverse vaccinomic approaches for effective design. Computational Approaches for Novel Therapeutic and Diagnostic Designing to Mitigate SARS-CoV-2 Infection, 357–378. [CrossRef]
- Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012 Jan 27;36(1):142-52.
- Chaplin, D.D. (2010). Overview of the immune response. The Journal of allergy and clinical immunology, 125(2 Suppl 2), S3–S23. [CrossRef]
- Kantari, C., Pederzoli-Ribeil, M., Witko-Sarsat, V. (2008). The role of neutrophils and monocytes in innate immunity. Contributions to microbiology, 15, 118–146. [CrossRef]
- Cano RLE, Lopera HDE. Introduction to T and B lymphocytes. In: Anaya JM, Shoenfeld Y, Rojas-Villarraga A, et al., editors. Autoimmunity: From Bench to Bedside Bogota (Colombia): El Rosario University Press; 2013 Jul 18. Chapter 5. (accessed on 10 July 2023).
- Wilkinson, T.M.; Li, C.K.F.; Chui, C.S.C.; Huang, A.K.Y.; Perkins, M.; Liebner, J.C.; Lambkin-Williams, R.; Gilbert, A.S.; Oxford, J.; Nicholas, B.; et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012, 18, 274–280. [Google Scholar] [CrossRef]
- Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, Peters B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019 Jan 8;47(D1):D339-D343. [CrossRef]
- Loyal, L.; Braun, J.; Henze, L.; Kruse, B.; Dingeldey, M.; Reimer, U.; Kern, F.; Schwarz, T.; Mangold, M.; Unger, C.; et al. Cross-reactive CD4+ T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science 2021, 374, eabh1823. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Garcia-Ibanez, L.; Ulbricht, C.; Lok, L.S.C.; Pike, J.A.; Mueller-Winkler, J.; Dennison, T.W.; Ferdinand, J.R.; Burnett, C.J.M.; Yam-Puc, J.C.; et al. Recycling of memory B cells between germinal center and lymph node subcapsular sinus supports affinity maturation to antigenic drift. Nat. Commun. 2022, 13, 2460. [Google Scholar] [CrossRef] [PubMed]
- Liu J, Chandrashekar A, Sellers D, Barrett J, Jacob-Dolan C, Lifton M, McMahan K, Sciacca M, VanWyk H, Wu C, Yu J, Collier AY, Barouch DH. Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 Omicron. Nature. 2022 Mar;603(7901):493-496. [CrossRef]
- Janeway, C. A., Travers, P., Walport, M., & Shlomchik, M. J. (2001). Immunobiology: The Immune System in Health and Disease (5th ed.). Garland Science.
- Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., & Walter, P. (2002). Molecular Biology of the Cell (4th ed.). Garland Science.
- Murphy, K., Travers, P., Walport, M., & Janeway, C. (2012). Janeway's Immunobiology (8th ed.). Garland Science.
- Harty, J.T.; Tvinnereim, A.R.; White, D.W. CD8+ T cell effector mechanisms in resistance to infection. Annu. Rev. Immunol. 2000, 18, 275–308. [Google Scholar] [CrossRef]
- Marrack, P., McKee, A. S. (2017) MHC class II antigen processing in immune tolerance and inflammation. Annual Review of Immunology, 35, 13-27.
- GAVI: COVAX explained. Retrieved from. https://www.gavi.org/vaccineswork/covax-explained (accessed on 10 July 2023).
- Ura, T., Takeuchi, M., Kawagoe, T., Mizuki, N., Okuda, K., Shimada, M. (2022). Current Vaccine Platforms in Enhancing T-Cell Response. Vaccines, 10(8), 1367. [CrossRef]
- Crotty, S. (2011) Follicular helper CD4 T-cells (TFH). Annual Review of Immunology, 29, 621-663.
- Plotkin, S. A. (2008) Vaccines: correlates of vaccine-induced immunity. Clinical Infectious Diseases, 47(3), 401-409.
- Gilbert, S.C. T-cell-inducing vaccines—What’s the future. Immunology 2012, 135, 19–26. [Google Scholar] [CrossRef]
- Colditz, G.A.; Brewer, T.F.; Berkey, C.S.; Wilson, M.E.; Burdick, E.; Fineberg, H.V.; Mosteller, F. Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. JAMA 1994, 271, 698–702. [Google Scholar] [CrossRef]
- Mishra R, Hannebelle M, Patil VP, Dubois A, Garcia-Mouton C, Kirsch GM, Jan M, Sharma K, Guex N, Sordet-Dessimoz J, er al., Mechanopathology of biofilm-like Mycobacterium tuberculosis cords. Cell. 2023 Oct 11:S0092-8674(23)01037-1. [CrossRef]
- Prabhudas, M.; Bonney, E.; Caron, K.; Dey, S.; Erlebacher, A.; Fazleabas, A.; Fisher, S.; Golos, T.; Matzuk, M.; McCune, J.M.; et al. Immune mechanisms at the maternal-fetal interface: Perspectives and challenges. Nat. Immunol. 2015, 16, 328–334. [Google Scholar] [CrossRef]
- Guthmiller, J.J.; Stovicek, O.; Wang, J.; Changrob, S.; Li, L.; Halfmann, P.; Zheng, N.Y.; Utset, H.; Stamper, C.T.; Dugan, H.L.; et al. SARS-CoV-2 Infection Severity Is Linked to Superior Humoral Immunity against the Spike. MBio 2021, 12, e02940-20. [Google Scholar] [CrossRef]
- Turner JS, O'Halloran JA, Kalaidina E, Kim W, Schmitz AJ, Zhou JQ, Lei T, Thapa M, Chen RE, Case JB, er al., SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021 Aug;596(7870):109-113. [CrossRef]
- Goel RR, Painter MM, Apostolidis SA, Mathew D, Meng W, Rosenfeld AM, Lundgreen KA, Reynaldi A, Khoury DS, Pattekar A, Gouma S, Kuri-Cervantes L, Hicks P, Dysinger S, Hicks A, Sharma H, Herring S, Korte S, Baxter AE, Oldridge DA, Giles JR, Weirick ME, McAllister CM, Awofolaju M, Tanenbaum N, Drapeau EM, Dougherty J, Long S, D'Andrea K, Hamilton JT, McLaughlin M, Williams JC, Adamski S, Kuthuru O; UPenn COVID Processing Unit‡; Frank I, Betts MR, Vella LA, Grifoni A, Weiskopf D, Sette A, Hensley SE, Davenport MP, Bates P, Luning Prak ET, Greenplate AR, Wherry EJ. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021 Dec 3;374(6572):abm0829. [CrossRef]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020, 130, 2620–2629. [Google Scholar] [CrossRef]
- Leung, S.; Liu, X.; Fang, L.; Chen, X.; Guo, T.; Zhang, J. The cytokine milieu in the interplay of pathogenic Th1/Th17 cells and regulatory T cells in autoimmune disease. Cell Mol. Immunol. 2010, 7, 182–189. [Google Scholar] [CrossRef]
- Khader, S.A.; Gaffen, S.L.; Kolls, J.K. Th17 cells at the crossroads of innate and adaptive immunity against infectious diseases at the mucosa. Mucosal. Immunol. 2009, 2, 403–411. [Google Scholar] [CrossRef]
- Ndure, J.; Flanagan, K.L. Targeting regulatory T cells to improve vaccine immunogenicity in early life. Front. Microbiol. 2014, 5, 477. [Google Scholar] [CrossRef]
- Amanat F, Krammer F. SARS-CoV-2 Vaccines: Status Report. Immunity. 2020;52(4):583–9.
- Chen J, Ting N. Design Considerations for Vaccine Trials with a Special Focus on COVID-19 Vaccine Development. Data Sci J. 2021;18(3):550–80. [CrossRef]
- Borah P, Deb PK, Deka S, Venugopala KN, Singh V, Mailavaram RP, Kalia K, Tekade RK. Current Scenario and Future Prospect in the Management of COVID-19. Curr Med Chem. 2021;28(2):284-307. [CrossRef]
- Kim MH, Kim HJ, Chang J. Superior immune responses induced by intranasal immunization with recombinant adenovirus-based vaccine expressing full-length Spike protein of Middle East respiratory syndrome Coronavirus. PLoS One. 2019;14(7). e0220196. [CrossRef]
- Bouhaddou M, Reuschl A-K, Polacco BJ, Thorne LG, Ummadi MR, Ye C, et al. SARS-CoV-2 variants evolve convergent strategies to remodel the host response. Cell. 2023;186(21):4597-614.e26.
- Plotkin, S. A. (2010) Correlates of protection induced by vaccination. Clinical and Vaccine Immunology, 17(7), 1055-1065. [CrossRef]
- Palucka, K., Banchereau, J. (2012) Cancer immunotherapy via dendritic cells. Nature Reviews Cancer, 12(4), 265-277. [CrossRef]
- Zhen, A., Kamata, M. (2020) Viral vectors for the delivery of human genes. Journal of Cellular Physiology, 235(4), 3235-3249. [CrossRef]
- Sadoff, J.; Le Gars, M.; Shukarev, G.; Heerwegh, D.; Truyers, C.; de Groot, A.M.; Stoop, J.; Tete, S.; Damme, W.V.; Leroux-Roels, I.; et al. Interim Results of a Phase 1–2a Trial of Ad26.COV2.S COVID-19 Vaccine. N. Engl. J. Med. 2021, 384, 1824–1835. [Google Scholar] [CrossRef]
- Pollard, A.J.; Bijker, E.M. A guide to vaccinology: From basic principles to new developments. Nat. Rev. Immunol. 2021, 21, 83–100. [Google Scholar] [CrossRef]
- Hokello, J.; Sharma, A.L.; Tyagi, M. An Update on the HIV DNA Vaccine Strategy. Vaccines 2021, 9, 605. [Google Scholar] [CrossRef]
- Schenkel, J.M.; Masopust, D. Tissue-resident memory T cells. Immunity 2014, 41, 886–897. [Google Scholar] [CrossRef]
- Yusuf, H.; Kett, V. Current prospects and future challenges for nasal vaccine delivery. Hum. Vaccin Immunother. 2017, 13, 34–45. [Google Scholar] [CrossRef]
- Menon, I.; Bagwe, P.; Gomes, K.; Bajaj, L.; Gala, R.; Uddin, M.; D’Souza, M.; Zughaier, S. Microneedles: A New Generation Vaccine Delivery System. Micromachines 2021, 12, 435. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos, O.; James, E.; Fernandez, A.; Lopes, V.; Rosas, L.A.; Cervantes-Medina, A.; Cleath, J.; Edwards, K.; Neitzey, D.; Gu, W.; et al. Efficacy of FLU-v, a broad-spectrum influenza vaccine, in a randomized phase IIb human influenza challenge study. NPJ Vaccines 2020, 5, 22. [Google Scholar] [CrossRef]
- Atsmon, J.; Kate-Ilovitz, E.; Shaikevich, D.; Singer, Y.; Volokhov, I.; Haim, K.Y.; Ben-Yedidia, T. Safety and immunogenicity of multimeric-001—A novel universal influenza vaccine. J. Clin. Immunol. 2012, 32, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Coffman, R. L., Sher, A., Seder, R. A. (2010). Vaccine adjuvants: Putting innate immunity to work. Immunity, 33(4), 492-503. [CrossRef]
- Krieg, A. M. (2002) CpG motifs in bacterial DNA and their immune effects. Annual Review of Immunology, 20, 709-760. [CrossRef]
- Lynn GM, Laga R, Darrah PA, Ishizuka AS, Balaci AJ, Dulcey AE, Pechar M, Pola R, Gerner MY, Yamamoto A. In vivo characterization of the physicochemical properties of polymer-linked TLR agonists that enhance vaccine immunogenicity. Nat Biotechnol. 2015 Nov;33(11):1201-10. [CrossRef]
- Graham, B. S., Sullivan, N. J. (2018) Emerging viral diseases from a vaccinology perspective: preparing for the next pandemic. Nature Immunology, 19(1), 20-28. [CrossRef]
- Shah, N.J.; Najibi, A.J.; Shih, T.-Y.; Mao, A.S.; Sharda, A.; Scadden, D.T.; Mooney, D.J. A biomaterial-based vaccine eliciting durable tumour-specific responses against acute myeloid leukaemia. Nat. Biomed. Eng. 2020, 4, 40–51. [Google Scholar] [CrossRef]
- Liu, H.; Moynihan, K.; Zheng, Y.; Szeto, G.; Li, A.V.; Huang, B.; Van Egeren, D.; Park, C.; Irvine, D.J. Structure-based programming of lymph-node targeting in molecular vaccines. Nature 2014, 507, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Pollard C, De Koker S, Saelens X, Vanham G, Grooten J. Challenges and advances towards the rational design of mRNA vaccines. Trends Mol Med. 2013;19(12):705–13. [CrossRef]
- Kanampalliwar, AM. Reverse Vaccinology and Its Applications. Methods Mol Biol. 2020;2131:1–16. [CrossRef]
- Cheung YK, Cheng SC, Sin FW, Chan KT, Xie Y. Induction of T- cell response by a DNA vaccine encoding a novel HLA-A*0201 severe acute respiratory syndrome coronavirus epitope. Vaccine. 2007;25(32):6070–7. [CrossRef]
- Wang X, Xu W, Tong D, Ni J, Gao H, Wang Y, Chu Y, Li P, Yang X, Xiong S. A chimeric multi-epitope DNA vaccine elicited specific antibody response against severe acute respiratory syndrome-associated coronavirus which attenuated the virulence of SARS-CoV in vitro. Immunol Lett. 2008 Aug 15;119(1-2):71-7. [CrossRef]
- Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE,; Oxford COVID Vaccine Trial Group. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021 Jan 9;397(10269):99-111. [CrossRef]
- Alarcon JB, Waine GW, McManus DP (1999). "DNA Vaccines: Technology and Application as Anti-parasite and Anti-microbial Agents". Advances in Parasitology Volume 42. Vol. 42. pp. 343–410. [CrossRef]
- Robinson HL, Pertmer TM (2000). DNA vaccines for viral infections: basic studies and applications. Advances in Virus Research. Vol. 55. pp. 1–74. [CrossRef]
- Sheridan, C. First COVID-19 DNA vaccine approved, others in hot pursuit. Nat. Biotechnol. 2021, 39, 1479–1482. [Google Scholar] [CrossRef]
- India’s DNA COVID vaccine is a world first – more are coming. https://www.nature.com/articles/d41586-021-02385-x (accessed on 10 July 2023).
- Khobragade A, Bhate S, Ramaiah V, Deshpande S, Giri K, Phophle H, Supe P, Godara I, Revanna R, Nagarkar R, Sanmukhani J, Dey A, Rajanathan TMC, Kansagra K, Koradia P; ZyCoV-D phase 3 Study Investigator Group. Efficacy, safety, and immunogenicity of the DNA SARS-CoV-2 vaccine (ZyCoV-D): the interim efficacy results of a phase 3, randomised, double-blind, placebo-controlled study in India. Lancet. 2022 Apr 2;399(10332):1313-1321. [CrossRef]
- Wolff, J.A.; Malone, R.W.; Williams, P.; Chong, W.; Acsadi, G.; Jani, A.; Felgner, P.L. Direct gene transfer into mouse muscle in vivo. Science 1990, 247, 1465–1468. [Google Scholar] [CrossRef]
- Mothe, B., Brander, C. (2016) HIV T-cell vaccines. Human Vaccines Immunotherapeutics, 12(8), 2013-2016.
- Colombani T, Haudebourg T, Pitard B. 704/DNA vaccines leverage cytoplasmic DNA stimulation to promote anti-HIV neutralizing antibody production in mice and strong immune response against alpha-fetoprotein in non-human primates. Mol Ther Nucleic Acids. 2023 May 4;32:743-757. [CrossRef]
- Lim, W. A., June, C. H. (2017) The principles of engineering immune cells to treat cancer. Cell, 168(4), 724-740. [CrossRef] [PubMed]
- Twyman-Saint Victor C, Rech AJ, Maity A, Rengan R, Pauken KE, Stelekati E, Benci JL, Xu B, Dada H, Odorizzi PM, Herati RS, et al., Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015 Apr 16;520(7547):373-7. [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Buddy Creech, C.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.B.; Kanevsky, I.; Che, Y.; Swanson, K.A.; Muik, A.; Vormehr, M.; Kranz, L.M.; Walzer, K.C.; Hein, S.; Güler, A.; et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 2021, 592, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Niazi, SK. RNA Therapeutics: A Healthcare Paradigm Shift. Biomedicines. 2023 Apr 25;11(5):1275. [CrossRef]
- Oleszycka, E., Lavelle, E. C. (2014) Immunomodulatory properties of the vaccine adjuvant alum. Current Opinion in Immunology, 28, 1-5. 28, 1–5. [CrossRef] [PubMed]
- Agnandji ST, Lell B, Fernandes JF, Abossolo BP, Methogo BG, Kabwende AL, Adegnika AA, Mordmüller B, Issifou S, Kremsner PG, A phase 3 trial of RTS,S/AS01 malaria vaccine in African infants. N Engl J Med. 2012 Dec 13;367(24):2284-95. [CrossRef]
- Khurana, S., Verma, N. (2021) MF59 adjuvant: The best insurance against influenza strain diversity. Expert Review of Vaccines, 20(8), 947-951. [CrossRef]
- Gustiananda M, Sulistyo BP, Agustriawan D, Andarini S. Immunoinformatics Analysis of SARS-CoV-2 ORF1ab Polyproteins to Identify Promiscuous and Highly Conserved T-Cell Epitopes to Formulate Vaccine for Indonesia and the World Population. Vaccines (Basel). 2021 Dec 9;9(12):1459. [CrossRef]
- Mitra R, Das J, Kamruzzaman M. Application of TOPSIS method for flood susceptibility mapping using Excel and GIS. MethodsX. 2023 Jun 15;11:102263. [CrossRef]
- Bagherzadeh MA, Izadi M, Baesi K, Jahromi MAM, Pirestani M. Considering epitopes conservity in targeting SARS-CoV-2 mutations in variants: a novel immunoinformatics approach to vaccine design. Sci Rep. 2022 Aug 18;12(1):14017. [CrossRef]
- Moss P. The T cell immune response against SARS-CoV-2. Nat Immunol. 2022;23:186–193. [CrossRef]
- Olukitibi TA, Ao Z, Warner B, Unat R, Kobasa D, Yao X. Significance of Conserved Regions in Coronavirus Spike Protein for Developing a Novel Vaccine against SARS-CoV-2 Infection. Vaccines (Basel). 2023 Feb 24;11(3):545. [CrossRef]
- Heinson, A. I., Woelk, C. H., Newell, M. L. (2015). The promise of reverse vaccinology. International health, 7(2), 85–89. [CrossRef] [PubMed]
- Pritam M, Singh G, Swaroop S, Singh AK, Singh SP. Exploitation of reverse vaccinology and immunoinformatics as promising platform for genome-wide screening of new effective vaccine candidates against Plasmodium falciparum. BMC Bioinformatics. 2019 Feb 4;19(Suppl 13):468. [CrossRef]
- Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017 Jul 3;45(W1):W24-W29. [CrossRef]
- Larsen JE, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2. [CrossRef]
- Saha S, Raghava GP. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65(1):40–8. [CrossRef]
- Rahman Kh S, Chowdhury EU, Sachse K, Kaltenboeck B. Inadequate Reference Datasets Biased toward Short Non-epitopes Confound B-cell Epitope Prediction. J Biol Chem. 2016;291(28):14585–99. [CrossRef]
- Ansari HR, Raghava GP. Identification of conformational B-cell Epitopes in an antigen from its primary sequence. Immunome Res. 2010;6:6. [CrossRef]
- Parvizpour S, Razmara J, Pourseif MM, Omidi Y. In silico design of a triple-negative breast cancer vaccine by targeting cancer testis antigens. Bioimpacts. 2019;9(1):45–56. [CrossRef]
- Potocnakova L, Bhide M, Pulzova LB. An Introduction to B-Cell Epitope Mapping and In Silico Epitope Prediction. J Immunol Res. 2016;2016:6760830. [CrossRef]
- Lundegaard C, Hoof I, Lund O, Nielsen M. State of the art and challenges in sequence based T-cell epitope prediction. Immunome Res. 2010;6 Suppl 2. S3. [CrossRef]
- Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L,. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017 Jul 13;547(7662):217-221. [CrossRef]
- Burgos RM, Badowski ME, Drwiega E, Ghassemi S, Griffith N, Herald F, Johnson M, Smith RO, Michienzi SM. The race to a COVID-19 vaccine: opportunities and challenges in development and distribution. Drugs Context. 2021 Feb 16;10:2020-12-2. [CrossRef]
- Mills MC, Salisbury D. The challenges of distributing COVID-19 vaccinations. EClinicalMedicine. 2021;31:100674. [CrossRef]
- Krause, P.R.; Fleming, T.R.; Longini, I.M.; Peto, R.; Briand, S.; Heymann, D.L.; Beral, V.; Snape, M.D.; Rees, H.; Ropero, A.M.; et al. SARS-CoV-2 Variants and Vaccines. N. Engl. J. Med. 2021, 385, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Accorsi, E.K.; Britton, A.; Fleming-Dutra, K.E.; Smith, Z.R.; Shang, N.; Derado, G.; Miller, J.; Schrag, S.J.; Verani, J.R. Association Between 3 Doses of mRNA COVID-19 Vaccine and Symptomatic Infection Caused by the SARS-CoV-2 Omicron and Delta Variants. JAMA 2022, 327, 639–651. [Google Scholar] [CrossRef]
- Hoffmann, M.; Krüger, N.; Schulz, S.; Cossmann, A.; Rocha, C.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Moldenhauer, A.-S.; Winkler, M.S.; et al. The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell 2021, 185, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Tarke, A.; Sidney, J.; Kidd, C.K.; Dan, J.M.; Ramirez, S.I.; Yu, E.D.; Mateus, J.; da Silva Antunes, R.; Moore, E.; Rubiro, P.; et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021, 2, 100204. [Google Scholar] [CrossRef]
- Swadling, L.; Diniz, M.O.; Schmidt, N.M.; Amin, O.E.; Chandran, A.; Shaw, E.; Pade, C.; Gibbons, J.M.; Le Bert, N.; Tan, A.T.; et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature 2022, 601, 110–117. [Google Scholar] [CrossRef]
- Mallajosyula, V.; Ganjavi, C.; Chakraborty, S.; McSween, A.M.; Pavlovitch-Bedzyk, A.J.; Wilhelmy, J.; Nau, A.; Manohar, M.; Nadeau, K.C.; Davis, M.M. CD8+ T cells specific for conserved coronavirus epitopes correlate with milder disease in COVID-19 patients. Sci. Immunol. 2021, 6, 61. [Google Scholar] [CrossRef] [PubMed]
- Gangaev, A.; Ketelaars, S.L.C.; Isaeva, O.I.; Patiwael, S.; Dopler, A.; Hoefakker, K.; Biasi, S.D.; Gibellini, L.; Mussini, C.; Guaraldi, G.; et al. Identification and characterization of a SARS-CoV-2 specific CD8+ T cell response with immunodominant features. Nat. Commun. 2021, 12, 2593. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov I, Flower DR, Doytchinova I. AllerTOP--a server for in silico prediction of allergens. BMC Bioinformatics. 2013;14 Suppl 6(Suppl 6):S4. [CrossRef]
- Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007 Jan 5;8:4. [CrossRef]
- http://www.ddg-pharmfac.net/vaxijen/ (accessed on 10 July 2023).
- https:// webs.iiitd.edu.in/raghava/toxinpred/design.php (accessed on 10 July 2023).
- http://tools.iedb.org/ellipro/ (accessed on 10 July 2023).
- Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics. 2006 Mar 17;7:153. [CrossRef]
- Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016 Jul 8;44(W1):W449-54. [CrossRef]
- Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W363-7. [CrossRef]
- Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W229-32. [CrossRef]
- https:// web.expasy.org/protparam (accessed on 10 July 2023).
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011 Sep 29;8(10):785-6. [CrossRef]
- https://zhanglab.ccmb.med.umich.edu/I-TASSER/ (accessed on 10 July 2023).
- https://zhanggroup.org/ModRefiner/ (accessed on 10 July 2023).
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W407-10. [CrossRef]
- https:// servicesn.mbi.ucla.edu/ERRAT/ (accessed on 10 July 2023).
- https:// servicesn.mbi.ucla.edu/PROCHECK/ (accessed on 10 July 2023).
- https://www.rcsb.org/ (accessed on 10 July 2023).
- https:// cluspro.bu.edu/login.php (accessed on 10 July 2023).
- https://pymol.org/2/ (accessed on 10 July 2023).
- https://kraken.iac.rm.cnr.it/CIMMSIM/ (accessed on 10 July 2023).
- http://apps.cytoscape.org/apps/cytohubb (accessed on 10 July 2023).
- https://services.healthtech.dtu.dk/services/NetMHCpan-4.1/ (accessed on 10 July 2023).
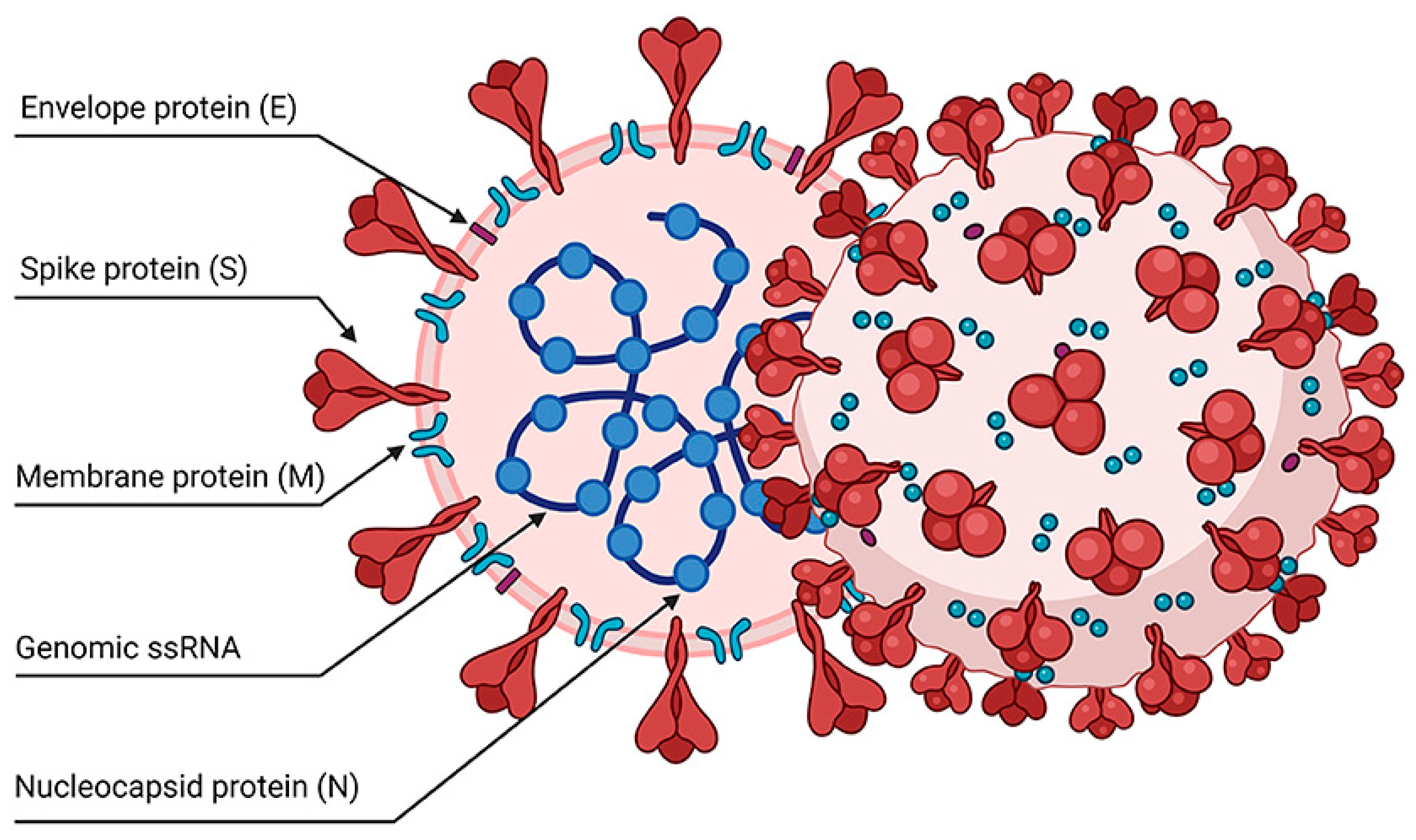
- Poonam, B., Gill, P. K. (2021). Coronavirus: History, genome structure and pathogenesis. Coronaviruses, 2(3), 325-338. [CrossRef]
- Kakavandi, S., Zare, I., Vaezjalali, M., Dadashi, M., Azarian, M., Akbari, A., Farani, M. R., Zalpoor, H., Hajikhani, B. (2023). Structural and non-structural proteins in SARS-CoV-2: potential aspects to COVID-19 treatment or prevention of progression of related diseases. Cell Communication and Signaling, 21(1). [CrossRef]
- Ong, E., Wong, M. U., Huffman, A., He, Y. (2020). COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. Frontiers in immunology, 11, 1581. [CrossRef]
- Zhang S, Gong C, Ruiz-Martinez A, Wang H, Davis-Marcisak E, Deshpande A, Popel AS, Fertig EJ. Integrating single cell sequencing with a spatial quantitative systems pharmacology model spQSP for personalized prediction of triple-negative breast cancer immunotherapy response. Immunoinformatics (Amst). 2021 Oct;1-2:100002. [CrossRef]
- Gustafson, C.E.; Kim, C.; Weyand, C.M.; Goronzy, J.J. Influence of immune aging on vaccine responses. J. Allergy Clin. Immunol. 2020, 145, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Allen H, Vusirikala A, Flannagan J, Twohig KA, Zaidi A, Chudasama D, Lamagni T, Groves N, Turner C, Rawlinson C, et al; COVID-19 Genomics UK (COG-UK Consortium). Household transmission of COVID-19 cases associated with SARS-CoV-2 delta variant (B.1.617.2): national case-control study. Lancet Reg Health Eur. 2022 Jan;12:100252. [CrossRef]
- Zhou B, Thao TTN, Hoffmann D, Taddeo A, Ebert N, Labroussaa F, Pohlmann A, King J, Steiner S, Kelly JN, Portmann J, SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021 Apr;592(7852):122-127.). [CrossRef]
- Jangra S, Ye C, Rathnasinghe R, Stadlbauer D; Personalized Virology Initiative study group; Krammer F, Simon V, Martinez-Sobrido L, García-Sastre A, Schotsaert M. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2021 Jul;2(7):e283-e284. [CrossRef]
- Almubaid, Z. Al-Mubaid, H. Analysis and comparison of genetic variants and mutations of the novel coronavirus SARS-CoV-2. Gene Rep. 23, 101064 (2021). [CrossRef]
- Hassan, S. S., Choudhury, P. P., Basu, P. Jana, S. S. Molecular conservation and differential mutation on ORF3a gene in Indian SARS-CoV2 genomes. Genomics 112, 3226–3237 (2020). [CrossRef]
- Dai Y, Chen H, Zhuang S, Feng X, Fang Y, Tang H, Dai R, Tang L, Liu J, Ma T, Zhong G. Immunodominant regions prediction of nucleocapsid protein for SARS-CoV-2 early diagnosis: a bioinformatics and immunoinformatics study. Pathog Glob Health. 2020 Dec;114(8):463-470.). [CrossRef]
- Garrett ME, Galloway J, Chu HY, Itell HL, Stoddard CI, Wolf CR, Logue JK, McDonald D, Weight H, Matsen FA 4th, Overbaugh J. High-resolution profiling of pathways of escape for SARS-CoV-2 spike-binding antibodies. Cell. 2021 May 27;184(11):2927-2938.e11.).
- Zeng, C.; Zhang, C.; Walker, P.G.; Dong, Y. Formulation and Delivery Technologies for mRNA Vaccines. In Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2020.
- Kenney, R.T.; Frech, S.A.; Muenz, L.R.; Villar, C.P.; Glenn, G.M. Dose sparing with intradermal injection of influenza vaccine. N. Engl. J. Med. 2004, 351, 2295–2301. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.; Naradikian, M.S.; Parkhouse, K.; Cain, D.W.; Jones, L.; Moody, M.A.; Verkerke, H.P.; Myles, A.; Willis, E.; et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018, 215, 1571–1588. [Google Scholar] [CrossRef] [PubMed]
- Holmgren, J.; Czerkinsky, C. Mucosal immunity and vaccines. Nat. Med. 2005, 11, S45–S53. [Google Scholar] [CrossRef]
- Xiong F, Zhang C, Shang B, Zheng M, Wang Q, Ding Y, Luo J, Li X. An mRNA-based broad-spectrum vaccine candidate confers cross-protection against heterosubtypic influenza A viruses. Emerg Microbes Infect. 2023 Dec;12(2):2256422. [CrossRef]
- https://investors.modernatx.com/news/news-details/2023/Moderna-Announces-First-Participant-Dosed-in-Phase-3-Study-of-mRNA-1083-a-Combination-Vaccine-Against-Influenza-and-COVID-19/default.aspx.
- Sahin, U., Derhovanessian, E., Miller, M., Kloke, B.P., Simon, P., Löwer, M., Omokoko, T. (2017). Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature, 547(7662), 222-226. [CrossRef]
- Berg MK, Yu Q, Salvador CE, Melani I, Kitayama S. Mandated Bacillus Calmette-Guerin (BCG) vaccination predicts flattened curves for the spread of COVID-19. Sci Adv. 2020;6(32):eabc1463. [CrossRef]
- Singh, S. BCG vaccines may not reduce COVID-19 mortality rates.; medRxiv 2020.04.11.20062232.
- June CH, O'Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T cell immunotherapy for human cancer. Science. 2018 Mar 23;359(6382):1361-1365. [CrossRef]
- Liu J, Fu M, Wang M, Wan D, Wei Y, Wei X. Cancer vaccines as promising immuno-therapeutics: platforms and current progress. J Hematol Oncol. 2022 Mar 18;15(1):28. [CrossRef]
- Apostolopoulos, V. (2019). Cancer Vaccines: Research and Applications. Cancers, 11(8), 1041. [CrossRef]
- Kübler, H., Scheel, B., Gnad-Vogt, U., Miller, K., Schultze-Seemann, W., Vom Dorp, F. Kallen, K.J. (2015). Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: a first-in-man phase I/IIa study. Journal of Immunotherapy of Cancer, 3(1), 26. [CrossRef]
- Boczkowski, D., Nair, S.K., Snyder, D., & Gilboa, E. (1996). Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. The Journal of Experimental Medicine, 184(2), 465-472. [CrossRef]
- Ribas, A., & Wolchok, J.D. (2018). Cancer immunotherapy using checkpoint blockade. Science, 359(6382), 1350-1355. [CrossRef] [PubMed]
- Reichmuth, A. M., Oberli, M. A., Jaklenec, A., Langer, R., & Blankschtein, D. (2016). mRNA vaccine delivery using lipid nanoparticles. Therapeutic Delivery, 7(5), 319-334. [CrossRef]
- Binnewies, M., Roberts, E. W., Kersten, K., Chan, V., Fearon, D. F., Merad, M., Krummel, M. F. (2018). Understanding the tumor immune microenvironment (TIME) for effective therapy. Nature Medicine, 24(5), 541-550. [CrossRef]
- Vanpouille-Box, C., Alard, A., Aryankalayil, M. J., Sarfraz, Y., Diamond, J. M., Schneider, R. J. Formenti, S. C. (2017). DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nature Communications, 8, 15618. [CrossRef]
- Galluzzi, L., Buqué, A., Kepp, O., Zitvogel, L., & Kroemer, G. (2017). Immunogenic cell death in cancer and infectious disease. Nature Reviews Immunology, 17(2), 97-111. [CrossRef]
- Slaoui, M., & Hepburn, M. (2020). Developing Safe and Effective COVID Vaccines—Operation Warp Speed's Strategy and Approach. The New England Journal of Medicine, 383(18), 1701-1703. [CrossRef]
- Bach, P. B. (2021). The Price of Progress—Cost of the COVID-19 Vaccine. JAMA Health Forum, 2(5), e210524-e210524. [CrossRef]
- Le, I., Dhandayuthapani, S., Chacon, J., Eiring, A. M., and Gadad, S. S. (2022). Harnessing the Immune System with Cancer Vaccines: From Prevention to Therapeutics. Vaccines, 10(5), 816. [CrossRef]
- Schumacher, T. N., Schreiber, R. D. (2015) Neoantigens in cancer immunotherapy. Science, 348(6230), 69-74. [CrossRef] [PubMed]
- Hilf N, Kuttruff-Coqui S, Frenzel K, Bukur V, Stevanović S, Gouttefangeas C, Platten M, Tabatabai G, Dutoit V, et al.,Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. 2019 Jan;565(7738):240-245. Erratum in: Nature. 2019 Feb;566(7745):E13. [CrossRef]
- Gorkhali, R., Koirala, P., Rijal, S., Mainali, A., Baral, A., Bhattarai, H. K. (2021). Structure and function of major SARS-CoV-2 and SARS-CoV proteins. Bioinformatics and biology insights, 15, 11779322211025876. [CrossRef]
| Type Abb. | Vaccine Type | # |
| PS | Protein subunit | 59 |
| RNA | RNA | 43 |
| VVnr | Viral Vector (non-replicating) | 25 |
| IV | Inactivated Virus | 22 |
| DNA | DNA | 17 |
| VLP | Virus-Like Particle | 7 |
| VVr | Viral Vector (replicating) | 4 |
| VVr + APC | VVr + Antigen Presenting Cell | 2 |
| LAV | Live Attenuated Virus | 2 |
| VVnr + APC | VVnr + Antigen Presenting Cell | 1 |
| BacAg-SpV | Bacterial antigen-spore expression vector | 1 |
| Conserved Epitopes | Position | Immune Response Induced | Type of Study | |
| B cells/Neutralizing antibodies | T cells | |||
| YLTPGDSSSGWTAGAAAYYV | 248–267 aa | Yes | Yes | Mathematically (in-house developed PERL scripts), in vivo |
| YYVGYLQPRTFLLKY | 264–278 aa | NT | Yes | Web-based analytic tools |
| VRFPNITNL | 327–335 aa | NT | Yes | Immunoinformatic, in vivo |
| FNATRFASVYAWNRK | 342–356 aa | Yes | Yes | In silico, T-cell epitope mapping, molecular dynamics simulations, immunoinformatic |
| TFKCYGVSPTKLNDL | 376–390 aa | Yes | Yes | Mathematically (in-house developed PERL scripts), bioinformatics, monoclonal antibody targeting |
| PYRVVVLSF | 507–515 aa | NT | Yes | Immunoinformatic |
| LPFQQFGRDIADT | 560–572 aa | Yes | Yes | PepSeq Analysis |
| Testing | Tools |
| Antigenic, non-toxic, and non-allergenic epitopes | AllerTop v2. [130] VaxiJen v.2.0 [131,132] and ToxinPred [133] |
| Linear and discontinuous B-cell epitope prediction | Ellipro [134] |
| Overlapping epitopes | IEDB. [135] |
| Conservancy of epitopes analysis | Infectious Disease Epidemiology Bureau conservancy tool. [135] |
| 3-D structure of screened-out best epitope sequences. | PEPFOLD 3l [136] |
| Epitope and allele interaction pattern | PatchDockr [137] |
| Complex Refining | FireDock [138] |
| Antigenicity anticipation | ANTIGENpro and VaxiJen v2.0 |
| Protein allergens based on auto and cross-covariance (ACC) transformation | AllerTOP v2.0 |
| Theoretical pI and molecular weight | ProtParam [139] |
| Existence of any signal peptide | SignalP4.1 [140] |
| Secondary structure. | SOPMA [44] |
| 3D structure prediction | I-TASSER [141] |
| Protein structure refining from Ca traces | ModRefiner [142] |
| 3D structure validation | ProSA-web, [143] The ERRAT server [144] |
| Ramachandran plot | ProCheck server [145] |
| TLR2 and TLR4 Crystal structures | PDB [146] |
| TLR2 and TLR4 molecular interactions | ClusPro 2.0 server [147] |
| Molecular interaction visualization | PyMOL [148] |
| Immunogenicity | C-ImmSim server [149] |
| Hub residues | CytoHubba [150] |
| Predict potential T cell epitopes | NetMHC [151] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




